Abstract
Background
Navigating the complexity of chronic myeloid leukemia (CML) diagnosis and management poses significant challenges, including the need for accurate prediction of disease progression and response to treatment. Artificial intelligence (AI) presents a transformative approach that enables the development of sophisticated predictive models and personalized treatment strategies that enhance early detection and improve therapeutic interventions for better patient outcomes.
Methods
An extensive search was conducted to retrieve relevant articles from PubMed, Scopus, and Web of Science databases up to April 24, 2023. Data were collected using a standardized extraction form, and the results are presented in tables and graphs, showing frequencies and percentages. The authors adhered to the PRISMA-ScR checklist to ensure transparent reporting of the study.
Results
Of the 176 articles initially identified, 12 were selected for our study after removing duplicates and applying the inclusion and exclusion criteria. AI’s primary applications of AI in managing CML included tumor diagnosis/classification (n = 9, 75%), prediction/prognosis (n = 2, 17%), and treatment (n = 1, 8%). For tumor diagnosis, AI is categorized into blood smear image-based (n = 5), clinical parameter-based (n = 2), and gene profiling-based (n = 2) approaches. The most commonly employed AI models include Support Vector Machine (SVM) (n = 5), eXtreme Gradient Boosting (XGBoost) (n = 4), and various neural network methods, such as Artificial Neural Network (ANN) (n = 3). Furthermore, Hybrid Convolutional Neural Network with Interactive Autodidactic School (HCNN-IAS) achieved 100% accuracy and sensitivity in organizing leukemia data types, whereas MayGAN attained 99.8% accuracy and high performance in diagnosing CML from blood smear images.
Conclusions
AI offers groundbreaking insights and tools for enhancing prediction, prognosis, and personalized treatment in chronic myeloid leukemia. Integrated AI systems empower healthcare practitioners with advanced analytics, optimizing patient care and improving clinical outcomes in CML management.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Leukemia is a cancer that affects blood cells and ranks among the most common and deadly malignancies worldwide [1, 2]. It constitutes 4% of all cancers and contributes to 4% of cancer-related mortality [3]. In this disease, abnormal blood cells proliferate in the bone marrow, outnumbering normal cells and leading to life-threatening infections and, in some cases, premature death [4, 5]. Individuals with leukemia often have fewer normal red blood cells, causing anemia, which manifests as paleness, weakness, and fatigue [6]. Additionally, leukemia can result in low platelet counts, impairing blood clotting and causing easy bruising, bleeding from the nose and gums, and purplish spots on the skin. Routine blood tests, like a complete blood count (CBC), typically reveal elevated white blood cell (WBC) counts, decreased red blood cells (RBC), and reduced platelets [7, 8].
Leukemia, if left undiagnosed and untreated, inevitably leads to an imbalance where the number of leukemic cells exceeds that of normal blood cells, disrupting systemic function. Physicians, crucially, use blood smears on microscopic slides to analyze cell morphology for diagnosing leukemia. The potential automation of this method suggests the use of image-processing techniques. In contrast, advanced techniques such as flow cytometry are employed to characterize the phenotype of leukemia and lymphoma cells before and after treatment [9, 10], using immunotyping or imaging with molecular probes. However, to minimize manual errors and variations, a robust system for automatic leukemia diagnosis is essential [11, 12]. In addition, some hematologists use interventional radiology as the primary alternative for diagnosing leukemia, employing techniques such as percutaneous aspiration and catheter drainage. However, these methods may be limited by the sensitivity of the imaging modality and challenges in achieving high-resolution radiographic images [13]. Smear examinations, lumbar punctures to study cerebrospinal fluid, bone marrow analysis, and myelography were performed manually by a pathologist. Therefore, the reliability of these tests depends on the pathologist’s experience and fatigue [14]. In addition, various techniques, such as molecular cytogenetics, long-range inverse polymerase chain reaction (LDI-PCR), and array-based comparative genomic hybridization (aCGH) require additional methods for better performance. In this regard, the objective decision-making capability of artificial intelligence (AI) has improved the sensitivity and specificity of leukemia diagnosis.
In recent years, there has been a significant increase in the clinical use of AI for all three conventional medical tasks: diagnosis, treatment, and prognosis [15, 16]. According to Sasaki et al. [17], optimal and effective machine learning (ML) classifiers are required to improve treatment outcomes and increase the CML patients’ life expectancy and survival [17]. Although AI is still in its infancy for CML, preliminary studies have shown promising results in several key areas, such as diagnosis, prognosis, and personalized therapy [18]. AI-based techniques have demonstrated high accuracy in identifying leukemia subtypes, including CML, using blood and bone marrow samples. For instance, a study by Huang et al. [19]. , showed that convolutional neural networks (CNNs) achieved over 95% accuracy in classifying CML cells from microscopic images. This high level of accuracy underscores the potential of AI in improving diagnostic precision and speed. Moreover, several AI models have been developed that outperform conventional prognostic scores in predicting disease progression and treatment response. These models utilize large datasets and advanced algorithms to analyze clinical, molecular, and hematological parameters, offering more accurate prognoses and tailored treatment recommendations [20]. For example, AI algorithms have been designed to optimize tyrosine kinase inhibitor (TKI) therapy in patients with CML. These models can suggest treatment plans that yield better survival outcomes than standard approaches by integrating a wide range of clinical, molecular, and blood factors [17].
Artificial intelligence has revolutionized disease prognosis, diagnosis, and management, including Chronic Myeloid Leukemia (CML), through the development of guideline-based clinical systems (expert systems), machine learning (ML), and deep learning (DL) methods in data processing and clinical image analysis. Machine learning algorithms enable early diagnosis of CML based on clinical and laboratory data, while deep learning algorithms, such as Convolutional Neural Networks (CNNs), automate the classification and diagnosis of CML using medical images. These advancements facilitate early detection, prompt treatment, and improved patient outcomes. Compared to traditional statistical and experimental prediction methods, AI offers profound, practical, and non-invasive analytical capabilities in complex and ambiguous situations, such as predicting cancer outcomes and survival [21, 22].
To our knowledge, no review study has yet explored disease prediction in myeloid leukemia using artificial intelligence (AI). Only a limited number of original studies have investigated ML and DL approaches specifically for the classification and prediction of leukemia [12, 23,24,25]. Therefore, in this study, artificial intelligence-based technologies such as machine learning can effectively and non-invasively be compared to traditional and experimental prediction methods for complex and ambiguous situations, such as predicting, early diagnosis, and managing the treatment of this disease.
Materials and methods
This study’s review process is based on the PRISMA Extension for Scoping Reviews (PRISMA-ScR) checklist [26]. The process of conducting this study is reported on the basis of this checklist.
Search Strategy
This scoping review was conducted to investigate the role of artificial intelligence in managing chronic myeloid leukemia by examining all relevant articles published up to April 24, 2023. It involved a comprehensive search for related keywords in PubMed, Scopus, and Web of Science databases without imposing a time limit. Keywords in the first category included Chronic Myelocytic Leukemia, Chronic Myelogenous Leukemia, Philadelphia-Positive Myeloid Leukemia, and Ph1-Positive Myelogenous Leukemia. The second category included Decision Support Techniques, Data Mining, and Artificial Intelligence. The search strategy used in this study was as follows:
Inclusion and exclusion criteria
The inclusion criteria for the study were original articles written in English that explored the application of artificial intelligence and its algorithms in the prevention, diagnosis, and treatment of chronic myeloid leukemia. Conversely, the exclusion criteria included studies that did not align with the study’s objectives, articles published in languages other than English, review articles, conference paper abstracts, and book chapters.
Data extraction process
Initially, two researchers (EE and SH) independently reviewed the titles and abstracts of the articles. They reached a strong agreement, and any disagreements that arose were discussed and resolved by a third researcher (AS). Subsequently, the full texts of the screened articles that met the exclusion criteria were downloaded for further investigation. Finally, the relevant data were extracted using a meticulously designed standardized extraction form developed by the researchers. This comprehensive form included several critical elements: the publication year and country, the first author’s name, the aim of the study, the types and applications of the algorithms used in disease management, the number of algorithms employed, and the main outcomes. The form’s validity was confirmed by two medical informatics experts. Additionally, it was designed in Excel to facilitate efficient data entry and analysis.
Risk of bias and quality assessment
Three independent appraisers (MR, ZK, and AS) evaluated the risk of bias for the included studies using the Prediction Model Study Risk of Bias Assessment Tool (PROBAST). This quality assessment tool consists of four domains (participant selection, predictors, outcome, and analysis) and includes 20 signaling questions as described in PROBAST [27].
Synthesis of results
Based on the study variables, descriptive analysis, including frequency and percentage parameters, was calculated and presented in the form of graphs and tables. In the results section, the authors employed a narrative synthesis to describe and compare the study findings.
Results
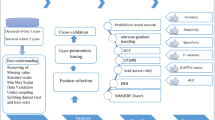
One hundred seventy-six potentially relevant articles were identified from the PubMed, Scopus, and Web of Science databases, and duplicates (n = 7) were removed. An additional search on Google Scholar yielded one study relevant to the aim, which was included in the review process. We excluded 145 articles based on the title and abstract due to their low relevance, and screened 25 full-text articles. The PRISMA diagram illustrates the characteristics of the excluded studies. The current scoping review included 12 articles after applying all eligibility criteria (Fig. 1).
Attributes of the included studies
Table 1 summarizes the included studies.
Figure 2 illustrates the frequency of articles published in different countries. The USA had the highest frequency of articles [13, 17, 31], with three, followed by Iran [6, 33], India [29, 34], and China [28, 36], each with two.
Figure 3 shows the distribution of article publication frequency for different years. The articles were published between 2011 and 2023. The most frequently published articles in this field were from 2021 [17, 30, 31], 2022 [13, 32, 33], and 2023 [34,35,36], each with three articles.
The most common use of AI in the management of CML, as depicted in Fig. 4, was in tumor diagnosis and classification (n = 9), prediction and prognosis (n = 2), and treatment (n = 1). Classification in this study refers to categorizing leukemia types and diagnosing the disease based on various criteria. It involves differentiating between malignant and normal cells, identifying disease phases and stages, and using automated methods for detection. (further details are provided in Table 2).
The use of artificial intelligence in the field of tumor diagnosis and classification was divided into three parts: disease diagnosis using blood smear images (n = 5), disease diagnosis using clinical parameters (n = 2), and disease diagnosis using gene profiling (n = 2).It is necessary to mention that ‘disease diagnosis using clinical parameters’ refers to the process of diagnosing CML based on specific clinical data and measurements obtained from patients. These clinical parameters can include a variety of diagnostic indicators such as blood counts, genetic markers (like the BCR-ABL1 fusion gene), bone marrow biopsy results, and other laboratory test results.
According to Table 3, the most widely used models of artificial intelligence in these articles include various algorithms of Support Vector Machine (SVM) (n = 5) [28, 30, 33, 34, 36], XGBoost (n = 4) [17, 31, 3337], and different neural network methods (ANN ) (n = 3). Algorithms such as the Convolutional Neural Network (CNN) were also used for feature selection.
Table 4 demonstrates the effectiveness of various methods for diagnosing and classifying CML using blood smear images. Among these methods, only the hybrid convolutional neural network method with the interactive self-learning school algorithm (HCNN-IAS) achieved 100% accuracy and sensitivity in diagnosing and classifying the disease using blood smear images. The generative adversarial network (MayGAN) method also achieved 99.8% accuracy, 98.5% sensitivity, 99.7% recall, and a 97.4% F1 score in classifying blood smear images as leukemia. Studies using various support vector machine methods obtained an average efficiency of 91.6%.
Discussion
Early detection of chronic myeloid leukemia (CML) is paramount for providing adequate patient care and treatment. Researchers have endeavored to develop advanced machine learning-based diagnostic systems to expedite CML identification. This scoping review sought to investigate the scholarly literature concerning artificial intelligence techniques in CML management. The study findings were classified into three functional categories: diagnosis and classification, prediction and prognosis, and therapeutic approach. Common AI methods in these studies include SVM, DTj48, XGBoost, RF, neural networks (ANN, CNN), LASSO, and KNN. Among these methods, only the hybrid convolutional neural network method with the interactive self-learning school algorithm (HCNN-IAS) achieved 100% accuracy and sensitivity in diagnosing and classifying leukemia types using blood smear images. The findings are discussed in the following sections.
Image-based diagnosis and classification of cml through hematological smear visualization
Several studies have utilized deep learning algorithms, such as Convolutional Neural Networks (CNNs), and machine learning methods, such as SVMs, to diagnose and classify chronic myeloid leukemia (CML) via blood smear image analysis. A deeper investigation revealed that models rooted in deep learning paradigms consistently achieved superior diagnostic accuracy compared with machine learning approaches. In particular, Abhishek et al. [34] conducted research titled “Automated detection and classification of leukemia on a subject-independent test dataset using deep transfer learning supported by Grad-CAM visualization,” which microscopically identified leukemia subtypes. Their study divided the entire dataset into unequal training and testing subsets. Pre-trained CNN ‘convolutional bases, the frozen lower convolutional layers that extract visual features, were employed to derive representations from microscopic images of the blood smears. The dataset contained 750 smear images of chronic lymphocytic leukemia, acute lymphoblastic leukemia, CML, and acute myeloid leukemia. A merged 500-image set of acute lymphoblastic and myeloid leukemias was constructed. These merged data serve as the basis for automatic leukemia detection and classification via deep transfer learning, which is the central aim of the proposed work. We applied SVMs, Random Forests, and new Fully Connected Layers as classifiers on various pre-trained CNN convolutional bases, such as MobileNet, DenseNet121, ResNet152V2, VGG16, Xception, and InceptionV3 models, pre-trained on massive datasets such as ImageNet, with high accuracy and availability. The study employed a pre-trained VGG-16 convolutional neural network model for leukemia detection and classification. Three classification algorithms were applied to the extracted representations of the CNN: support vector machine (SVM), random forest (RF), and a new fully connected layer (FCL). Compared with individual applications, using these algorithms in conjunction with the combined dataset improved the overall performance of the classifiers. Specifically, FCL achieved 80% classification accuracy, whereas SVM attained 84% accuracy, which was the highest among the approaches. The Grad-CAM visualization method creates class-specific activation heatmaps for each image. These heatmaps showed researchers the distinguishing image regions and helped them determine which visual features placed an image into a certain diagnostic class.
Dese et al. [30] obtained 250 peripheral blood smears from the Department of Hematology at the Jimma University Medical Center in Ethiopia for their study. The smears were stained with eosin, methylene blue (Wright stain), and Sudan black B to aid in the identification of leukemia subtypes. Two experienced hematologists independently examined the smears under a microscope and agreed to the leukemia classification. K-means clustering was used to segment white blood cells from red blood cells and platelets in the digitized images. Multi-class support vector machines (MCSVM) were compared with ANNs, KNNs, and binary SVMs for leukemic subgroup categorization. Using K-means clustering to divide the blood smears into sections before MCSVM classification greatly improves the accuracy, sensitivity, and diagnostic sensitivity of leukemia. Compared with manual examination, digital image-based leukemia diagnosis is more straightforward and faster, eliminating human bias and error while requiring minimal clinical expertise. This computer-assisted system achieved 97.69% accuracy, 97.86% sensitivity, and 100% specificity in diagnosing and categorizing leukemia subtypes.
In a study by Khosla et al. [29] entitled “Phase classification of chronic myeloid leukemia using convolutional neural networks,” TensorFlow was applied to develop a CNN model for classifying histopathological images of CML. Using a sophisticated CNN architecture, researchers have determined the phases and stages of CML progression. Identifying the specific phase of CML a patient is experiencing is paramount, as it dictates the most suitable treatment approach. The findings demonstrated that the CNN could predict different CML stages with 99.6% accuracy. By developing a robust computational model using CNNs, this study provides an automated method for clinicians to accurately assess the CML phase from histopathological samples, informing personalized and optimized therapeutic management for affected patients.
Sakthiraj et al. [13] developed and evaluated an autonomous machine-learning leukemia diagnosis and classification model. A hybrid CNN was designed to extract features from the leukemia dataset and classify samples using a SoftMax-CNN layer. Additionally, a HCNN-IAS was integrated to increase the classification accuracy through the iterative optimization of network weights. The leukemia dataset was divided into subgroups before model training and evaluation. The IASO technique optimizes the CNN hyperparameters to maximize diagnostic performance. Based on confusion matrix analysis and comparison with prior methods, the hybrid model demonstrated superior efficiency, achieving over 99% accuracy and recall. The hybrid model also reduced the diagnosis time compared with alternative approaches. The machine learning model was subsequently deployed on the Internet of Medical Things (IoMT) platform to facilitate remote patient monitoring and management. Clinicians can then securely receive medical data and diagnostic predictions from home environments for review and follow-up care. This telehealth integration aimed to diagnose and monitor leukemia more conveniently, while minimizing health risks through relaxed remote care.
Upon review of the relevant literature, the study conducted by Veeraiah et al. [35] was the sole investigation to employ a novel methodology for identifying four leukemia subgroups (ALL, AML, CLL, and CML) using blood smear images. The authors introduced a Mayfly optimization algorithm with a generative adversarial network (MayGAN) to enhance feature extraction and classification pragmatically. The generative adversarial system (GAS) can also classify leukemia typologies using the derived model’s principal component analysis (PCA). The results demonstrated that the proposed system (dubbed MayGAN) achieved 99.8% precision, 98.5% accuracy, 99.7% recall, 97.4% F1 score, and 98.5% Dice similarity coefficient in categorizing blood smear images as indicative of leukemia, thus enabling the diagnosis of this disease. Subject to approval, the suggested approach could be utilized in the day-to-day clinical management of leukemia patients and could assist medical professionals and individuals in the expedited diagnosis of the affliction.
Comparable results have been observed in other peer-reviewed studies employing convolutional neural networks (CNNs) for identification and categorization tasks. One study by Baig et al. [38] utilized a CNN architecture to identify leukemia subtypes from blood-smear images. In this investigation, we leveraged a CNN model founded on the principles of deep learning consisting of two distinct CNN blocks denoted CNN-1 and CNN-2 to classify samples as acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), or multiple myeloma (MM) based on microscopic smear imagery. The proposed framework demonstrated the ability to detect malignant leukemia cells via microscopic analysis. The team compiled a dataset of approximately 4,150 images sourced from publicly available repositories. Preprocessing procedures included removing background elements and isolating diagnostically relevant hematological components via segmentation, while minimizing noise and blurring effects. Images were initially converted from RGB color space to an 8-bit grayscale format to prepare for preprocessing and segmentation routines. Subsequently, the processed images served as inputs to train the parallel CNN models to extract the deep semantic features. Feature vectors extracted by CNN-1 and CNN-2 were then fused using canonical correlation analysis (CCA) to accentuate the salient patterns. Five classification algorithms — support vector machines (SVM), Bagging Ensemble, AdaBoost, RUSBoost, and k-nearest neighbors (KNN) — were employed to evaluate the performance of the feature extraction pipeline. Among the classification algorithms tested, the bagging ensemble approach yielded superior performance relative to the other techniques, attaining the highest measured accuracy rate of 97.04%.
Ahmed et al. [39] leveraged publicly available ALL-IDB and ASH Image Bank datasets in a recent study to identify leukemia subtypes from microscopic imagery using convolutional neural networks. Seven distinct image transformation techniques were employed for data augmentation. Furthermore, a CNN architecture design was devised with the capability of detecting all leukemia varieties. In addition to exploring CNN modeling, conventional machine learning algorithms, such as Naive Bayes, Support Vector Machine, K-Nearest Neighbor, and Decision Tree, were investigated. A 5-fold cross-validation procedure was adopted for performance evaluation. The findings demonstrated that the CNN model achieved accuracies of 88.25% and 81.74% for leukemia versus healthy classification and multiclass categorization of all subgroups, respectively. Finally, the validation metrics demonstrated that the CNN framework outperformed other well-established machine learning methods.
In the study “IoMT-based automated detection and classification of leukemia using deep learning, “Bibi et al. [40] proposed utilizing a dense convolutional neural network (DenseNet-121) and a residual convolutional neural network (ResNet-34) to identify leukemia subtypes. This study leveraged two publicly accessible datasets on leukemia: ALL-IDB and ASH image banks. The findings demonstrate that the proposed models outperformed well-established machine learning algorithms when discerning healthy subtypes versus leukemia classifications. Specifically, according to the diagnostic accuracy metrics, the DenseNet-121 and ResNet-34 frameworks surpassed conventional machine learning techniques previously applied to leukemia subtype identification.
Diagnosis and classification of cml through clinical parameters
Two studies out of the available 12 utilized clinical parameters to predict and diagnose diseases via artificial neural network (ANN) modeling. In Afshar et al.‘s research [6] entitled “Recognition and prediction of leukemia with Artificial Neural Network (ANN),” eight out of forty-one clinical and laboratory features exhibiting significant differences between cancerous and non-cancerous patient groups (n = 131, 63 with confirmed pathology from Sina Hospital of Hamedan records) were input into the ANN analysis. These characteristics included sex, fever, bleeding, lymphadenopathy, enlarged liver/spleen, hematocrit, hemoglobin, and platelets. The randomly selected samples were divided into training (80%), cross-validation (10%), and test (10%) datasets. The training data were provided to the network for learning. Cross-validation ensures proper model fitting during training or avoidance of overfitting. The independent test data, excluded from modeling, were used to assess the training performance. The area under the receiver operating characteristic curve was 0.967, demonstrating a solid predictive ability. The Phi coefficient yielded a statistically significant (p = 0.005) moderate-strong correlation of 0.778 between the predicted and actual diagnoses.
In a study titled “Beyond the in-practice CBC: the research CBC parameters-driven machine learning predictive modeling for early differentiation among leukemias,” Haider et al. [32] Initially conducted differential count analysis on 200 cells from samples of each participant using optical microscopy at 100x magnification by two experienced hematologists to collect raw cell data. These data were analyzed using SPSS version 23.0 and visualized with ClustVis, a web tool for visualizing multivariate data clustering. An artificial neural network was selected as a machine learning tool for predictive modeling. To test predictive modeling capabilities for the retrieval, discrimination, classification, and determination of data patterns, they initially evaluated the performance of two suitable modeling algorithms: Radial Basis Function Network and Multiple Perceptron Networking. The results demonstrated that in practice, CBC parameters can detect the presence of leukemia and predict cancer origin and type.
Additionally, the results indicated that an artificial intelligence approach utilizing machine learning trained on routine CBC parameters and expected test results could reliably distinguish leukemia histopathology (myeloid or lymphoid) and predict the leukemia type (acute, chronic, or other) and related disorders. The Radial Basis Function Network model achieved high classification accuracy and successfully differentiated the studied groups with a significant correction rate of 10.6%. The prediction model based on the routine CBC parameters proposed in this study achieved 83.1% and 89.4% accuracy for the training and test datasets, respectively. Consequently, researchers have suggested utilizing CBC parameter-driven predictive modeling as an assistant predictive tool in hematology laboratory/clinic decision support systems.
Gene expression profiling as a tool for cml diagnosis and classification
Twelve relevant articles examined the diagnostic and classification methods for chronic myeloid leukemia (CML) using gene expression profiling. Both studies employed support vector machine (SVM) modeling, which demonstrated high diagnostic accuracy. Zhong et al. [36] analyzed CML’s biological characteristics of CML and identified diagnostic markers. Gene expression profiles obtained from the Gene Expression Omnibus database revealed 210 differentially expressed genes between CML and normal samples. Recursive vector machine feature removal, LASSO, and Random Forest algorithms were used to identify four diagnostic CML genes (HDC, SMPDL3A, IRF4, and AQP3). A risk-score model was developed. Comparatively, downregulated genes outweighed upregulated genes in CML samples, with most downregulated genes being related to immune signaling pathways, suggesting immunosuppression. Multiple machine learning methods have been used to identify high-value diagnostic biomarkers. Diagnostic efficiency was improved using a lasso regression risk score model, with significantly higher risk scores observed in CML patients than in healthy individuals across cohorts. Using these four identified genes, researchers constructed a risk score model using LASSO. Comparing the results, the HDC biomarker achieved AUC/ROC values of 98% and 96% in the two databases, indicating a superior diagnostic ability.
Ni et al. [28] attempted to expand flow cytometry applications to distinguish malignant from normal chronic myeloid leukemia (CML) neutrophils using a support vector machine (SVM) predictive model. This study introduces a novel flow cytometry method for differentiating mature CML patients from normal neutrophils. Only four antibodies were used to detect CD45, CD65s, CD15, and CD11b. Mature neutrophils from CML patients and healthy controls uniformly expressed these markers at similar levels using classical two-dimensional flow cytometry analysis. Researchers have used SVM (LIBLINEAR software) and a four-color detection panel to differentiate mature neutrophils. The receiver operating characteristic curve for source differentiation reached 79.51%. The sensitivity and specificity of this technique were 95.80% and 95.30%, respectively. Despite statistically equivalent antigen expression, these results demonstrated a superior ability to discriminate between mature CML patients and normal neutrophils. The predictive model, combining a four-color panel, SVM algorithm, and LIBLINEAR library, detected healthy versus malignant neutrophil differences in all CML disease phases with over 95% specificity and sensitivity. Given these promising findings, as current methods struggle to determine CML disease versus healthy neutrophil status, artificial intelligence-derived predictive models have potential utility for CML diagnosis.
Prediction and prognosis
We identified two studies that used machine learning techniques to predict patient outcomes. The first study by Shanbehzadeh et al. [33] compared machine learning algorithms for predicting the 5-year survival rates in patients with chronic myeloid leukemia. Following the Cross-Industry Standard Process (CRISP) for data mining, the researchers employed a five-step approach involving data understanding, preprocessing, feature selection, modeling, and evaluation. Researchers identified important prognostic variables associated with CML survival and used them as inputs to develop predictive models using various machine learning techniques, such as eXtreme Gradient Boosting (XGBoost), K-Nearest Neighbors (KNN), Probabilistic Neural Network (PNN), MultiLayer Perceptron (MLP), Support Vector Machine (SVM) with linear and Radial Basis Function (RBF) kernels, and J-48 decision trees. Their findings suggest that machine learning approaches hold promise as novel, non-invasive methods for predicting 5-year CML survival and enhancing healthcare quality by facilitating personalized treatment and mitigating complications. Specifically, after selecting the top 12 predictive variables and implementing the classification algorithms, the SVM model with an RBF kernel achieved the highest performance with 85.7% accuracy, 85% specificity, 86% sensitivity, an F-score of 87%, an Area Under the Curve (AUC) of 0.85, and a kappa statistic of 86.1%. Effectively distinguishing high-risk patients and forecasting disease progression could help clinicians optimize resource allocation and safety, potentially extending patient lifespans.
Hauser et al. [31] aimed to ascertain whether blood cell counts from different time intervals could predict chronic myeloid leukemia diagnosis among patients later subjected to definitive BCR-ABL1 mutation testing. They employed machine learning methods using hematological parameters collected over five years to distinguish patients who developed CML from those who did not. Two standard machine learning modeling techniques, eXtreme Gradient Boosting (XGBoost) and Least Absolute Shrinkage and Selection Operator (LASSO), were implemented to model the intricate nonlinear relationships between blood cell quantities and CML diagnosis. The variables examined included laboratory results, demographic data, and clinical encounter information, including cell counts from complete blood counts with differentials (e.g., red blood cells, leukocytes, hemoglobin, hematocrit, platelets, monocytes, eosinophils, and basophils). Cell counts were gathered 1, 3, and 5 years prior. When multiple values existed, five aggregation methods were utilized (i.e., maximum, minimum, difference between maximum and minimum, standard deviation, and count). The demographic factors included age and sex. The clinical encounter factors involved several outpatient and inpatient visits before the initiation of the study. The model was trained on 80% of the positive and negative patients, with the remaining 20% per group reserved for testing. Data analysis was conducted in R Version 3.6.3 using the “XGBoost” and “Glmnet” packages. The findings indicated that blood cell counts up to five years before BCR-ABL1 testing can predict CML status, supporting the hypothesis that predictive models may enable earlier CML detection compared to current approaches.
Therapeutic approach
Sasaki et al. [17] conducted the sole study identified in this field, entitled “The LEukemia Artificial Intelligence Program (LEAP) in Chronic Myeloid Leukemia in Chronic Phase: A Model to Improve Patient Outcomes.” This study analyzed data from 630 consecutive patients with newly diagnosed chronic phase chronic myeloid leukemia (CML-CP) enrolled in seven prospective clinical trials from July 30, 2000, to November 25, 2014, at a single institution. The findings indicated that treatment selection based on personalized recommendations provided through the LEAP artificial intelligence program, compared to treatment without LEAP guidance, conferred a higher probability of survival and improved therapeutic outcomes in patients with myeloid leukemia. Specifically, tailoring treatment decisions to individualized LEAP suggestions rather than opting for alternatives without such algorithmic support enhanced survival prospects and resulted in superior management of CML-CP compared with approaches lacking this personalized, data-driven selection model. Thus, this study demonstrated the potential of artificial intelligence to optimize treatment selection and outcomes for those afflicted with this hematologic malignancy.
Study limitations
This study has several limitations. Firstly, it only included studies written in English, thereby excluding potentially relevant research published in other languages. This language restriction may have led to the omission of significant findings from non-English studies, thereby limiting the comprehensiveness of our review. Additionally, our examination was confined to three major databases: PubMed, Scopus, and Web of Science. While these databases are extensive, they do not cover all available research. Furthermore, selection bias could arise from the inclusion criteria of the studies, which might not accurately represent the general population.
To enhance the robustness and validity of future research, it is recommended to include studies published in multiple languages and expand the scope to additional databases. Incorporating studies from additional databases such as Embase, Cochrane Library, and others could provide a more exhaustive overview of the existing literature. Furthermore, careful consideration of the inclusion criteria in future studies is recommended to minimize the risk of selection bias and ensure accurate representation of the diversity and characteristics of the general population.
Conclusions
In this scoping review, we explored the multifaceted role of artificial intelligence (AI) in chronic myeloid leukemia (CML) prediction and management. Our findings underscore the transformative potential of AI to enable precise disease diagnosis, prognostication, and treatment selection. From tumor classification to treatment response prediction, AI-based approaches offer novel insights and tools to navigate the complexities of CML, heralding a new era of personalized medicine and improving patient care. The use of artificial intelligence for CML prediction and management demonstrates promising avenues for enhancing disease prognosis and treatment strategies. Through sophisticated predictive models and personalized interventions, AI facilitates early detection, accurate classification, and effective therapeutic interventions, ultimately improving patient outcomes.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Kumar V, Ailawadhi S, Bojanini L, Mehta A, Biswas S, Sher T, et al. Trends in the risk of second primary malignancies among survivors of chronic lymphocytic leukemia. Blood cancer J. 2019;9(10):75.
Chung YN, Kim H-N, Lee S-R, Sung HJ, Nam M-H. Usefulness of chromosomal microarray in hematologic malignancies: a case of aggressive NK-cell leukemia with 1q abnormality. Lab Med Online. 2019;9(3):189–93.
Breccia M, Celant S, Olimpieri PP, Olimpieri OM, Pane F, Iurlo A, et al. Mortality rate in patients with chronic myeloid leukemia in chronic phase treated with frontline second generation tyrosine kinase inhibitors: a retrospective analysis by the monitoring registries of the Italian Medicines Agency (AIFA). Ann Hematol. 2021;100:481–5.
Vogado LH, Veras RM, Araujo FH, Silva RR, Aires KR. Leukemia diagnosis in blood slides using transfer learning in CNNs and SVM for classification. Eng Appl Artif Intell. 2018;72:415–22.
Pefani E, Panoskaltsis N, Mantalaris A, Georgiadis MC, Pistikopoulos EN. Chemotherapy drug scheduling for the induction treatment of patients with acute myeloid leukemia. IEEE Trans Biomed Eng. 2014;61(7):2049–56.
Afshar S, Abdolrahmani F, Vakili Tanha F, Zohdi Seif M, Taheri K. Recognition and prediction of leukemia with Artificial neural network (ANN). Med J Islamic Repub Iran. 2011;25(1):35–9.
Shanafelt TD, Call TG, editors. Current approach to diagnosis and management of chronic lymphocytic leukemia. Mayo Clinic Proceedings; 2004: Elsevier.
Khonglah Y, Basu D, Dutta TK. Bone marrow trephine biopsy findings in chronic myeloid leukemia. Malays J Pathol. 2002;24(1):37–43.
Mishra S, Majhi B, Sa PK, Sharma L. Gray level co-occurrence matrix and random forest based acute lymphoblastic leukemia detection. Biomed Signal Process Control. 2017;33:272–80.
Moraes LO, Pedreira CE, Barrena S, Lopez A, Orfao A. A decision-tree approach for the differential diagnosis of chronic lymphoid leukemias and peripheral B-cell lymphomas. Comput Methods Programs Biomed. 2019;178:85–90.
Abdelsalam EMN, Hussain KF, Omar NM, Ali QT. Computer Aided Leukemia Detection using microscopic blood image based machine learning convolutional neural network. Clin Lymphoma Myeloma Leuk. 2018;18(1):S297.
Rehman A, Abbas N, Saba T, Rahman SIu, Mehmood Z, Kolivand H. Classification of acute lymphoblastic leukemia using deep learning. Microsc Res Tech. 2018;81(11):1310–7.
Sakthiraj FSK. Autonomous leukemia detection scheme based on hybrid convolutional neural network model using learning algorithm. Wireless Pers Commun. 2022;126(3):2191–206.
Scotti F, editor. Automatic morphological analysis for acute leukemia identification in peripheral blood microscope images. CIMSA 2005 IEEE International Conference on Computational Intelligence for Measurement Systems and Applications, 2005; 2005: IEEE.
Farrugia A, Al-Jumeily D, Al-Jumaily M, Hussain A, Lamb D, editors. Medical diagnosis: Are artificial intelligence systems able to diagnose the underlying causes of specific headaches? 2013 Sixth International Conference on Developments in eSystems Engineering; 2013: IEEE.
Osmani N, Rezayi S, Esmaeeli E, Karimi A. Transfer learning from non-medical images to medical images using deep learning algorithms. Front Health Inf. 2024;13:177.
Sasaki K, Jabbour EJ, Ravandi F, Konopleva M, Borthakur G, Wierda WG, et al. The LEukemia Artificial Intelligence Program (LEAP) in chronic myeloid leukemia in chronic phase: a model to improve patient outcomes. Am J Hematol. 2021;96(2):241–50.
Bernardi S, Vallati M, Gatta R. Artificial Intelligence-based management of adult chronic myeloid leukemia: where are we and where are we going? Cancers. 2024;16(5):848.
Huang F, Guang P, Li F, Liu X, Zhang W, Huang W. AML, ALL, and CML classification and diagnosis based on bone marrow cell morphology combined with convolutional neural network: a STARD compliant diagnosis research. Medicine. 2020;99(45):e23154.
Dey P, Lamba A, Kumari S, Marwaha N. Application of an artificial neural network in the prognosis of chronic myeloid leukemia. Anal Quant Cytol Histol. 2011;33(6):335–9.
Simmons CP, McMillan DC, McWilliams K, Sande TA, Fearon KC, Tuck S, et al. Prognostic tools in patients with advanced cancer: a systematic review. J Pain Symptom Manag. 2017;53(5):962–70.
Koteluk O, Wartecki A, Mazurek S, Kołodziejczak I, Mackiewicz A. How do machines learn? Artificial intelligence as a new era in medicine. J Personalized Med. 2021;11(1):32–54.
Jagadev P, Virani H, editors. Detection of leukemia and its types using image processing and machine learning. 2017 International Conference on Trends in Electronics and Informatics (ICEI); 2017: IEEE.
Maria IJ, Devi T, Ravi D. Machine learning algorithms for diagnosis of leukemia. Int J Sci Technol Res. 2020;9(1):267–70.
Arivuselvam B, Sudha S. Leukemia classification using the deep learning method of CNN. J X-Ray Sci Technol. 2022;30(3):567–85.
Moher D, Shamseer L, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015 statement. Syst Reviews. 2015;4:1–9.
Moons KG, Wolff RF, Riley RD, Whiting PF, Westwood M, Collins GS, et al. PROBAST: a tool to assess risk of bias and applicability of prediction model studies: explanation and elaboration. Ann Intern Med. 2019;170(1):W1–33.
Ni W, Tong X, Qian W, Jin J, Zhao H. Discrimination of malignant neutrophils of chronic myelogenous leukemia from normal neutrophils by support vector machine. Comput Biol Med. 2013;43(9):1192–5.
Khosla E, Ramesh D, editors. Phase classification of chronic myeloid leukemia using convolution neural networks. 2018 4th International Conference on Recent Advances in Information Technology (RAIT); 2018: IEEE.
Dese K, Raj H, Ayana G, Yemane T, Adissu W, Krishnamoorthy J, et al. Accurate machine-learning-based classification of leukemia from blood smear images. Clin Lymphoma Myeloma Leuk. 2021;21(11):e903–14.
Hauser RG, Esserman D, Beste LA, Ong SY, Colomb DG Jr, Bhargava A, et al. A machine learning model to successfully predict future diagnosis of chronic myelogenous leukemia with retrospective electronic health records data. Am J Clin Pathol. 2021;156(6):1142–8.
Haider RZ, Ujjan IU, Khan NA, Urrechaga E, Shamsi TS. Beyond the in-practice CBC: the research CBC parameters-driven machine learning predictive modeling for early differentiation among leukemias. Diagnostics. 2022;12(1):138–49.
Shanbehzadeh M, Afrash MR, Mirani N, Kazemi-Arpanahi H. Comparing machine learning algorithms to predict 5-year survival in patients with chronic myeloid leukemia. BMC Med Inf Decis Mak. 2022;22(1):236–49.
Abhishek A, Jha RK, Sinha R, Jha K. Automated detection and classification of leukemia on a subject-independent test dataset using deep transfer learning supported by Grad-CAM visualization. Biomed Signal Process Control. 2023;83(7):104722.
Veeraiah N, Alotaibi Y, Subahi AF. MayGAN: Mayfly optimization with Generative Adversarial Network-based Deep Learning Method to classify leukemia form blood smear images. Comput Syst Sci Eng. 2023;46(2):2039–58.
Zhong F-M, Yao F-Y, Yang Y-L, Liu J, Li M-Y, Jiang J-Y, et al. Molecular subtypes predict therapeutic responses and identifying and validating diagnostic signatures based on machine learning in chronic myeloid leukemia. Cancer Cell Int. 2023;23(1):1–14.
Zhong P, Hong M, He H, Zhang J, Chen Y, Wang Z, et al. Diagnosis of acute leukemia by multiparameter flow cytometry with the assistance of artificial intelligence. Diagnostics. 2022;12(4):827.
Baig R, Rehman A, Almuhaimeed A, Alzahrani A, Rauf HT. Detecting malignant leukemia cells using microscopic blood smear images: a deep learning approach. Appl Sci. 2022;12(13):6317.
Ahmed N, Yigit A, Isik Z, Alpkocak A. Identification of leukemia subtypes from microscopic images using convolutional neural network. Diagnostics. 2019;9(3):104–15.
Bibi N, Sikandar M, Ud Din I, Almogren A, Ali S. IoMT-based automated detection and classification of leukemia using deep learning. J Healthc Eng. 2020:1–12.
Funding
In this article, we didn’t have any financial sponsor.
Author information
Authors and Affiliations
Contributions
MR, MRA, MP, ZK, AS: Conceptualization, Methodology, Software. MR, EE, SH, KM, MP, and AS: Data curation, Writing- Original draft preparation. MRA, KM, and ZK: Visualization, Investigation. KM, and AS: Supervision.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ram, M., Afrash, M.R., Moulaei, K. et al. Application of artificial intelligence in chronic myeloid leukemia (CML) disease prediction and management: a scoping review. BMC Cancer 24, 1026 (2024). https://doi.org/10.1186/s12885-024-12764-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12764-y