Abstract
The importance of 3D reconstruction of coronary arteries using multiple coronary angiography (CAG) images has been increasingly recognized in the field of cardiovascular disease management. This process relies on the camera matrix’s optimization, needing correspondence info for identical point positions across two images. Therefore, an automatic method for determining correspondence between two CAG images is highly desirable. Despite this need, there is a paucity of research focusing on image matching in the CAG images. Additionally, standard deep learning image matching techniques often degrade due to unique features and noise in CAG images. This study aims to fill this gap by applying a deep learning-based image matching method specifically tailored for the CAG images. We have improved the structure of our point detector and redesigned loss function to better handle sparse labeling and indistinct local features specific to CAG images. Our method include changes to training loss and introduction of a multi-head descriptor structure leading to an approximate 6% improvement. We anticipate that our work will provide valuable insights into adapting techniques from general domains to more specialized ones like medical imaging and serve as an improved benchmark for future endeavors in X-ray image-based correspondence matching.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Coronary angiography (CAG) is an invasive technique for evaluation of coronary arteries. A small catheter is inserted via the radial or femoral artery and selectively cannulated into the coronary artery ostium. Radiocontrast dye is injected into the lumen, which is visualized by the X-ray cine system. Cardiac anatomy is complex and thus interpreting CAG images is notoriously difficult even for trained specialists. One of the factors is projection of complicated 3-dimensional (3D) structures into 2-dimensional (2D) images. Coronary artery travels in a tortuous rather than a straight way.
The coronary artery system, which originates from the two aortic ostia, has numerous branches in order to supply blood flow to every corner of myocardium. Therefore, some segments may thus be displayed shortened, overlapped, or superimposed on a 2D image. In addition, coronary artery stenosis, which causes myocardial ischemia, is not always concentric. Eccentric stenosis, which does not involve the entire artery circumference, may be under- or over-estimated when assessed with a single 2D image. In order to overcome such limitations, CAG images are acquired from many different angles ranging from three to nine for each coronary artery depending on the complexity of the anatomy, cranial or caudal and left anterior oblique, anteroposterior, or right anterior oblique. It is still challenging to human operators to reconstruct 3D coronary system from many different 2D images in their own mind. Beginners often find it frustrating to identify each vessel or to recognize the sense of forward versus backward direction of the arterial course. A Solution that enable automatic 3D reconstruction would help operators understand coronary anatomy and perform coronary intervention more efficiently and safely1,2,3.
Currently, software that performs 3D reconstruction based on 2D CAG images requires user input for operation4,5,6,7. However, in real-time surgical environments like the cath lab, manual data input by the operator poses practical challenges. This highlights the importance of eliminating the need for user input for automating the 3D reconstruction process. Moreover, handling intra- and inter- operator variability in medical environment is a noteworthy concern, and automation is expected to help address these issues. However, at present, there is no known software or equipment capable of performing 3D reconstruction without requiring user input. When performing 3D reconstruction through triangulation from multiple projected 2D images, it is essential to have accurate calibration parameter information of the cameras used to during image capture. In the case of CAG images captured with a C-Arm, the calibration parameter information can be obtained based on the metadata stored in the DICOM files. However, due to mechanical noise and errors caused by gravity acting on the equipment, obtaining the accurate values of the camera matrix is practically impossible. Hence, to achieve reliable 3D reconstruction, the camera matrix correction process is crucial to obtain precise values8,9. This process typically forces users to input the locations of corresponding points on two images, as it relies on triangulation using matching information from those two images10,11. If a method that automatically detects and matches the same points in two images can be applied, the camera matrix correction process can be performed without user input. The field of image matching has been significant evolution with the advancement of deep learning. It now encompasses both descriptor-based models and descriptor-free models, each being explored through various research approaches. A descriptor-based model typically consists of components such as a point detector, descriptor extractor, and feature matcher. These functions can be addressed using relatively classical algorithms12,13,14. Nevertheless, recent research efforts have been directed towards enhancing these components through deep learning15,16,17,18,19. Furthermore, one-stage descriptor-free models are gaining significant attention20,21,22. These models are appealing because they enable end-to-end learning without separately training point detectors or descriptor extractors. Yet there is a downside: since there is no feature point extraction process in these models, they require matching across the entire image which results in a higher computational workload23. Furthermore, it’s been reported that in cases with significant spatial distortion these models may even perform worse24.
Image matching tasks are receiving significant attention in the field of deep learning, but their primary applicability is often found in more general domains. Research efforts in this area aren’t as actively pursued within specific domains that exhibit atypical characteristics. This is particularly true in CAG iamges, where research has predominantly focused on vessel classification and vessel segmentation25,26,27. The field of feature matching remains relatively underexplored here. Unlike general domains, images within the CAG images exhibit significant morphological variations and are characterized by noisy X-ray images. Therefore, methods that work well in general domains may not guarantee similar performance when applied to CAG due to these unique characteristics. Therefore, it’s essential to consider these domain-specific characteristics when applying deep learning methods to tasks within the CAG images. This principle holds true for research into deep learning-based image matching.
This study aims to establish a baseline for future research on image matching within the CAG images. Descriptor-free models may not be suitable for the CAG images because images in this field often depict target objects like blood vessels, which undergo significant morphological distortions due to cardiac motion. Furthermore, there are typically very limited numbers of feature points available for image matching in these cases. Therefore, in this study, we focus on image matching using SuperPoint and SuperGlue28,29. Both of these are known for their high performance in descriptor-based model applications. Our aim is to improve them so they become more suitable for use with CAG images.
The base detector is trained on the Synthetic Shapes dataset as shown in (a). This dataset, proposed in the paper that introduced the SuperPoint, consists of artificially generated shapes with distinct feature points28. However, real-world CAG images, like those in our labeled dataset shown in (b), often contain anatomical landmarks that are not as prominently featured or distinctive.
Related works
Feature detector and descriptor
A feature descriptor is a high-dimensional vector that effectively represents the visual characteristics of a specific part of an image. Traditionally, methods such as the SIFT algorithm have been widely used for detecting feature points and extracting feature descriptors13. More recently, a method known as SuperPoint has been introduced. This approach utilizes deep learning models based on self-supervised learning to generate feature descriptors28. SuperPoint is renowned for its exceptional matching performance, both when used independently and in combination with various feature matchers.
SuperPoint utilizes a model known as the base detector for training the point detector through self-supervised learning. The base detector is trained on a dataset composed of synthetic images with distinct feature points, termed as Synthetic Shapes. Examples of Synthetic Shapes images can be seen in Fig. 1a. The trained base detector is capable of detecting high-frequency components in images, identifying them as feature points. This allows the assignment of pseudo feature point labels to unlabeled images. However, CAG images comprises X-ray images that have high-frequency components distributed throughout the entire image. Furthermore, the target feature points are primarily located within central regions of blood vessels where local features may not be clearly discernible. For instance, within the red box shown in Fig. 1b, the local feature lacks distinctiveness. Consequently, the base detector does not perform optimally within this domain. In SuperPoint, to train a descriptor extractor using self-supervised learning, it is necessary to generate pseudo image pairs for each image. Essentially, a single image with pseudo feature points labeled by the base detector is subjected to a homographic transformation to create a pseudo image pair. The descriptor extractor is trained to bring descriptors closer for the same point pair in two images and push them further apart for different point pairs in those images. Homographic transformations are generally effective when two images are taken from different angles, as they simulate various viewpoints of an object, as illustrated in Fig. 2a. However, the inherent dynamics of the heart and the unique characteristics of X-ray imaging render homographic transformation unsuitable for CAG images. This domain is characterized by X-ray images with significant overlap and foreshortening. The comparison between Fig. 2b and c clearly illustrates the challenges in creating a feasible image pair using homographic transformations. Furthermore, it’s common within CAG images for points at identical locations to exhibit significant differences in their local features as shown in Fig. 3. This further emphasizes that images transformed using homographic transformations do not accurately represent actual image pairs within this specific domain.
Figure (a) demonstrates that homographic transformation can generate images similar to those captured from various angles in a generic domain. However, Figure (b) and (c) demonstrate that images generated through homographic transformation can be significantly different from practical image pairs in the CAG images.
Feature matcher
A feature matcher is a model that takes both point location information and feature descriptor information as input, then outputs pair matching results. Recently, in the field of feature matching, a method known as SuperGlue has achieved high performance. This success is attributed to its use of an advanced deep learning model called the Attentional Graph Neural Network29. However, it’s important to note that a feature matcher on its own cannot generate feature point locations or feature descriptors from images. Therefore, it necessitates usage in combination with point detectors and descriptor extractors such as SuperPoint. Indeed, an image matcher that integrates SuperPoint and SuperGlue is steadily earning a reputation as one of the most potent descriptor-based models in this field.
SuperGlue, as a feature matcher, manipulates descriptor vectors and point locations instead of directly interacting with the images themselves. Intriguingly, despite SuperGlue’s lack of direct engagement with these elements, the outcome of the matching process can vary greatly depending on the domain or unique characteristics of input images This underscores that SuperGlue’s performance is fundamentally tied to the quality of point detectors and descriptor extractors used in conjunction with it. As we mentioned before, if SuperGlue is utilized with point detectors or descriptor extractors that are designed for general domains, a significant deterioration in the quality of matches can occur when dealing with CAG images. This situation underlines the necessity of prioritizing advancements in the accuracy and efficiency of detection and extraction processes specifically tailored for unique domains like CAG images over just focusing on improving feature matching capabilities.
In this study, we utilize SuperPoint and SuperGlue for the image matching task within CAG images. As we mentioned, the CAG images consists of highly distorted spatial images, and there are very few potential matching points compared to those in general domains. Our study primarily focuses on enhancing the performance of detector and descriptor extractors, while considering the limitations that arise when applying SuperPoint to the specific domain of CAG images.
Methods
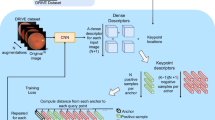
This section outlines the primary methods and key findings of our study. We first provide an overview of the main contributions and activities, followed by detailed descriptions of data preparation and network structure. Figure 4 illustrates the architecture of our proposed model.
Illustrated here is the architecture of our proposed model. Two angiograms are processed through a multi-head descriptor feature extractor, generating features for each image. These features are then fed into an Attentional Graph Neural Network (AGNN), resulting in a score matrix that enables correspondence prediction.
The key findings and activities in this study are summarized as follows:
-
We annotated a set of CAG images for training point detectors and descriptor extractors using supervised learning. This approach was necessary because the self-supervised learning method used in SuperPoint is ineffective for detecting candidate points in CAG images.
-
We employed the HigherHRNet architecture to train our point detector and integrated a Descriptor Head with a structure similar to SuperPoint for descriptor extraction. HigherHRNet, known for its high performance in keypoint detection tasks like pose estimation, was particularly suitable for detecting sparse and indistinct local features in CAG images.
-
We redesigned the loss function originally used in SuperPoint to better align with the characteristics of CAG images. The model was trained to bring descriptors closer for the same point pairs and push them further apart for different pairs, categorizing pairs into positive, sub-positive, negative, and hard-negative categories.
-
We implemented a multi-head architecture for the descriptor head to better capture the unique characteristics of CAG images. Descriptors from smaller patches contained more detailed information, while those from larger patches provided broader context. The multi-head architecture utilized these varying scales effectively.
Data preparation
Feature points in CAG images are typically located at the center of blood vessels. However, these points often display non-distinguishing local features, This complicates the process of generating pseudo feature points with a feature-based detector, a commonly used in general domains other than CAG images. Additionally, pseudo pair images generated through homographic transformation cannot realistically represent actual pair images. Furthermore, feature points often exhibit highly different local features in the CAG images. For these reasons, the self-supervised learning approach employed by the conventional SuperPoint does not work as intended in the CAG images. Consequently, in this study, we conducted research by creating a supervised learning dataset to effectively apply deep learning-based feature matching methods while considering the characteristics of CAG images. We annotated all images with the names and location information of anatomical landmarks. Examples of our annotation can be seen in Fig. 5. Moreover, we grouped these images by patient and target vessel type. Specifically, we categorized LAD (Left Anterior Descending artery), LCx (Left Circumflex artery), LM (Left Main artery) as the LCA (Left Coronary Artery) group and the remainng vessels as the RCA(Right Coronary Artery) group to create pairs for 3D reconstruction.
The network structure
Distinct from SuperPoint, our approach involved annotationg a dataset specifically designed for the supervised learning of extremely sparse anatomical landmarks. For the detectection of these anatomical landmark points, we employed the HigherHRNet, well-known for its high performance in keypoint detection such as pose estimation30. The HigherHRNet is an enhanced model based on HRNet that previously demonstrated high performance in human pose estimation tasks by applying gaussian map to points to generate heatmaps31. It reportedly maintain the performance of large-scale features while incorporating smaller-scale features, leading to significant performance improvements.
In this study, when training our keypoint detector with multi-head descriptor(Fig. 4, 6) for anatomical landmarks, a single CAG image was utilized as the input to the model. After sufficient training, it can be expected that the feature map fed into the point detecting head will effectively represent the image. Therefore, for efficient descriptor extractor training, all parameters from the trained keypoint detector were frozen and descriptor extractor head was added to this network.
Redesigned loss function
In the original method to train descriptor detector, the input image was segmented into patches of \(8\times 8\) pixels from which a single descriptor vector was extracted per patch. After obtaining descriptor vectors from all patches, interpolation was performed to ensure every pixel whithin these patches possessed an individual descriptor vector. Consider D and \(D'\) be sets of descriptor vectors (\(d, d'\)) extracted from all patches across each image, with S defined as a function returning 1 for positive pairs and 0 for negative pairs of these descriptor vectors. The original loss function utilized to train the Descriptor extractor is as follows28:
d and \(d'\) are normalized in advance, so \(d^T d'\) represents the cosine similarity between the two vectors. Essentially, (1) is devised to increase the cosine similarity for positive descriptor pairs while decrease it for negative descriptor pairs. However, through our empirical studies, we found that directly applying the original loss function was inefficient for training within CAG images, which is characterized by limited labeling points per images and insufficient number of images. As such, modifications were made to this loss function in order to better leverage constructed data as well as adapt more effectively to training process for CAG images. each alteration is explained in detail below.
Exponential function and constant terms
The conventional loss function uses cosine similarity represented as \(d^T d'\). However, through our experimental trials applying this function directly onto CAG images led us to observe that the model consistently converged towards a dysfunctional state. In such conditions it generated descriptor vectors only for two particular instances: when an candidate point existed and when there were none present. To tackle this problem, we incorporated the convex exponential function into the cosine similarity. Additionally, we added a constant term to our loss function to ensure that its minimum value would always be zero; with increasing dot product values under theses adjustments we ensured that any resultant losses would converge progressively closer towards zero instead of increasing indefinitely. In other words, instead of \(d^T d'\) in the conventional SuperPoint loss function, we proposed an improved version denoted as \(similarity(d^T d')\) , and the corresponding formulation is provided as follows:
Focal loss for class imbalance
For CAG images specifically, extreme class imbalance can significantly undermine training efficiency. To tackle this challenge, we have incorporated the concept of FocalLoss into our methodology32. FocalLoss is well-known for its effectiveness in mitigating performance degradation typically associated with training on imbalanced data sets. It achieves this by assigning less weight to easy samples, i.e., samples with smaller losses during training, to focus more on samples that do not achieve a sufficiently low loss. Essentially, FocalLoss is a modification of the cross-entropy loss, where the weight is scaled by an exponential term \((1-p)^{\gamma }\). The specific formulation for this adjustment as follows:
The objective of FocalLoss is to ensure a sharper decline in loss fuction as the probability value p nears 1. When integrating FocalLoss to (2), we add a weight that encourages a steeper reduction in loss as cosine similarity approaches 1. In this study, instead of the previously proposed term \(similarity(d^T d')\), we adopt \(FocalSimilarity(d^T d')\) as follows:
Sub-positive pairs
Feature points may be located on the boundaries of a patch or at the center of a vessel that is wider than the patch size. In these situations, it can be challenging for descriptors to fully represent the features of the patch. However, in CAG images, where feature point labels are extremely sparse, efficient learning from these instances is essential. Despite this need, conventional SuperPoint loss, which designates pairs of patches containing the same points in two images as positive pairs to encourage their descriptors to become closer during training, finds it difficult to effectively learn from these scenarios. This is primarily due to the fact that conventional SuperPoint loss was originally designed for general domains in mind, and thus, it struggles to effectively address these unique challenges that are inherent in CAG images To address these issues, we propose the concept of sub-positive pairs, assuming that neighboring patches may share some characteristics. These sub-positive pairs are incorporated into the our loss function as follows:
Hard-negative pairs
In conventional SuperPoint loss, all instances that are not positive pairs are deemed as negative pairs, and the descriptors are trained to diverge from each other. However, even among non-positive pairs, there is a difference in importance between patches where detected feature points and those without. If the point detector operates properly, descriptors of patches without feature points would less frequently serve as input for the feature matcher. Thus, we introduced hard-negative pairs to assign greater weight to descriptors of patches where feature points are detected in the loss calculation. In contrast to positive pairs where an increase in similarity is sought after, for hard-negative pairs we aim for a decrease in similarity. Hence, we invert the sign of \(d^T d'\) during similarity function computation as follows:
Combined
Finally, we trained the descriptor extractor utilizing a modified loss function that incorporates four types of image patches. These four types of image patches incorporated in the loss function are illustrated in Fig. 7. Here, \(S_{pos}\) denotes a function that returns 1 for positive pairs and 0 otherwise. Functions \(S_{sub}\), \(S_{neg}\) and \(S_{hard}\) are defined analogously.
Examples illustrating four types of pairs for the proposed loss. When considering the red patch on the left image as the reference, the red patch on the right image becomes a positive pair, the orange patch becomes a sub-positive pair, the blue patch becomes a hard-negative pair, and all unmarked patches become negative pairs.
Multi-head descriptor structure
The conventional SuperPoint employs a fixed patch size of \(8\times 8\) for descriptor computation. However, in the CAG image dataset, as illustrated in Fig. 6, the features that an \(8\times 8\) patch can represent are substantially limited. This necessitates adjusting the patch size used by descriptor head for better feature representation. As indicated by the red patch in Fig. 6, when a size of \(32\times 32\) is used, it adequately captures surrounding blood vessels at a point, but it may also include unnecessary parts of the vascular shape or position the patch too close to patches associated with other points. Conversely, as shown by the orange patch in Fig. 6, using a size of \(16\times 16\) enables more detailed feature representation within a single image, but may not completely capture features specific to an individual point. Moreover, determining appropriate patch size can fluctuate depending on factors such as image capture magnification or variations in vascular shape among individual patients. To mitigate this trade-off, we designed our model with a multi-head structure that accommodates both \(8\times 8\) and \(16\times 16\) scales of patches. This structure yields two descriptors for each point as its final output. The similarity is then computed by summing up similarities obtained from each descriptor.
Examples of inference results from the test set are displayed, where red dotted lines indicate the highest confidence matching point pairs for corresponding anatomical landmarks, while lime-colored dotted lines represent the output of our proposed model. The top 4 figures represent results from the in-house test set, and the bottom 6 figures represent results from the public dataset.
Results
Data
This study was approved by the Institutional Review Board of the Seoul National University Bundang Hospital (Seongnam-si, Republic of Korea), with written informed consent being waived due to its retrospective nature, and was conducted in compliance with the Declaration of Helsinki. A retrospective data collection was conducted on a total of 920 cases sampled from consecutive patients who underwent CAG between 2017 and 2020 at Seoul National University Bundang Hospital in 2022. The necessary labeling procedures, such as selecting suitable frames for vascular analysis and annotating anatomical landmarks, were carried out by three radiologic technologists. These procedures were then confirmed by two cardiology specialists at Seoul National University Bundang Hospital. Our CAG image dataset comprises paired images captured at different angles, each pair targeting the same vessel within the single patient. This dataset was assembled from a total of 6,320 images gathered from 920 patients. All images were initially categorized by patient and further categorized based on the target vessel, resulting in 4,981 pairs. Roughly 10% of the patients (90 individuals) were randomly chosen and their corresponding 200 pairs were set aside as the test set. The remaining pairs were used for training. Each image includes labeled anatomical landmarks with their repective positions and names. On average, each pair of images contains approximately 3.67 pair labeled points. To establish a fair basis for comparision in future studies, we curated an additional test set using the publicly available Stenosis Detection Dataset33. This dataset comprises CAG images sourced from 100 patients at various angles. We categorized theses images into LAD, LCx, RCA and LM groups, and selected frames with adequate radiocontrast dye filling. As a result, we assembled a public test dataset that includes 56 pairs of images taken at different angles, with each pair targeting the same vessel within a single patient, similar to our in-house dataset. The pair information of this test set is available for download at: https://github.com/medipixel/Angiography-Image-Pairs-for-Correspondence-Matching
Training environments
We assigned to \(\lambda _{pos}, \lambda _{sub}, \lambda _{neg},\) and \(\lambda _{hard}\) were determined experimentally and set to 0.2, 0.2, 0.8, and 0.8 respectively. These values were found to optimize the model’s performance through experimentation. An Adam optimizer was utilized with a learning rate of \(5 \times 10^{-5}\) and a weight decay of 0.9, along with a cosine annealing scheduler for training acorss 200 epochs. The SuperGlue feature matcher was configured using identical settings as those in the original paper29. All training procedures were conducted within an AWS EC2 g5.12xlarge instance environment.
Evaluation and discussions
In this study, we enhanced the SuperPoint method for both point detection and descriptor extraction. However, as independent evaluation of the point detector and descriptor extractor is unfeasible, we trained the SuperGlue model as a feature matcher and established an image matcher for the final evaluation. This image matcher model takes image pairs as input and generates multiple matching point pairs along with their corresponding confidences levels. Nonetheless, since correcting the camera matrix can be achieved with just one pair, it is not unnecessary to evaluate whether all output pairs match in 3D reconstruction10,11. Consequently, we conducted a binary evaluation to ascertain whether the matching point pair with the highest confidence from the model’s output was the correct. Moreover, due to practical impossibility of having dense labels for all pixel points in an image there could be instances where the model’s output point pairs are evaluated as incorrect due to a lack of corresponding labels. Therefore, manual evaluation was carried out by radiologic technologists specializing in CAG domain knowledge because automated evaluation was not feasible.
In order to evaluate the effectiveness of our proposed methods, we conducted an ablation study and trained SuperPoint and SuperGlue using several different approaches as outlined in Table 1. The table summarizes the impact of incrementally adding each proposed method, including annotation with specific loss terms (\(\lambda _{pos}\) and \(\lambda _{neg}\)), the multi-head descriptor structure, and the additional loss terms (\(\lambda _{sub}\) and \(\lambda _{hard}\)). In the initial experiment, self-supervised training was conducted utilizing SuperPoint and SuperGlue on the CAG images, followed by an evaluation of the outcomes. It was observed that the majority of the predictions were unsuccessful due to several issues discussed earlier, such as the model erroneously matching background regions or local features that are not related to blood vessels. This typically occurs because the background and unrelated features can sometimes have similar visual characteristics to the target features, leading to incorrect correspondences. Furthermore, the inherent noise and variations in X-ray images can exacerbate this problem, causing the model to misidentify the correct matching points. Figure 8 provides visual instances of the model’s failure in predicting correspondence matching, illustrating how these errors manifest in practical scenarios. In the second experiment, supervised learning was implemented based on SuperPoint and SuperGlue using our annotated in-house dataset. Notably, we did not employ our proposed methods in this experiment, such as a redesigned loss function or multi-head descriptor, during this training. It was found that the matching pair with the highest confidence was correct in 66.5% of the pairs. In the final experiment, we trained the correspondence matching model using supervised learning, similar to the second experiment. However, we also applied our proposed methods such as the multi-head descriptor structure and the redesigned loss function. As a result, the matching pairs with the highest confidence score were found to be correct in approximately 73% of the cases. Figure 9 offers visual examples where the model accurately identified the matching pair with highest confidence from test data set. This held true even when there were significant differences in overall vessel appearance between image pairs or drastic disparities in shape and angle of anatomical landmarks. Following these initial experiments conducted on our in-house dataset, we sought to further validate our methods using a public dataset. This additional dataset was constructed based on the public Stenosis Detection Dataset. We executed the same three experiments on this public dataset. However, consistent with our experiencewith the in-house dataset, the first experiment failed to yield any meaningful results. As outlined in Table 1, the matching pair with the highest confidence was correct in 64.3% of the cases for the second experiment, and 73.2% for the third experiment, where our proposed methods were integrated with the conventional approach. These results further underscore the effectiveness of our proposed methods in enhancing correspondence matching accuracy.
Figure 9 showcases visual instances where the model accurately identified the matching pair with the highest confidence, taken from both the in-house and public test datasets. The successful outcomes from these experiment underscore the robustness of our proposed methods, demonstrating their effectiveness diverse data sources. These results suggest that our methods wide applicability across various sets of coronary angiography images, further reinforcing their practical utility in the field.
Conclusion
CAG images, unlike typical image domains, pose significant challenges for image matching due to their extreme label sparsity, the presence of morphological variations and high-frequency components in X-ray images. These factors made it challenging to apply image matching methods that are commonly studied in the field of deep learning. To overcome these challenges, this study proposed several enhancements including modifications to the point detector architecture, enhanced loss functions, and the implementation of a multi-head architecture for descriptor. The proposed enhancements resulted in a matching success rate approximately 6% higher than that of conventional method. However, a limitation of our study is the lack of comparative analysis of the proposed method’s effectiveness in natural domains with sparse ground truth data. Future research could benefit from including such comparisons to further explore the robustness and applicability of our method across different image domains. This demonstrates that our approaches effectively enhance image matching method for CAG images by better accommodating its unique characteristics. This research represents an inaugural attempt to apply deep learning for image matching within CAG images and is anticipated to establish a new baseline in this field.
Data availability
The In-house datasets for training and testing during the current study are not publicly available due to the protection of patient’s personal information as per the opinion of the Institutional Review Board of the hospital that provided the data, but are available from the corresponding author on reasonable request. The additional test datasets for analysed during the current study are available in the medipixel github repository, https://github.com/medipixel/Angiography-Image-Pairs-for-Correspondence-Matching.
References
Nishi, T. et al. Comparison of 3-dimensional and 2-dimensional quantitative coronary angiography and intravascular ultrasound for functional assessment of coronary lesions. J. Cardiol. 69, 280–286 (2017).
Tu, S., Huang, Z., Koning, G., Cui, K. & Reiber, J. H. A novel three-dimensional quantitative coronary angiography system: In-vivo comparison with intravascular ultrasound for assessing arterial segment length. Catheter. Cardiovasc. Interv. 76, 291–298 (2010).
Girasis, C. et al. Advanced three-dimensional quantitative coronary angiographic assessment of bifurcation lesions: Methodology and phantom validation. EuroIntervention 8, 1451–1460 (2013).
Masdjedi, K. et al. Vessel fractional flow reserve (VFFR) for the assessment of stenosis severity: The fast ii study. EuroIntervention 17, 1498 (2022).
Xu, B. et al. Angiographic quantitative flow ratio-guided coronary intervention (FAVOR III China): A multicentre, randomised, sham-controlled trial. Lancet 398, 2149–2159 (2021).
Fearon, W. F. et al. Accuracy of fractional flow reserve derived from coronary angiography. Circulation 139, 477–484 (2019).
Li, J. et al. Accuracy of computational pressure-fluid dynamics applied to coronary angiography to derive fractional flow reserve: Flash FFR. Cardiovasc. Res. 116, 1349–1356 (2020).
Tu, S., Koning, G., Jukema, W. & Reiber, J. H. Assessment of obstruction length and optimal viewing angle from biplane x-ray angiograms. Int. J. Cardiovasc. Imaging 26, 5–17 (2010).
De Molina, C., Pascau, J., Desco, M. & Abella, M. Calibration of a c-arm x-ray system for its use in tomography. In XIII Mediterranean Conference on Medical and Biological Engineering and Computing 2013: MEDICON 2013, 25-28 September 2013, Seville, Spain, 245–248 (Springer, 2014).
Onuma, Y. et al. A novel dedicated 3-dimensional quantitative coronary analysis methodology for bifurcation lesions. EuroIntervention 7, 629–635 (2011).
Kim, Y. et al. Quantitative evaluation and comparison of coronary artery characteristics by 3d coronary volume reconstruction. Sci. Rep. 11, 1170 (2021).
Harris, C. et al. A combined corner and edge detector. In Alvey Vision Conference, Vol. 15, 10–5244 (Citeseer, 1988).
Lowe, D. G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60, 91–110 (2004).
Bay, H., Tuytelaars, T. & Van Gool, L. Surf: Speeded up robust features. In Computer Vision–ECCV 2006: 9th European Conference on Computer Vision, Graz, Austria, May 7-13, 2006. Proceedings, Part I 9, 404–417 (Springer, 2006).
Revaud, J. et al. R2d2: repeatable and reliable detector and descriptor. arXiv preprint arXiv:1906.06195 (2019).
Dusmanu, M. et al. D2-net: A trainable CNN for joint detection and description of local features. arXiv preprint arXiv:1905.03561 (2019).
Liu, Y. et al. Gift: Learning transformation-invariant dense visual descriptors via group CNNs. Adv. Neural Inf. Process. Syst. 32 (2019).
Luo, Z. et al. Aslfeat: Learning local features of accurate shape and localization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 6589–6598 (2020).
Tyszkiewicz, M., Fua, P. & Trulls, E. Disk: Learning local features with policy gradient. Adv. Neural. Inf. Process. Syst. 33, 14254–14265 (2020).
Sun, J., Shen, Z., Wang, Y., Bao, H. & Zhou, X. Loftr: Detector-free local feature matching with transformers. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 8922–8931 (2021).
Tang, S., Zhang, J., Zhu, S. & Tan, P. Quadtree attention for vision transformers. arXiv preprint arXiv:2201.02767 (2022).
Chen, H. et al. Aspanformer: Detector-free image matching with adaptive span transformer. In European Conference on Computer Vision, 20–36 (Springer, 2022).
Xue, F., Budvytis, I. & Cipolla, R. Sfd2: Semantic-guided feature detection and description. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 5206–5216 (2023).
Zhang, H., Yi, H., Jia, H., Wang, W. & Odamaki, M. Panopoint: Self-supervised feature points detection and description for 360deg panorama. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 6448–6457 (2023).
Liu, W. et al. Full-resolution network and dual-threshold iteration for retinal vessel and coronary angiograph segmentation. IEEE J. Biomed. Health Inform. 26, 4623–4634 (2022).
Shi, T. et al. Affinity feature strengthening for accurate, complete and robust vessel segmentation. IEEE J. Biomedical Health Inform. (2023).
Zhang, Y. et al. Centerline-supervision multi-task learning network for coronary angiography segmentation. Biomed. Signal Process. Control 82, 104510 (2023).
DeTone, D., Malisiewicz, T. & Rabinovich, A. Superpoint: Self-supervised interest point detection and description. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, 224–236 (2018).
Sarlin, P.-E., DeTone, D., Malisiewicz, T. & Rabinovich, A. Superglue: Learning feature matching with graph neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4938–4947 (2020).
Cheng, B. et al. Higherhrnet: Scale-aware representation learning for bottom-up human pose estimation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, 5386–5395 (2020).
Wang, J. et al. Deep high-resolution representation learning for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 43, 3349–3364 (2020).
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, 2980–2988 (2017).
Danilov, V. V. et al. Real-time coronary artery stenosis detection based on modern neural networks. Sci. Rep. 11, 7582 (2021).
Acknowledgements
We would like to appreciate Sung-Wook Kim, Kahyuk Park, Seong Geol Soo (Cardiovascular Center, Seoul National University Bundang Hospital) for their time and effort in data labeling. Additionally, we extend our thanks to Won-Ji Shin(Medipixel Inc.) for the time dedicated towards data review. This work was supported by the Bio &Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government (MSIT) (NO. RS-2023-00222910).
Author information
Authors and Affiliations
Contributions
H.-W.K. and S.-C.N. contributed to conceptualization and methodology. H.-W.K., S.-C.N. and S.-H.Kang contributed to the preparation of the original draft. H.-W.C. and S.-H.Kang supervised data selection and labeling. S.-H.Kim conducted formal analysis. H.-W.K. and S.-H.Kang provided review and editing. S.-H.Kang and C.-H.J. provided supervision. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.-H.Kang. owns stock options and receives counseling fee from Medipixel Inc. Others declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kim, HW., Noh, SC., Kim, SH. et al. Effective descriptor extraction strategies for correspondence matching in coronary angiography images. Sci Rep 14, 18630 (2024). https://doi.org/10.1038/s41598-024-69153-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69153-5
- Springer Nature Limited