Abstract
Urease inhibitors can inhibit the decomposition rate of urea, and decrease the air pollution caused by ammonia. In this paper, three new copper(II) and nickel(II) complexes [Cu2L2(μ1,1-N3)2] (1), [Cu(HL)2]Br2 (2), and [Ni3L2(DMF)2(μ2–η1:η1-CH3COO)2(μ1,1-N3)2] (3), where L = 5-bromo-2-(((2-isopropylamino) ethyl)imino)methyl)phenolate, HL = 5-bromo-2-(((2-isopropylammonio)ethyl)imino)methyl)phenolate are synthesized and characterized. The complexes are characterized by elemental analyses, IR, UV-Vis spectra, molar conductivity, and single crystal X-ray diffraction. The X-ray analysis indicates that Cu atoms in complexes 1 and 2 are in square pyramidal and square planar coordination, respectively. The Ni atoms in complex 3 are in octahedral coordination. The molecules of the complexes are linked through hydrogen bonds and π⋯π interactions. The inhibitory effects of the complexes on jack bean urease are studied, which show that the copper complexes have a strong inhibitory effect on urease.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
INTRODUCTION
Urea is a major nitrogen-containing soil fertilizer, with an annual production projected to reach 226 million tons in 2021 [1]. Once deposited in soil, urea quickly hydrolyzes by urease to yield NH3 [2]. This reaction causes a number of agronomic, environmental and economic problems and affects the global nitrogen cycle [3-5]. In particular, a too rapid increase in soil pH upon urea hydrolysis catalyzed by the urease activity causes the loss of urea nitrogen as gaseous ammonia which is toxic to plants and contributes to the production of fine inorganic particulate matter [6, 7]. This process causes the tropospheric pollution by NO, NO2, and N2O, which is a greenhouse gas with 300 times the heat trapping capacity of CO2 [8]. Urease occurs widely in most bacteria, plants, algae, fungi, and invertebrates [9, 10]. The urease enzyme catalyzes the highly efficient decomposition of urea into ammonia with a rate 1014 times faster than that of the non-catalyzed reaction [11]. This process is harmful for the health of human beings and environment [12]. The use of inhibitors proves to be a good way to solve this problem [13-17]. Recent research indicated that metal complexes had interesting activities on urease [18-20]. However, the study on this topic is limited, and no definite relationship between structures and properties is given. Schiff bases are a kind of interesting ligands in the formation of metal complexes, which have received particular attention due to their facile synthesis, versatile structures, and good biological activities [21-24]. Schiff base complexes of copper and nickel are reported to have urease inhibitory activities [25-27]. In pursuit of exploring novel urease inhibitors, three new copper and nickel complexes [Cu2L2(μ1,1-N3)2] (1), [Cu(HL)2]Br2 (2), and [Ni3L2(DMF)2(μ2–η1:η1-CH3COO)2(μ1,1-N3)2] (3), where L = 5-bromo-2-(((2-isopropylamino)ethyl)imino)methyl)phenolate, HL = 5-bromo-2-(((2-isopropylammonio)ethyl)imino) methyl)phenolate, are presented.
EXPERIMENTAL
General methods and materials. 4-Bromosalicylaldehyde and N-isopropylethane-1,2-diamine were purchased (Lancaster) and used as received. All other reagents were of analytical grade. The C, H, and N elemental analyses were carried out in a 2400 Series-II CHN analyzer. FTIR spectra were obtained on a Jasco FT/IR-4000 spectrometer with samples prepared as KBr pellets. Electronic spectra were obtained with a LAMBDA 35 spectrophotometer. Single crystal X-ray diffraction was carried out with a Bruker Apex II CCD diffractometer. Molar conductance was measured with a Shanghai DDS-11A conductometer.
Caution! Although our samples never exploded during handling, azide compounds are potentially explosive. Only a small amount of an azide compound should be prepared and it should be handled with care.
Synthesis of complex 1. 4-Bromosalicylaldehyde (0.20 g, 1.0 mmol) and N-isopropylethane-1,2-diamine (0.10 g, 1.0 mmol) were dissolved and mixed in methanol (30 mL). The mixture was stirred at room temperature for 10 min to give a yellow solution. A methanol solution (20 mL) containing copper bromide (0.22 g, 1.0 mmol) and sodium azide (0.065 g, 1.0 mmol) was added dropwise to the former solution The color immediately changed to blue. The mixture was further stirred at room temperature for 30 min and filtered. The filtrate was kept at ambient temperature. Single crystals of the complex, suitable for X-ray diffraction, were grown from the filtrate upon slow evaporation within a few days. The crystals were isolated by filtration, washed with methanol, and dried in air. Yield: 0.27 g (69%). Anal. calcd for C12H16BrCuN5O (%): C 36.98, H 4.14, N 17.97. Found (%): C 37.21, H 4.22, N 17.86. IR data (KBr, cm–1): 3237, 2046, 1638, 1583, 1515, 1468, 1455, 1425, 1388, 1367, 1333, 1290, 1201, 1188, 1133, 1062, 988, 933, 917, 862, 783, 732, 621, 602, 536, 506, 463, 447. UV-Vis data in methanol (λmax (nm), ε (L·mol–1·cm–1)): 230, 17230; 248, 16010; 272, 14633; 362, 7025.
Synthesis of complex 2. 4-Bromosalicylaldehyde (0.20 g, 1.0 mmol) and N-isopropylethane-1,2-diamine (0.10 g, 1.0 mmol) were dissolved and mixed in methanol (30 mL). The mixture was stirred at room temperature for 10 min to give a yellow solution. The solution was added dropwise with a methanol solution (20 mL) containing copper bromide (0.22 g, 1.0 mmol). The color immediately changed to blue. The mixture was further stirred at room temperature for 30 min and filtered. The filtrate was kept at ambient temperature. Single crystals of the complex, suitable for X-ray diffraction, were grown from the filtrate upon slow evaporation within a few days. The crystals were isolated by filtration, washed with methanol, and dried in air. Yield: 0.16 g (52%). Anal. calcd for C24H34Br4CuN4O2 (%): C 36.32, H 4.32, N 7.06. Found (%): C 36.45, H 4.37, N 6.95. IR data (KBr, cm–1): 3215, 1625, 1583, 1519, 1467, 1425, 1410, 1393, 1333, 1299, 1278, 1254, 1201, 1156, 1130, 1071, 1037, 999, 930, 915, 885, 854, 781, 740, 675, 621, 602, 542, 455, 412. UV-Vis data in methanol (λmax (nm), ε (L·mol–1·cm–1)): 225, 18350; 250, 15670; 270, 13245; 364, 4710.
Synthesis of complex 3. 4-Bromosalicylaldehyde (0.20 g, 1.0 mmol) and N-isopropylethane-1,2-diamine (0.10 g, 1.0 mmol) were dissolved and mixed in methanol (30 mL). The mixture was stirred at room temperature for 10 min to give a yellow solution. The solution was added dropwise with a methanol solution (20 mL) containing nickel acetate tetrahydrate (0.25 g, 1.0 mmol). The color immediately changed to green. Then a few drops of DMF were added, and the mixture was further stirred at room temperature for 30 min and filtered. The filtrate was kept at ambient temperature. Single crystals of the complex, suitable for X-ray diffraction, were grown from the filtrate upon slow evaporation within a few days. The crystals were isolated by filtration, washed with methanol, and dried in air. Yield: 0.15 g (41%). Anal. calcd for C34H52Br2N12Ni3O8 (%): C 37.37, H 4.80, N 15.38. Found (%): C 37.23, H 4.87, N 15.26. IR data (KBr, cm–1): 3271, 2068, 1656, 1585, 1572, 1525, 1427, 1395, 1342, 1290, 1254, 1205, 1190, 1133, 1105, 1073, 996, 979, 968, 925, 911, 864, 804, 785, 730, 672, 615, 600, 575, 467, 431. UV-Vis data in methanol (λmax (nm), ε (L·mol–1·cm–1)): 228, 16380; 246, 16870; 307, 5127; 358, 2623; 408, 1712.
X-ray structure determination. Single crystal X-ray diffraction data for the complexes were collected on a Bruker Apex II CCD diffractometer at 298(2) K with MoKα radiation (λ = 0.71073 Å) by the ω scan mode. The SAINT program was used for the integration of the diffraction profiles [28]. The structures were solved by direct methods using the SHELXS program of the SHELXTL package and refined by full-matrix least-squares methods with SHELXL (semi-empirical absorption corrections were applied using the SADABS program) [29, 30]. The positions of the non-hydrogen atoms were located in difference Fourier maps and least-squares refinement cycles, and finally refined anisotropically. All hydrogen atoms of the complexes were placed theoretically onto the specific atoms and refined isotropically as riding atoms. Crystallographic data and experimental details for structural analyses are summarized in Table 1. Selected bond lengths and angles for the complex are listed in Table 2.
Measurement of the urease inhibitory activity. The assay mixture, containing 25 μL (10U) of jack bean urease which was replaced by 25 μL of a cell suspension (4.0×107 CFU/mL) for the urease assay of intact cells and 25 μL of the test compound, was pre-incubated for 1.5 h at room temperature in a 96-well assay plate. The urease activity was determined by measuring the ammonia production using the indophenol method as described by Weatherburn [31].
RESULTS AND DISCUSSION
General chemistry. The Schiff base ligand was prepared by the reaction of 4-bromosalicylaldehyde and N-isopropylethane-1,2-diamine in methanol. The ligand was not isolated and purified, and was used to prepare complexes with copper and nickel salts. Elemental analyses of the complexes are in accordance with the molecular structures proposed by the X-ray analysis. The complexes are stable in air at room temperature. The molar conductivity of complexes 1 and 3 measured in methanol at concentration of 10–3 mol/L is 15-37 Ω–1·cm2/mol, indicating their non-electrolytic nature in the solution [32]. However, the molar conductivity of complex 2 is 205 Ω–1·cm2/mol, indicating it is a 1:2 electrolyte in the solution [32].
IR and UV-vis spectra. In the IR spectra, weak and sharp absorption bands in the range of 3215-3271 cm–1 are assigned to the N–H vibrations of the Schiff base ligands. The characteristic C=N stretching is observed at 1625-1656 cm–1 as intense signals [33]. The νasym(COO) and νsym(COO) vibrations of the acetate ligands in complex 3 are observed at 1572 cm–1 and 1427 cm–1, respectively. The difference between the two bands (Δν = 145 cm–1) is smaller than 164 cm–1 observed in ionic acetate, indicating the bidentate bridging coordination mode [34, 35]. The IR spectra of complexes 1 and 3 show strong bands at 2046-2068 cm–1 characteristic of the azide ligands [36, 37]. The Ar–O stretching bands are located at 1156- 1190 cm–1 [38].
In the UV-Vis spectra, the bands at 225-250 nm and 270-307 nm are attributed to the π–π* and n–π* transitions [39]. The bands at 358-408 nm can be assigned to the ligand-to-metal charge transition (LMCT) [40].
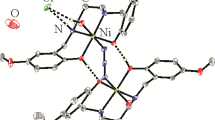
Structural description of complex 1. The molecular structure of the end-on azido-bridged dinuclear copper complex is shown in Fig. 1. The molecule possesses the crystallographic inversion center of symmetry. The Cu atoms are bridged by two azide ligand, with a distance of 3.210(1) Å. The Cu atom is coordinated in a square pyramidal geometry, with the basal plane being defined by phenolate oxygen atoms, imino and amino nitrogen atoms of the Schiff base ligand, and one azide N atom, with the apical position being occupied by another azide N atom. The Cu atom deviates from the basal plane by 0.074(2) Å. The square pyramidal coordination is distorted from the ideal model, as evidenced by the bond angles. The cis and trans angles in the basal plane are in the ranges of 84.35(13)-93.08(12)° and 173.79(14)-176.10(12)°, respectively. The bond angles among the apical and basal donor atoms are in the range of 88.13(12) -97.19(13)°. The Cu–O and Cu–N bond lengths are comparable to those observed in Schiff base copper complexes with azide ligands [41, 42].
In the crystal structure of the complex, the molecules are stacked along the a axis via π⋯π interactions (Cg1⋯Cg2#1 4.484(5) Å, Cg2⋯Cg2#1 3.820(5) Å, symmetry code: #1 1–x, 1–y, –z; Cg1 and Cg2 are the centroids of Cu1–O1–C2–C1–C7–N1 and C1–C2–C3–C4–C5–C6, respectively) (Fig. 2).
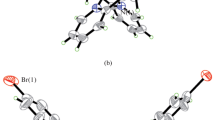
Structural description of complex 2. The molecular structure of complex 2 is shown in Fig. 3. The asymmetric unit of the complex contains two [Cu(HL)2]2+ cations and four bromide anions. The molecules possess crystallographic inversion centers of symmetry. The Cu atom in each cation is coordinated by two phenolate oxygen atoms and two imino nitrogen atoms from two Schiff base ligands, forming a square planar geometry. The amino nitrogen atom is protonated, and does not participate in coordination. The Cu–O and Cu–N bond lengths are comparable to those observed in Schiff base copper complexes [38, 39].
As shown in Fig. 4, the complex cations and the bromide anions are linked through N–H⋯O, N–H⋯Br, C–H⋯O, and C–H⋯Br hydrogen bonds (Table 3), to form a network. In addition, there are π⋯π interactions (Cg3⋯Cg3#3 3.840(5) Å, Cg4⋯Cg4#4 4.403(5) Å, symmetry codes: #3 –x, y, 1/2–z; #4 1–x, y, 3/2–z; Cg3 and Cg4 are the centroids of C1–C2–C3–C4–C5–C6 and C13–C14–C15–C16–C17–C18, respectively) among the molecules.
Structural description of complex 3. The molecular structure of phenolate, azide, and acetate bridged complex 3 is shown in Fig. 5. The molecule possesses the crystallographic inversion center of symmetry. The Ni⋯Ni distance is 3.069(1) Å. The outer Ni atom is coordinated in the octahedral geometry, with the equatorial plane being defined by the phenolate oxygen atom, imino and amino nitrogen atoms of the Schiff base ligand, and one terminal N atom of the end-on azide ligand. The axial positions are occupied by one O atom of the μ2–η1:η1-acetate ligand and one O atom of the DMF ligand. The Ni atom deviates from the equatorial plane by 0.011(2) Å. The octahedral coordination is distorted from the ideal model, as evidenced by the bond angles. The cis and trans angles in the equatorial plane are in the ranges of 82.28(17)-107.42(19)° and 169.90(17)-170.07(19)°, respectively. The bond angles among the axial and equatorial donor atoms are in the ranges of 83.80(18)-93.60(16)°. The inner Ni atom is coordinated in the octahedral geometry. The equatorial plane is defined by two phenolate oxygen from two Schiff base ligands, and two N atoms from two azide ligands. The axial positions are occupied by two O atoms from two acetate ligands. The octahedral coordination is distorted from ideal model, as evidenced by the bond angles. The cis angles in the equatorial plane are in the range of 81.15(17)-98.85(17)°. The bond angles among the axial and equatorial donor atoms are in the range of 89.38(18)-90.77(16)°. The Ni–O and Ni–N bond lengths are comparable to those observed in Schiff base nickel complexes with acetate and azide ligands [40, 41].
As shown in Fig. 6, the complex molecules of complex 3 are linked through N–H⋯Br hydrogen bonds, to form chains along the b axis. In addition, there are π⋯π interactions among the molecules (Cg5⋯Cg5#5 4.690(5) Å, symmetry code: #5 –x, 1–y, 1–z; Cg5 is the centroid of C1–C2–C3–C4–C5–C6).
Urease inhibitory activity. We investigated the urease inhibition of the three complexes in an effort to find more potent compounds in comparison with the previously reported compounds. The IC50 values of complexes 1 and 2 are 1.23±0.12 μmol/L and 0.87±0.09 μmol/L, respectively. However, complex 3 has a weak activity, with the IC50 value of 31.3±1.85 μmol/L. Thus, it is obvious that copper complexes have more potent urease inhibition than nickel complexes. This trend is in accordance with those reported in the literature [42-46]. Moreover, the nickel complex is larger than the copper complexes, illustrating that the hydrophobic area of the enzyme cannot comfortably accommodate the large molecule. From the comparison of these copper complexes with Schiff base copper complexes found in the literature, it can be seen that when a bromine atom was introduced to the Schiff base ligand, the resulting complexes had increased activities. Acetohydroxamic acid and copper perchlorate were used as references, with IC50 values of 27.8±2.12 μmol/L and 9.1±1.5 μmol/L, respectively. The urease inhibition of copper perchlorate is weaker than that of the copper complexes. Thus, the two copper complexes would be potential urease inhibitors that deserve further study.
CONCLUSIONS
Aiming at obtaining new and efficient urease inhibitors, three new copper and nickel complexes with Schiff base ligands have been synthesized and characterized. Single crystal structures of the complexes were confirmed by X-ray diffraction and described. The copper complexes have a strong urease inhibitory activity, which deserves further study to explore novel and efficient urease inhibitors.
Supplementary materials. X-ray crystallographic data for the complexes have been deposited with the Cambridge Crystallographic Data Centre (The Director, CCDC, 12 Union Road, Cambridge, CB2 1 EZ, UK; e-mail: deposit@ccdc.cam.ac.uk; http://www.ccdc.cam.ac.uk; fax: +44-(0)1223–336033) and are available free of charge on request, quoting the deposition numbers CCDC 2062918-2062920.
ACKNOWLEDGMENTS
This project was supported by the Heilongjiang Province Qiqihar Ecological Environment Monitoring Center.
REFERENCES
L. Casali, L. Mazzei, O. Shemchuk, L. Sharma, K. Honer, F. Grepioni, S. Ciurli, D. Braga, and J. Baltrusaitis. ACS Sustainable Chem. Eng., 2019, 7, 2852-2859. https://doi.org/10.1021/acssuschemeng.8b06293
P. A. Karplus, M. A. Pearson, and R. P. Hausinger. Acc. Chem. Res., 1997, 30, 330-337. https://doi.org/10.1021/ar960022j
B. Bano, Kanwal, K. M. Khan, A. Lodhi, U. Salar, F. Begum, M. Ali, M. Taha, and S. Perveen. Bioorg. Chem., 2018, 80, 129-144. https://doi.org/10.1016/j.bioorg.2018.06.007
P. Y. Oikawa, C. Ge, J. Wang, J. R. Eberwein, L. L. Liang, L. A. Allsman, D. A. Grantz, and G. D. Jenerette. Nat. Commun., 2015, 6, 8753. https://doi.org/10.1038/ncomms9753
D. Coskun, D. T. Britto, W. Shi, and H. J. Kronzucker. Nat. Plants, 2017, 3, 17074. https://doi.org/10.1038/nplants.2017.74
S. V. Krupa. Environ. Pollut., 2003, 124, 179-221. https://doi.org/10.1016/S0269-7491(02)00434-7
J. L. Hand, B. A. Schichtel, M. Pitchford, W. C. Malm, and N. H. Frank. J. Geophys. Res. Atmos., 2012, 117, D05209. https://doi.org/10.1029/2011JD017122
J. N. Galloway, F. J. Dentener, D. G. Capone, E. W. Boyer, R. W. Howarth, S. P. Seitzinger, G. P. Asner,C. C. Cleveland, P. A. Green, E. A. Holland, D. M. Karl, A. F. Michaels, J. H. Porter, A. R. Townsend,and C. J. Vorosmarty. Biogeochemistry, 2004, 70, 153-226. https://doi.org/10.1007/s10533-004-0370-0
Z.-P. Xiao, Z.-Y. Peng, J.-J. Dong, J. He, H. Ouyang, Y.-T. Peng, C.-L. Lu, W.-Q. Lin, J.-X. Wang, Y.-P. Xiang,and H.-L. Zhu. Eur. J. Med. Chem., 2013, 63, 685-695. https://doi.org/10.1016/j.ejmech.2013.03.016
A. Hameed, K. M. Khan, S. T. Zehra, R. Ahmed, Z. Shafiq, S. M. Bakht, M. Yaqub, M. Hussain, A. de la Vega de Leon, N. Furtmann, J. Bajorath, H. A. Shad, M. N. Tahir, and J. Iqbal. Bioorg. Chem., 2015, 61, 51-57. https://doi.org/10.1016/j.bioorg.2015.06.004
F. Iftikhar, Y. Ali, F. A. Kiani, S. F. Hassan, T. Fatima, A. Khan, B. Niaz, A. Hassan, F. L. Ansari, and U. Rashid. Bioorg. Chem., 2017, 74, 53-65. https://doi.org/10.1016/j.bioorg.2017.07.003
W.-W. Ni, Q. Liu, S.-Z. Ren, W.-Y. Li, L.-L. Yi, H. Jing, L.-X. Sheng, Q. Wan, P.-F. Zhong, H.-L. Fang, H. Ouyang,Z.-P. Xiao, and H.-L. Zhu. Bioorg. Med. Chem., 2018, 26, 4145-4152. https://doi.org/10.1016/j.bmc.2018.07.003
Q. Liu, W.-K. Shi, S.-Z. Ren, W.-W. Ni, W.-Y. Li, H.-M. Chen, P. Liu, J. Yuan, X.-S. He, J.-J. Liu, P. Cao, P.-Z. Yang, Z.-P. Xiao, and H.-L. Zhu. Eur. J. Med. Chem., 2018, 156, 126-136. https://doi.org/10.1016/j.ejmech.2018.06.065
W.-W. Ni, H.-L. Fang, Y.-X. Ye, W.-Y. Li, C.-P. Yuan, D.-D. Li, S.-J. Mao, S.-E. Li, Q.-H. Zhu, H. Ouyang, Z.-P. Xiao, and H.-L. Zhu. Future Med. Chem., 2020, 12, 1633-1645. https://doi.org/10.4155/fmc-2020-0048
W.-K. Shi, R.-C. Deng, P.-F. Wang, Q.-Q. Yue, Q. Liu, K.-L. Ding, M.-H. Yang, H.-Y. Zhang, S.-H. Gong, M. Deng, W.-R. Liu, Q.-J. Feng, Z.-P. Xiao, and H.-L. Zhu. Bioorg. Med. Chem., 2016, 24, 4519-4527. https://doi.org/10.1016/j.bmc.2016.07.052
Z.-P. Xiao, W.-K. Shi, P.-F. Wang, W. Wei, X.-T. Zeng, J.-R. Zhang, N. Zhu, M. Peng, B. Peng, X.-Y. Lin, H. Ouyang,X.-C. Peng, G.-C. Wang, and H.-L. Zhu. Bioorg. Med. Chem., 2015, 23, 4508-4513. https://doi.org/10.1016/j.bmc.2015.06.014
Z.-P. Xiao, Z.-Y. Peng, J.-J. Dong, R.-C. Deng, X.-D. Wang, H. Ouyang, P. Yang, J. He, Y.-F. Wang, M. Zhu,X.-C. Peng, W.-X. Peng, and H.-L. Zhu. Eur. J. Med. Chem., 2013, 68, 212-221. https://doi.org/10.1016/j.ejmech.2013.07.047
M. K. Rauf, S. Yaseen, A. Badshah, S. Zaib, R. Arshad, Imtiaz-ud-Din, M. N. Tahir, and J. Iqbal. J. Biol. Inorg. Chem., 2015, 20, 541-554. https://doi.org/10.1007/s00775-015-1239-5
X. Dong, Y. Li, Z. Li, Y. Cui, and H. Zhu. J. Inorg. Biochem., 2012, 108, 22-29. https://doi.org/10.1016/j.jinorgbio.2011.12.006
Y. Gou, M. Yu, Y. Li, Y. Peng, and W. Chen. Inorg. Chim. Acta, 2013, 404, 224-229. https://doi.org/10.1016/j.ica.2013.03.045
S. Najm, H. Naureen, K. Sultana, F. Anwar, M. M. Khan, H. Nadeem, and M. Saeed. ACS Omega, 2021, 6, 7719-7730. https://doi.org/10.1021/acsomega.1c00027
T. Chandrasekar, A. Arunadevi, and N. Raman. J. Coord. Chem., 2021, 74, 804-822. https://doi.org/10.1080/00958972.2020.1870967
A. Arunadevi and N. Raman. J. Coord. Chem., 2020, 73, 2095-2116. https://doi.org/10.1080/00958972.2020.1824293
C. J. Dhanaraj and S. S. S. Raj. Inorg. Chem. Commun., 2020, 119, 108087. https://doi.org/10.1016/j.inoche.2020.108087
H. Zhu, Z.-Z. Wang, B. Qi, T. Huang, and H.-L. Zhu. J. Coord. Chem., 2013, 66, 2980-2991. https://doi.org/10.1080/00958972.2013.821198
L. Habala, A. Roller, M. Matuska, J. Valentova, A. Rompel, and F. Devinsky. Inorg. Chim. Acta, 2014, 421, 423-426. https://doi.org/10.1016/j.ica.2014.06.035
Y.-G. Li, D.-H. Shi, H.-L. Zhu, H. Yan, and S. W. Ng. Inorg. Chim. Acta, 2007, 360, 2881-2889. https://doi.org/10.1016/j.ica.2007.02.019
G. M. Sheldrick. SAINT (version 6.02), SADABS (version 2.03). Madison, WI, USA: Bruker AXS, 2002.
G. M. Sheldrick. Acta Crystallogr., Sect. C, 2015, 71, 3-8. https://doi.org/10.1107/S2053229614024218
G. M. Sheldrick. SADABS Program for Empirical Absorption Correction of Area Detector. Gottingen, Germany: University of Gottingen, 1996.
M. W. Weatherburn. Anal. Chem., 1967, 39, 971-974. https://doi.org/10.1021/ac60252a045
W. J. Geary. Coord. Chem. Rev., 1971, 7, 81-122. https://doi.org/10.1016/S0010-8545(00)80009-0
K. R. Surati and B. T. Thaker. Spectrochim. Acta, Part A, 2010, 75, 235-242. https://doi.org/10.1016/j.saa.2009.10.018
B. Sarkar, M. G. B. Drew, M. Estrader, C. Diaz, and A. Ghosh. Polyhedron, 2008, 27, 2625-2633. https://doi.org/10.1016/j.poly.2008.05.004
U. Kumar, J. Thomas, and N. Thirupathi. Inorg. Chem., 2010, 49, 62-72. https://doi.org/10.1021/ic901100z
S. S. Massoud and F. A. Mautner. Inorg. Chim. Acta, 2005, 358, 3334-3340. https://doi.org/10.1016/j.ica.2005.05.007
A. Ray, S. Banerjee, R. J. Butcher, C. Desplanches, and S. Mitra. Polyhedron, 2008, 27, 2409-2415. https://doi.org/10.1016/j.poly.2008.04.018
Y.-M. Zhou, X.-R. Ye, F.-B. Xin, and X.-Q. Xin. Transition Met. Chem., 1999, 24, 118-120. https://doi.org/10.1023/A:1006989707001
L. Pogany, J. Moncol, M. Gal, I. Salitros, and R. Boca. Inorg. Chim. Acta, 2017, 462, 23-29. https://doi.org/10.1016/j.ica.2017.03.001
A. Jayamani, M. Sethupathi, S. O. Ojwach, and N. Sengottuvelan. Inorg. Chem. Commun., 2017, 84, 144-149. https://doi.org/10.1016/j.inoche.2017.08.013
P. K. Bhaumik, K. Harms, and S. Chattopadhyay. Polyhedron, 2014, 68, 346-356. https://doi.org/10.1016/j.poly.2013.10.031
J. Wang, Y. Luo, Y. Zhang, Y. Chen, F. Gao, Y. Ma, D. Xian, and Z. You. J. Coord. Chem., 2021, 74, 1028-1038. https://doi.org/10.1080/00958972.2020.1861603
M. Duan, Y. Li, L. Xu, H. Yang, F. Luo, Y. Guan, B. Zhang, C. Jing, and Z. You. Inorg. Chem. Commun., 2019, 100, 27-31. https://doi.org/10.1016/j.inoche.2018.12.009
Z. You, H. Yu, Z. Li, W. Zhai, Y. Jiang, A. Li, S. Guo, K. Li, C. Lv, and C. Zhang. Inorg. Chim. Acta, 2018, 480, 120-126. https://doi.org/10.1016/j.ica.2018.05.020
L. Pan, C. Wang, K. Yan, K. Zhao, G. Sheng, H. Zhu, X. Zhao, D. Qu, F. Niu, and Z. You. J. Inorg. Biochem., 2016, 159, 22-28. https://doi.org/10.1016/j.jinorgbio.2016.02.017
Y. Luo, J. Wang, B. Zhang, Y. Guan, T. Yang, X. Li, L. Xu, J. Wang, and Z. You. J. Coord. Chem., 2020, 73, 1765-1777. https://doi.org/10.1080/00958972.2020.1795645
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares that he has no conflict of interests.
Additional information
Russian Text © The Author(s), 2021, published in Zhurnal Strukturnoi Khimii, 2021, Vol. 62, No. 11, pp. 1780-1790.https://doi.org/10.26902/JSC_id81674
Supplementary material
Rights and permissions
About this article
Cite this article
Wang, Y. SYNTHESIS, CRYSTAL STRUCTURES, AND UREASE INHIBITORY ACTIVITY OF SCHIFF BASE COPPER AND NICKEL COMPLEXES. J Struct Chem 62, 1667–1677 (2021). https://doi.org/10.1134/S0022476621110020
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0022476621110020