Abstract
Two new zinc(II) and nickel(II) complexes, [Zn2L(μ2-η1:η1-CH3COO)(μ2-η1:η2-CH3COO)]n (1) and [Ni2(HL)2(H2O)2(μ1,3-N3)]Cl·2H2O (2·2H2O), where L is the doubly deprotonated form of 4,4′-dimethoxy-2,2′-(propane-1,3-diyldiiminodimethylene)diphenol (H2L), were synthesized and characterized by elemental analyses, infrared and electronic spectroscopy, as well as single crystal X-ray diffraction. Complex 1 is a bidentate bridging and chelating bridging acetate bridged polymeric zinc complex. In the asymmetric unit of the complex, one Zn atom is in distorted trigonal bipyramidal coordination, and the other one is in octahedral coordination. Complex 2·2H2O is an end-to-end azide bridged dinuclear nickel complex. The Ni atoms are in octahedral coordination. The inhibitory property on Jack bean urease of the complexes was studied, and the nickel complex has effective activity.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Urease (amidohydrolase; EC 3.5.1.5) is a nickel-containing enzyme that catalyzes the hydrolysis of urea to NH3 and CO2. The catalyzed reaction rate is about 1014 times faster than that uncatalyzed. Urease enzyme is widely found in fungi, bacteria and plants [1-3]. The high efficiency of urease increased the hydrolysis of urea into NH3, which deprives plants from essential nutrients and leads to severe toxicity in air and disgusting economic damages [4-6]. In human, urease may produce several health concerns including hepatic coma, pyelonephritis, gastric and peptic ulcer [7-10]. In recent years, a various kinds of urease inhibitors are reported [11-15]. However, most of them are not applicable. Therefore, it is of great interest to explore new urease inhibitors.
Schiff bases show fascinating pharmaceutical and biological activities, such as antibacterial, antifungi, antitumor, and urease inhibition [16-18]. Metal complexes of Schiff base ligands have attracted much attention because of their important role in bioinorganic chemistry [19-21]. Zinc and nickel complexes with Schiff base ligands have shown urease inhibitory activities [22-25]. However, Schiff bases and their complexes are sensitive to pH, and unstable at acid condition.
Instead, reduced Schiff bases with CH–NH groups are more suitable for biological applications [26-28]. Polynuclear complexes have been intensively studied over the last few years, not only for their versatile molecular topologies but also for their widely functionalities such as catalysts [29, 30], magnetism [31, 32] and biological applications [33, 34]. The main strategy for preparing the polynuclear metal complexes is to introduce bridging ligands. Acetate, arising from the inorganic metal acetate salts, is a good bridging ligand containing two oxygen atoms [31, 35]. Azide anion is also a magical building block in the construction of polynuclear complexes, with various bridging modes [36, 37]. Some complexes with the ligands 2,2′-(propane-1,3-diyldiiminodimethylene)diphenolate have been reported with interesting structures [38] and properties like catecholase [39], magnetic [40, 41], and redox potential [42]. Recently, we have reported some zinc, cobalt, nickel and copper complexes derived from reduced Schiff base ligands, which have interesting urease inhibitory activities [24, 28]. In pursuit of new urease inhibitors, two zinc and nickel complexes [Zn2L(μ2-η1:η1-CH3COO)(μ2-η1:η2-CH3COO)]n (1) and [Ni2(HL)2(H2O)2(μ1,3-N3)]Cl·2H2O (2·2H2O), where L is the doubly deprotonated form of 4,4′-dimethoxy-2,2′-(propane-1,3-diyldiiminodimethylene)diphenol (H2L; Scheme 1), are presented.
EXPERIMENTAL
Materials and measurements. 5-Methoxysalicylaldehyde, propane-1,3-diamine, sodium borohydride, sodium azide, nickel acetate and zinc chloride were purchased from Aladdin Co. Ltd. The remaining chemicals and solvents used were of analytical grade and purchased from chemical stores. Elemental analyses for C, H and N were performed on a PerkinElmer 240C elemental analyzer. Infrared spectra were recorded on a Nicolet AVATAR 360 spectrophotometer as KBr pellets in the 4000-400 cm–1 region. Electronic spectra were recorded on a LAMBDA 35 spectrophotometer. 1H NMR data were determined with a Bruker 500 MHz instrument. Single crystal structures were determined with a Bruker Apex II CCD diffractometer.
Synthesis of H2L. To an ethanol solution (100 mL) of 5-methoxysalicylaldehyde (3.0 g, 0.020 mol) was added dropwise an ethanol solution (50 mL) of 1,3-diaminopropane (0.74 g, 0.010 mol) over 1 h. Then the reaction mixture was heated to reflux for 2 h. After the solution was cooled, NaBH4 (1.9 g, 0.05 mol) was carefully added. The reaction solution was heated to reflux for a further 30 min, filtered, and the solvent removed under reduced pressure by using a rotary evaporator. The residue was treated with 1 mol/L NaOH (100 mL). Then it was extracted into chloroform (4×50 mL). The resulting chloroform solution was extracted by shaking with 3 mol/L HCl (4×50 mL). The acid phase was made basic (pH > 12) by the addition of 1 mol/L NaOH and the crude product was then back extracted into chloroform. The chloroform extracts were combined and dried over anhydrous Na2SO4, filtered, and the solvent removed under vacuum by using a rotary evaporator to yield the product as oil. Yield 2.7 g (78%). Anal. calc. for C19H26N2O4: C 65.87; H 7.56; N 8.09. Found (%): C 65.68; H 7.67; N 7.98. IR data (cm–1): ν(OH) 3387, ν(NH) 3289, ν(CHaromatic) 3005, νas(CH2) 2936, νs(CH2) 2831, ν(C–N) 1045. UV-Vis data (methanol, λ, nm (ε, L·mol–1·cm–1)): 260 (6,370), 293 (24,130). 1H NMR (500 MHz, d6-DMSO, ppm) δ 6.68-6.59 (m, 6H, ArH), 3.75 (s, 4H, CH2), 3.64 (s, 6H, CH3), 2.54 (m, 4H, CH2), 1.62 (t, 2H, CH2).
Synthesis of complex 1. The ligand H2L (1.0 mmol, 0.35 g) was dissolved in ethanol (15 mL), to which was added dropwise an ethanol solution (10 mL) of zinc acetate dihydrate (2.0 mmol, 0.44 g). The mixture was magnetic stirred for 30 min at room temperature and filtered. The filtrate was kept in air for a few days, to form colorless crystals suitable for single crystal X-ray diffraction. The crystals were isolated and dried in a vacuum desiccator containing anhydrous CaCl2. Yield: 0.23 g (39%). Anal. calc. for C23H30N2O8Zn2: C 46.56; H 5.10; N 4.72. Found (%): C 46.37; H 5.22; N 4.79. IR data (KBr, cm–1): ν(NH) 3271, 3241, ν(CH3COO) 1572, 1425, ν(C–N) 1085. UV-Vis data (methanol, λ, nm (ε, L·mol–1·cm–1)): 240 (18,260), 305 (8,670). ΛM (10–3 M in methanol): 36 Ω–1·cm2·mol–1.
Synthesis of complex 2·2H2O. The ligand H2L (1.0 mmol, 0.35 g) was dissolved in ethanol (15 mL), to which was added dropwise an ethanol solution (10 mL) of nickel chloride hexahydrate (2.0 mmol, 0.48 g) and an aqueous solution of sodium azide (2.0 mmol, 0.13 g). The mixture was magnetic stirred for 30 min at room temperature and filtered. The filtrate was kept in air for a few days, to form green crystals suitable for single crystal X-ray diffraction. The crystals were isolated and dried in a vacuum desiccator containing anhydrous CaCl2. Yield: 0.25 g (51%). Anal. calc. for C38H58ClN7Ni2O12: C 47.65; H 6.10; N 10.24. Found (%): C 47.91; H 6.23; N 10.09. IR data (KBr, cm–1): ν(OH) 3423, ν(NH) 3262, ν(N3) 2065, ν(C–N) 1078. UV-Vis data (methanol, λ, nm (ε, L·mol–1·cm–1)): 238 (17,525), 310 (9,730). ΛM (10–3 M in methanol): 127 Ω–1·cm2·mol–1.
Crystal structure determination. Single crystal X-ray diffraction was performed with a Bruker Apex II CCD diffractometer using graphite-monochromated MoKα radiation (λ = 0.71073 Å). Diffraction intensities for the complexes were collected at 298(2) K. The collected data were reduced with the SAINT [43], and multi-scan absorption correction was performed using the SADABS [44]. The structures of the complexes were solved by direct method and refined against F2 by full-matrix least-squares method using the SHELXT [45] and SHELXL programs [46]. All of the non-hydrogen atoms were refined anisotropically. The amino H atoms in both complexes and the H9A, H9B, H1A and H6A atoms in complex 2·2H2O were located from different Fourier maps and refined with N–H, O–H and H⋯H distances restrained to 0.90(1) Å, 0.85(1) Å and 1.37(2) Å, respectively. The other H atoms have been placed at geometrical positions with fixed thermal parameters. Crystallographic data of the complexes are summarized in Table 1. Selected bond lengths and angles are listed in Table 2.
Urease inhibitory activity assay. The measurement of urease inhibitory activity was carried out according to the literature method [47]. The assay mixture containing 75 μL of Jack bean urease and 75 μL of tested compounds with various concentrations (dissolved in DMSO) was pre-incubated for 15 min on a 96-well assay plate. Acetohydroxamic acid was used as a reference. Then 75 μL of phosphate buffer at pH 6.8 containing phenol red (0.18 mmol/L) and urea (400 mmol/L) were added and incubated at room temperature. The reaction time required for enough ammonium carbonate to form to raise the pH phosphate buffer from 6.8 to 7.7 was measured by micro-plate reader (560 nm) with end-point being determined by the color change of phenol-red indicator.
RESULTS AND DISCUSSION
Chemistry. The reduced Schiff base ligand H2L was facile synthesized by the condensation reaction of 5-methoxysalicylaldehyde with propane-1,3-diamine, followed by reduction with NaBH4. The zinc and nickel complexes were readily prepared by the reaction of H2L and inorganic metal salts. Single crystals of the complexes were obtained by slow evaporation of the solution containing the complexes. The complexes are stable in air at room temperature. The complexes are soluble in DMF, DMSO, MeOH, EtOH and MeCN, but insoluble in water. Molar conductivity values of the complexes measured in methanol at concentration of 10–3 mol/L are 36 Ω–1·cm2·mol–1 for complex 1, and 127 Ω–1·cm2·mol–1 for complex 2·2H2O, indicating the non-electrolytic nature of complex 1, and 1:1 electrolytic nature of complex 2 in such solution [48].
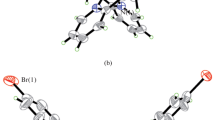
Structure description of complex 1. The perspective view of the zinc(II) complex is shown in Fig. 1. The asymmetric unit of the complex is [Zn2L(CH3COO)2], in which the two Zn atoms are bridged by a μ2-η1:η2-CH3COO ligand and two phenolate groups of the reduced Schiff base ligand. The Zn⋯Zn distance is 2.947(1) Å. The dihedral angle between the two benzene rings of the ligand is 63.1(6)°.
The Zn1 atom is in a distorted trigonal bipyramidal coordination, with the phenolate oxygen (O2) and acetate oxygen (O6 and O8) defining the equatorial plane, and with phenolate oxygen (O1) and acetate oxygen (O7) located at the axial positions. The definition of the distorted trigonal bipyramidal coordination can be calculated from the structural index τ [49] which represents the relative amount of trigonality (square pyramid, τ = 0; trigonal bipyramid, τ = 1); τ = (β – α)/60°, α and β being the two largest angles around the central atom. The value of τ is 0.65. Thus, the coordination of Zn1 is close to trigonal bipyramidal. The distortion is mainly arising from the strain created by the four-membered chelate rings Zn1–O7–C23–O8 and Zn1–O1–Zn2–O2. In the equatorial plane, the bond angles are 99.24(18)-131.05(19)°. The angles among the axial and equatorial donor atoms are 57.15(16)-113.01(18)°. The diagonal angle O1–Zn1–O7 is 170.15(16)°. The Zn2 atom is in an octahedral coordination, with the phenolate oxygen (O1, O2) and amino nitrogen (N1, N2) defining the equatorial plane, and with the acetate oxygen (O5) and ethanol oxygen (O7A) located at the axial positions. In the equatorial plane, the cis and trans bond angles are 79.58(16)-97.2(2)° and 169.02(19)-170.93(18)°, respectively. The angles among the axial and equatorial donor atoms are 80.26(17)-99.13(17)°. The diagonal angle O5–Zn2–O7 is 172.41(16)°. The bond distances Zn–O/N are comparable to those observed in reduced Schiff base complexes (1.90-2.10 Å for Zn–O, 2.00-2.19 Å for Zn–N) [24, 50, 51].
In the crystal structure of the complex, the asymmetric dinuclear Zn units are bridged by μ2-η1:η1-CH3COO ligands, to form one-dimensional chain structure along the c axis. The chains are further linked through intermolecular C–H⋯O hydrogen bonds at the b axis (Fig. 2).
Structure description of complex 2·2H2O. The perspective view of the nickel(II) complex is shown in Fig. 3. The complex contains a dinuclear [Ni2(HL)2(H2O)2(μ1,3-N3)] cation, a chloride anion and two water molecules. The two Ni atoms in the cation are bridged by an end-to-end azide ligand, with the Zn⋯Zn distance of 4.681(1) Å. There are two intramolecular O–H⋯O hydrogen bonds between the two ligands. The dihedral angles between the two benzene rings of the ligands are 67.7(4)° and 51.2(4)°.
The Ni atoms are in an octahedral coordination, with the phenolate oxygen and amino nitrogen of the reduced Schiff base ligands defining the equatorial plane, and with the azide nitrogen (N5 for Ni1, and N7 for Ni2) and the ethanol oxygen respectively. The angles among the axial and equatorial donor atoms are 85.91(13)-95.48(14)° for Ni1 and 85.20(10)-93.20(12)° for Ni2. The diagonal angles O10–Ni1–N5 and O9–Ni2–N7 are 178.09(13)° and 177.95(12)°, respectively. The bond distances Ni–O/N are comparable to those observed in reduced Schiff base complexes (2.00-2.13 Å for Ni–O, 2.02-2.18 Å for Ni–N) [24, 52, 53].
In the crystal structure of the complex, the dinuclear nickel complex cations are linked through N–H⋯Cl, O–H⋯Cl and C–H⋯Cl hydrogen bonds, to form two-dimensional network along the ac plane (Fig. 4).
IR spectra. The IR spectra of the free Schiff base and the complexes provide information about the metal-ligand bonding. The assignments are based on the typical group frequencies. The weak and broad absorptions centered at 3387 cm–1 for H2L and 3423 cm–1 for complex 1 prove the presence of ν(O–H). The absence of strong absorption bands at about 1600 cm–1 for H2L and the complexes indicates that the C=N bonds of the Schiff base have been destroyed. Instead, bands at 1085 cm–1 for complex 1 and 1078 cm–1 for complex 2·2H2O prove the formation of C–N bonds. The weak and sharp bands indicative of the N–H groups of H2L and the complexes are located at 3241-3289 cm–1. The typical absorption for the azido ligand is observed at 2065 cm–1 for complex 2·2H2O [54]. Complex 1 reveals a band at 1572 cm–1 for νasym(OCO) and bands at 1425 cm–1 and 1394 cm–1 for νsym(OCO). The Δν of 147 cm–1 and 178 cm–1 of complex 1 indicates a syn-anti bidentate bridging coordination mode [55].
The UV-Vis spectra of H2L and the complexes were recorded in methanol with concentration of 10–5 mol/L. The free ligand H2L and the complexes displayed strong bands centered at 230-260 nm, which can be assigned to the intra-ligand π–π* transition of the aromatic rings. The charge transfer LMCT bands of the complexes are located at 305-310 nm.
Urease inhibition. Complex 1 has weak activity on urease, with percentage inhibition of (37±2.1)% at the concentration of 100 μmol/L. Complex 2·2H2O has effective urease inhibition with the inhibition rate of (91.3±2.6)% at the concentration of 50 μmol/L, and with IC50 value of (11±1.5) μmol/L (Fig. 5). As a comparison, acetohydroxamic acid was used as a reference with the inhibition rate of (85±3.9)%, and with IC50 value of (28±3.6) μmol/L. Zinc acetate, nickel chloride, sodium azide and H2L have no or weak activity on urease. Thus, complex 2·2H2O has stronger activities than the starting materials and the reference drug acetohydroxamic acid. The nickel complex has similar activity when compared with the nickel complexes with Schiff base ligands (IC50 = 8.7-9.4 μmol/L) [56] or reduced Schiff base ligands (IC50 = 11.6 μmol/L) [24], but weaker activity than the copper complex with reduced Schiff base ligand (IC50 = 1.6 μmol/L) [28].
CONCLUSIONS
The present study reports the syntheses, characterization and crystal structures of a new bidentate bridging and chelating bridging acetate bridged polymeric zinc(II) complex and a new end-to-end azide bridged dinuclear nickel complex with the ligand 4,4′-dimethoxy-2,2′-(propane-1,3-diyldiiminodimethylene)diphenol. The ligand coordinates to the metal atoms through phenolate oxygen and amino nitrogen. The inhibitory property on Jack bean urease of the complexes was studied, and the nickel complex has effective activity.
ACKNOWLEDGMENTS
This work was financially supported by the Education Office of Liaoning Province (LJKZ0984), and the high-level scientific research achievement funding program of Liaoning Normal University (21GDL003).
ADDITIONAL INFORMATION
CCDC 2128777 and 2128778 contain the supplementary crystallographic data for the complexes. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html, or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: (+44) 1223-336-033; or e-mail: deposit@ccdc.cam.ac.uk.
REFERENCES
G. Mohiuddin, K. M. Khan, U. Salar, Kanwal, M. A. Lodhi, A. Wadood, M. Riaz, and S. Perveen. Bioorg. Chem., 2019, 83, 29-46. https://doi.org/10.1016/j.bioorg.2018.10.021
W.-Q. Song, M.-L. Liu, S.-Y. Li, and Z.-P. Xiao. Curr. Top. Med. Chem., 2022, 22, 95-107. https://doi.org/10.2174/1568026621666211129095441
Q. Liu, W.-W. Ni, Z. Li, C.-F. Bai, D.-D. Tan, C.-J. Pu, D. Zhou, Q.-P. Tian, N. Luo, K.-L. Tan, L. Dai, Y. Yan, Y. Pei, X.-H. Li, Z.-P. Xiao, and H.-L. Zhu. Eur. J. Pharm. Sci., 2018, 121, 293-300. https://doi.org/10.1016/j.ejps.2018.05.029
F. S. Carlos, R. J. Kunde, R. O. de Sousa, C. Weinert, A. D. Ulguim, F. Viero, I. Rossi, M. P. Buchain, C. L. Boechat, and F. A. D. Camargo. Nutr. Cycling Agroecosyst., 2022, 122, 313-324. https://doi.org/10.1007/s10705-022-10203-7
T. Lan, X. Q. He, Q. Wang, O. P. Deng, W. Zhou, L. Luo, G. D. Chen, J. Zeng, S. Yuan, M. Zeng, H. H. Xiao, and X. S. Gao. Appl. Soil Ecol., 2022, 174, 104412. https://doi.org/10.1016/j.apsoil.2022.104412
L. A. R. Ferreira, S. R. Silva, and O. T. Kolln. Inter. J. Plant Prod., 2022, 16, 313-328. https://doi.org/10.1007/s42106-022-00191-7
Q. Liu, W.-K. Shi, S.-Z. Ren, W.-W. Ni, W.-Y. Li, H.-M. Chen, P. Liu, J. Yuan, X.-S. He, J.-J. Liu, P. Cao, P.-Z. Yang, Z.-P. Xiao, and H.-L. Zhu. Eur. J. Med. Chem., 2018, 156, 126-136. https://doi.org/10.1016/j.ejmech.2018.06.065
W.-W. Ni, Q. Liu, S.-Z. Ren, W.-Y. Li, L.-L. Yi, H. Jing, L.-X. Sheng, Q. Wan, P.-F. Zhong, H.-L. Fang, H. Ouyang, Z.-P. Xiao, and H.-L. Zhu. Bioorg. Med. Chem., 2018, 26, 4145-4152. https://doi.org/10.1016/j.bmc.2018.07.003
W.-K. Shi, R.-C. Deng, P.-F. Wang, Q.-Q. Yue, Q. Liu, K.-L. Ding, M.-H. Yang, H.-Y. Zhang, S.-H. Gong, M. Deng, W.-R. Liu, Q.-J. Feng, Z.-P. Xiao, and H.-L. Zhu. Bioorg. Med. Chem., 2016, 24, 4519-4527. https://doi.org/10.1016/j.bmc.2016.07.052
A. F. Uberti, N. Callai-Silva, M. V. C. Grahl, A. R. Piovesan, E. G. Nachtigall, C. R. G. Furini, and C. R. Carlini. Inter. J. Mol. Sci., 2022, 23, 3091. https://doi.org/10.3390/ijms23063091
M.-L. Liu, W.-Y. Li, H.-L. Fang, Y.-X. Ye, S.-Y. Li, W.-Q. Song, Z.-P. Xiao, H. Ouyang, and H.-L. Zhu. ChemMedChem, 2022, 17, e202100618. https://doi.org/10.1002/cmdc.202100618
W.-W. Ni, H.-L. Fang, Y.-X. Ye, W.-Y. Li, L. Liu, Z.-J. Fu, Dawalamu, W.-Y. Zhu, K. Li, F. Li, X. Zou, H. Ouyang, Z.-P. Xiao, and H.-L. Zhu. Med. Chem., 2021, 17, 1046-1059. https://doi.org/10.2174/1573406416999200818152440
W.-Y. Li, W.-W. Ni, Y.-X. Ye, H.-L. Fang, X.-M. Pan, J.-L. He, T.-L. Zhou, J. Yi, S.-S. Liu, M. Zhou, Z.-P. Xiao, and H.-L. Zhu. J. Enzyme Inhib. Med. Chem., 2020, 35, 404-413. https://doi.org/10.1080/14756366.2019.1706503
W.-W. Ni, H.-L. Fang, Y.-X. Ye, W.-Y. Li, C.-P. Yuan, D.-D. Li, S.-J. Mao, S.-E. Li, Q.-H. Zhu, H. Ouyang, Z.-P. Xiao, and H.-L. Zhu. Future Med. Chem., 2020, 12, 1633-1645. https://doi.org/10.4155/fmc-2020-0048
Z.-P. Xiao, W.-K. Shi, P.-F. Wang, W. Wei, X.-T. Zeng, J.-R. Zhang, N. Zhu, M. Peng, B. Peng, X.-Y. Lin, H. Ouyang, X.-C. Peng, G.-C. Wang, and H.-L. Zhu. Bioorg. Med. Chem., 2015, 23, 4508-4513. https://doi.org/10.1016/j.bmc.2015.06.014
J. Ceramella, D. Iacopetta, A. Catalano, F. Cirillo, R. Lappano, and M. S. Sinicropi. Antibiotics (Basel, Switz.), 2022, 11, 191. https://doi.org/10.3390/antibiotics11020191
N. Lolak, M. Boga, G. D. Sonmez, M. Tuneg, A. Dogan, and S. Akocak. Pharm. Chem. J., 2022, 55, 1338-1344. https://doi.org/10.1007/s11094-022-02581-7
K. Rafiq, M. Khan, N. Muhammed, A. Khan, N. U. Rehman, B. E. M. Al-Yahyaei, M. Khiat, S. A. Halim, Z. R. Shah, and R. Csuk. Med. Chem. Res., 2021, 30, 712-728. https://doi.org/10.1007/s00044-020-02696-0
C.-H. Dai and F.-L. Mao. J. Struct. Chem., 2013, 54, 624-629. https://doi.org/10.1134/S0022476613030244
S. Thalamuthu and M. A. Neelakantan. Inorg. Chim. Acta, 2021, 516, 120109. https://doi.org/10.1016/j.ica.2020.120109
A. Sudha and S. J. A. Ali. Inorg. Chim. Acta, 2022, 534, 120817. https://doi.org/10.1016/j.ica.2022.120817
H. Wang, T. X. Lan, X. Zhang, D. M. Zhang, C. F. Bi, and Y. H. Fan. J. Inorg. Biochem., 2016, 165, 18-24. https://doi.org/10.1016/j.jinorgbio.2016.10.006
A. de Fatima, C. D. Pereira, C. R. S. D. G. Olimpio, B. G. D. Oliveira, L. L. Franco, and P. H. C. da Silva. J. Adv. Res., 2018, 13, 113-126. https://doi.org/10.1016/j.jare.2018.03.007
Y. M. Li, L. Y. Xu, M. M. Duan, B. T. Zhang, Y. H. Wang, Y. X. Guan, J. H. Wu, C. L. Jing, and Y. L. You. Polyhedron, 2019, 166, 146-152. https://doi.org/10.1016/j.poly.2019.03.051
H. Wang, C. G. Xu, X. Zhang, D. M. Zhang, F. Jin, and Y. H. Fan. J. Inorg. Biochem., 2020, 204, 110959. https://doi.org/10.1016/j.jinorgbio.2019.110959
M. Wozniczka, M. Lichawska, M. Sutradhar, M. Chmiela, W. Gonciarz, and M. Pajak. Pharmaceuticals, 2021, 14, 1254. https://doi.org/10.3390/ph14121254
S. Belaid, O. Benali-Baitich, G. Bouet, and A. Landreau. Chem. Pap., 2015, 69, 1350-1360. https://doi.org/10.1515/chempap-2015-0132
M. M. Duan, Y. M. Li, L. Y. Xu, H. L. Yang, F. W. Luo, Y. X. Guan, B. T. Zhang, C. L. Jing, and Z. L. You. Inorg. Chem. Commun., 2019, 100, 27-31. https://doi.org/10.1016/j.inoche.2018.12.009
D. S. Nesterov and O. V. Nesterova. Catalysts, 2021, 11, 1148. https://doi.org/10.3390/catal11101148
Y. Isaka, K. Oyama, Y. Yamada, T. Suenobu, and S. Fukuzumi. Catal. Sci. Technol., 2016, 6, 681-684. https://doi.org/10.1039/C5CY01845E
A. Paul, A. Figuerola, H. Puschmann, and S. C. Manna. Polyhedron, 2019, 157, 39-48. https://doi.org/10.1016/j.poly.2018.09.023
Saswati, M. Mohanty, A. Banerjee, S. Biswal, A. Horn, G. Schenk, K. Brzezinski, E. Sinn, H. Reuter, and R. Dinda. J. Inorg. Biochem., 2020, 203, 110908. https://doi.org/10.1016/j.jinorgbio.2019.110908
V. G. Vlasenko, A. S. Burlov, Y. V. Koshchienko, A. A. Kolodina, B. V. Chaltsev, Y. V. Zubavichus, V. N. Khrustalev, T. N. Danilenko, A. A. Zubenko, L. N. Fetisov, and A. I. Klimenko. Inorg. Chim. Acta, 2021, 523, 120408. https://doi.org/10.1016/j.ica.2021.120408
E. S. Koumousi, G. Lazari, S. Grammatikopoulos, C. Papatriantafyllopoulou, M. J. Manos, S. P. Perlepes, A. J. Tasiopoulos, G. Christou, and T. C. Stamatatos. Polyhedron, 2021, 206, 115298. https://doi.org/10.1016/j.poly.2021.115298
P.-J. Huang and H. Miyasaka. Dalton Trans., 2020, 49, 16970-16978. https://doi.org/10.1039/D0DT03615C
H. Jeon, J. Kim, J. Kim, K.-B. Cho, and S. Hong. Chem. Commun., 2022, 58, 4623-4626. https://doi.org/10.1039/D2CC01129H
J. Q. Wang, Y. Y. Luo, Y. X. Zhang, Y. Chen, F. Gao, Y. Ma, D. M. Xian, and Z. L. You. J. Coord. Chem., 2021, 74, 1028-1038. https://doi.org/10.1080/00958972.2020.1861603
A. Akay, C. Arici, O. Atakol, H. Fuess, and I. Svoboda. J. Coord. Chem., 2006, 59, 933-938. https://doi.org/10.1080/00958970500410374
A. Hazari, L. K. Das, R. M. Kadam, A. Bauza, A. Frontera, and A. Ghosh. Dalton Trans., 2015, 44, 3862-3876. https://doi.org/10.1039/C4DT03446E
B. Liu, J. Chai, S. Feng, and B. Yang. Spectrochim. Acta, Part A, 2015, 140, 437-443. https://doi.org/10.1016/j.saa.2015.01.012
Y. Song, P. Gamez, O. Roubeau, I. Mutikainen, U. Turpeinen, and J. Reedijk. Inorg. Chim. Acta, 2005, 358, 109-115. https://doi.org/10.1016/j.ica.2004.07.033
M. K. Taylor, J. Reglinski, L. E. A. Berlouis, and A. R. Kennedy. Inorg. Chim. Acta, 2006, 359, 2455-2464. https://doi.org/10.1016/j.ica.2006.01.039
Bruker, SMART and SAINT. Madison, WI: Bruker AXS Inc., 2002.
G. M. Sheldrick. SADABS. Göttingen, Germany: University of Göttingen, 1996.
G. M. Sheldrick. Acta Crystallogr., Sect. A, 2015, 71, 3-8. https://doi.org/10.1107/S2053273314026370
G. M. Sheldrick. Acta Crystallogr., Sect. C, 2015, 71, 3-8. https://doi.org/10.1107/S2053229614024218
J. Meletiadis, J. F. G. M. Meis, J. W. Mouton, J. P. Donnelly, and P. E. Verweij. J. Clin. Microbiol., 2000, 38, 2949-2954. https://doi.org/10.1128/JCM.38.8.2949-2954.2000
W. J. Geary. Coord. Chem. Rev., 1971, 7, 81-122. https://doi.org/10.1016/S0010-8545(00)80009-0
A. W. Addison, T. N. Rao, J. Reedijk, J. van Rijn, and G. C. Verschoor. J. Chem. Soc., Dalton Trans., 1984, 7, 1349-1356. https://doi.org/10.1039/DT9840001349
S. R. Korupoju, N. Mangayarkarasi, P. S. Zacharias, J. Mizuthani, and H. Nishihara. Inorg. Chem., 2002, 41, 4099-4101. https://doi.org/10.1021/ic0201102
V. K. Bhardwaj, M. S. Hundal, M. Corbella, V. Gomez, and G. Hundal. Polyhedron, 2012, 38, 224-234. https://doi.org/10.1016/j.poly.2012.03.029
M. Dey, C. P. Rao, P. K. Saarenketo, and K. Rissanen. Inorg. Chem. Commun., 2002, 5, 924-928. https://doi.org/10.1016/S1387-7003(02)00602-0
J. Reglinski, M. K. Taylor, and A. R. Kennedy. Inorg. Chem. Commun., 2006, 9, 736-739. https://doi.org/10.1016/j.inoche.2006.04.013
P. K. Bhaumik, K. Harms, and S. Chattopadhyay. Polyhedron, 2014, 68, 346-356. https://doi.org/10.1016/j.poly.2013.10.031
U. Kumar, J. Thomas, and N. Thirupathi. Inorg. Chem., 2010, 49, 62-72. https://doi.org/10.1021/ic901100z
Y. Luo, J. Wang, B. Zhang, Y. Guan, T. Yang, X. Li, L. Xu, J. Wang, and Z. You. J. Coord. Chem., 2020, 73, 1765-1777. https://doi.org/10.1080/00958972.2020.1795645
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interests.
Additional information
Text © The Author(s), 2022, published in Zhurnal Strukturnoi Khimii, 2022, Vol. 63, No. 8, 99105.https://doi.org/10.26902/JSC_id99105
Rights and permissions
About this article
Cite this article
Jiang, J., Liu, B., Liu, Y. et al. SYNTHESES, CRYSTAL STRUCTURES AND UREASE INHIBITORY ACTIVITIES OF ZnII AND NiII COMPLEXES DERIVED FROM 4,4′-DIMETHOXY-2,2′-(PROPANE-1,3- DIYLDIIMINODIMETHYLENE)DIPHENOL. J Struct Chem 63, 1371–1381 (2022). https://doi.org/10.1134/S0022476622080182
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0022476622080182