Abstract
Background
Prostate Imaging Reporting and Data System (PI-RADS) 3 lesions, identified through multiparametric magnetic resonance imaging (mpMRI), present a clinical challenge due to their equivocal nature in predicting clinically significant prostate cancer (csPCa). Aim of the study is to improve risk stratification of patients with PI-RADS 3 lesions and candidates for prostate biopsy.
Methods
A cohort of 4841 consecutive patients who underwent MRI and subsequent MRI-targeted and systematic biopsies between January 2016 and April 2023 were retrospectively identified from independent prospectively maintained database. Only patients who have PI-RADS 3 lesions were included in the final analysis. A multivariable logistic regression analysis was performed to identify covariables associated with csPCa defined as International Society of Urological Pathology (ISUP) grade group ≥2. Performance of the model was evaluated using the area under the receiver operating characteristic curve (AUC), calibration, and net benefit. Significant predictors were then selected for further exploration using a Chi-squared Automatic Interaction Detection (CHAID) analysis.
Results
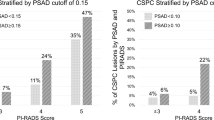
Overall, 790 patients had PI-RADS 3 lesions and 151 (19%) had csPCa. Significant associations were observed for age (OR: 1.1 [1.0–1.1]; p = 0.01) and PSA density (OR: 1643 [2717–41,997]; p < 0.01). The CHAID analysis identified PSAd as the sole significant factor influencing the decision tree. Cut-offs for PSAd were 0.13 ng/ml/cc (csPCa detection rate of 1% vs. 18%) for the two-nodes model and 0.09 ng/ml/cc and 0.16 ng/ml/cc for the three-nodes model (csPCa detection rate of 0.5% vs. 2% vs. 17%).
Conclusions
For individuals with PI-RADS 3 lesions on prostate mpMRI and a PSAd below 0.13, especially below 0.09, prostate biopsy can be omitted, in order to avoid unnecessary biopsy and overdiagnosis of non-csPCa.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
In the last decade, multiparametric magnetic resonance imaging (mpMRI) has emerged as a crucial instrument, empowering urologists with optimized diagnostic capabilities and improved triage methods for patients with prostate cancer (PCa). Prostate mpMRI has solidified its position as the foremost modality in PCa diagnosis, demonstrating outstanding sensitivity in clinically significant prostate cancer (csPCa) detection and a robust negative predictive value. This advancement has notably contributed to minimizing unnecessary biopsies in the management of PCa [1,2,3,4].
The Prostate Imaging Reporting & Data System (PI-RADS) initially described in 2011 has been widely used by radiologists as an objective score to standardize reporting prostate lesions [5]. The European Association of Urology (EAU) guidelines currently endorse MRI before prostate biopsies and recommend prostate biopsies in patients with PI-RADS ≥ 3 lesions, depending on the value of the Prostate Specific Antigen (PSA) density [6]. However, these values were developed in the pre-MRI-targeted biopsy era.
Approximately 45% of lesions with a PI-RADS ≥ 3 are found to have an International Society of Urological Pathology (ISUP) ≥ 2, but mostly in case of PI-RADS 4–5 tumors [7]. In case of equivocal MRI lesions defined as PI-RADS 3, only 20% of patients have csPCa highlighting the need for a better risk stratification to avoid unnecessary biopsy [8]. Moreover, the real occurrence of csPCa following an MRI-targeted biopsy in PI-RADS 3 lesions has been shown to differ among different patient subgroups depending on the lesion volume, ranging from 4% to 29% [9].
In the present study, we aimed to sub-stratify patients identified from a large European cohort of patients who underwent MRI-targeted and systematic biopsies for PI-RADS 3 lesions and to identify predictive factors of csPCa.
Materials and methods
Population
Data from 4841 consecutive patients who underwent MRI and subsequent MRI-targeted and systematic biopsies between January 2016 and April 2023 were retrospectively identified from an independent prospectively maintained and board-approved databases at fifteen European tertiary referral-centers.
MRI and biopsy procedures
All prebiopsy MRI were performed within 6 months before biopsy, following the European Society of Urogenital Radiology guidelines, and scored using the PI-RADS version 2.1 protocols by local dedicated genitourinary radiologists [10, 11]. MRIs done before 2019 were re-evaluated to ensure consistency. Prostate biopsies were carried out by dedicated urologists and exclusively with the KOELIS system (KOELIS®, La Tronche, France) allowing elastic MRI-tridimensional ultrasound images fusion. A minimum of three MRI-targeted cores per target were taken combined with concomitant bilateral systematic biopsy. A dedicated uropathologist evaluated the biopsy cores following the ISUP 2014 recommendation [12].
Selection criteria
Only patients who have PI-RADS 3 lesions on MRI were included in the final analysis. Patients with missing information on clinical, radiological, and biopsy data were excluded. Other missing data on demographic and secondary variables were managed by multiple imputations.
Data and outcomes
Covariables which were retrieved were: age, PSA, clinical stage at digital rectal exam (DRE), prostate volume calculated using ellipsoidal formula on prebiopsy MRI, PSA density (PSAd), maximum MRI lesion diameter, localization of the MRI lesion in the prostate, previous negative biopsy status, biopsy approach, and number of cores taken. The primary outcome was the identification of covariate significantly associated with a risk of csPCa defined as an ISUP grade group ≥2 on MRI-targeted and systematic biopsies.
Statistical analysis
Descriptive statistics were presented using frequency for categorical variables, and median with interquartile range (IQR) for continuous variables.
A multivariable logistic regression analysis was performed in order to identify covariables associated with csPCa within patients with PI-RADS 3 lesions on MRI. The minimum sample size for the multivariable logistic regression analysis was determined using two methods: (1) the first method, a classical approach, required a number of events equal to 10 subjects per included variable, and (2) the second method followed the criteria proposed by Riley et al. [13]. The more restrictive of the two methods was selected.
Three models were created. The first model focused solely on clinical factors, incorporating age, PSAd, and DRE status (normal vs. abnormal). The second model integrated both clinical and radiological parameters, introducing index lesion diameter and index lesion localization. The third model expanded further by incorporating the previous biopsy status (positive vs. negative previous biopsy). Performance of the models was evaluated in terms of discrimination using the area under the receiver operating characteristic curve (AUC). To reduce overfit bias and for internal validation, 1000 bootstrap resamples were performed. The extent of over- or under-estimation of the confirmed vs. predicted csPCa was analyzed graphically using a calibration plot. The net clinical benefit was evaluated on decision-curve analysis (DCA). Dependent covariables found to be significant on the logistic regression analysis were then selected for further exploration using a Chi-squared Automatic Interaction Detection (CHAID) analysis. CHAID constructs a decision tree by iteratively partitioning the patient dataset into subgroups based on variables identified as influential in predicting the studied outcome. It determines the variables most pivotal in creating meaningful subgroups within the PI-RADS 3 cohort. It then generates nodes in the decision tree, with each node representing a patient subgroup defined by specific characteristics. As the algorithm progresses, the tree branches into different paths, discerning key factors influencing the prediction of csPCa. The process continues until further splitting ceases to significantly enhance the prediction of csPCa within patients with PI-RADS 3 lesions. The terminal nodes, situated at the endpoints of the branches, represent the final, distinct subgroups with varying risks. The model was tested using a different number of nodes (two vs. three). The model with the highest discerning capability was retained. Missing data were considered missing at random, and a complete-case analysis was performed. All statistics were performed using STATA (StataCorp®, Texas, USA) software using packages pmsampsize, logit, and chaid. A p-value < 0.05 was considered as statistically significant.
Results
Patient characteristics
A total of 790 consecutive patients presented PI-RADS 3 lesions on MRI and were enrolled in the study (Fig. 1). Among them, 151 patients (19%) were diagnosed with csPCa. Comprehensive clinical, imaging, and biopsy parameters for the entire cohort and subgroups based on csPCa status are detailed in Table 1. Median PSAd was 0.19 ng/ml/cc (0.15–0.28) in patients with csPCa, compared to 0.11 ng/ml/cc (0.08–0.16) in patients with non-csPCa (Fig. 2).
Multivariable logistic regression
Table 2 displays the outcomes of the multivariable logistic regression analysis. In the first model, which focused solely on clinical factors, only PSAd emerged as a significant predictor of csPCa status (OR: 82.7 [9.7–699]; p < 0.001), yielding an AUC of 0.77 in the ROC curve. The second model, incorporating both clinical and radiological parameters, identified age (OR: 1.1 [1.0–1.1]; p = 0.01) and PSAd (OR: 1414 [63–31,957]; p < 0.001) as the sole significant predictors of csPCa, maintaining the AUC at 0.78 in the ROC curve. The third model, which included clinical, radiological, and previous biopsy status, affirmed that age (OR: 1.1 [1.0–1.1]; p = 0.009) and PSAd (OR: 1619 [66–39,535]; p < 0.001) remained the only significant predictors of csPCa, with an AUC of 0.79 in the ROC curve. Supplementary Fig. 1 shows the calibration plots, and Supplementary Fig. 2 illustrates the net benefit on DCA, demonstrating enhanced detection of csPCa within the 0–30% probability range.
The statistical analyses were repeated, excluding patients with a history of previous biopsies and focusing solely on biopsy-naïve individuals. The results remained consistent in the multivariate analysis, with only PSAd demonstrating statistical significance (OR: 466 [18–12,257]; p < 0.001), while age, DRE status, and index lesion localization were found to be non-statistically significant.
CHAID analysis
The CHAID analysis, considering only age and PSAd, identified PSAd as the sole significant factor influencing the decision tree (Supplementary Fig. 3). Cut-offs for PSAd were 0.13 ng/ml/cc for the two-nodes model and 0.09 ng/ml/cc and 0.16 ng/ml/cc for the three-nodes model. For the two-nodes model, the low-risk subgroup (PSAd < 0.13 ng/ml/cc) comprised 11 patients (1%) with csPCa, while the high-risk subgroup (PSAd ≥ 0.13 ng/ml/cc) included 140 patients (18%) with csPCa (Fig. 3). In the three-nodes model, the low-risk subgroup (PSAd < 0.09 ng/ml/cc) had 4 patients (0.5%) with csPCa, the intermediate-risk subgroup (PSAd between 0.09 ng/ml/cc and 0.16 ng/ml/cc) included 16 patients (2%), and the high-risk subgroup (PSAd ≥ 0.16 ng/ml/cc) encompassed 131 patients (17%) (Fig. 3).
For PSAd <0.09 ng/mL/cc, 13.6% of biopsies could be avoided. For PSAd <0.13 ng/mL/cc, 26% of biopsies could be avoided.
Discussion
Despite the efforts put in scoring suspicious prostate lesions, PI-RADS 3 lesions continue to present a challenge in the daily clinical practice due to their equivocal nature [11, 14]. The importance of MRI quality, as indicated by the PIQUAL classification, is underscored by the critical role played by the expertise of the uro-radiologist in interpretation [15]. PIRADS 3 lesions are particularly susceptible to variations in radiologists’ experience, given their indeterminate or equivocal nature for csPCa; expert uroradiologists are more inclined to confidently reclassify PIRADS 3 lesions as either PIRADS 1–2 or 4–5, leading to caution regarding immediate biopsy in cases classified as PIRADS 3 [15].
Previous studies have been conducted in the literature with the objective of refining biopsy indications specifically for these PI-RADS 3 lesions. To the best of our knowledge, this is the largest multi-institutional European study aimed at defining the association between clinical parameters of patients with PI-RADS 3 lesions and the presence of csPCa. Significant associations were observed for age and PSAd. In the CHAID analysis, PSAd emerged as the only statistically significant factor influencing the construction of the decision tree, with a cut-off of 0.13 in the two-nodes model and 0.09–0.16 in the three-nodes model. This study gives therefore an update of the PSA density cut-off to consider in the era of MRI-targeted biopsies when tackling PI-RADS 3 lesions.
According to the EAU-EANM-ESTRO-ESUR-ISUP-SIOG Guidelines on Prostate Cancer (2024), the decision to proceed with a biopsy based on MRI findings incorporates various thresholds for PSAd in PI-RADS 3 lesions [16]:
-
If PSAd <0.10 ng/mL/cc, a biopsy is generally not recommended.
-
If PSAd is between 0.10 and 0.15 ng/mL/cc, a biopsy should be considered.
-
If PSAd is between 0.15 and 0.20 ng/mL/cc, a biopsy should be highly considered.
-
If PSAd >0.20 ng/mL/cc, a biopsy should definitely be performed.
Our study proposes new PSAd cut-offs for patients with PI-RADS 3 lesions, suggesting that biopsies could be omitted for those with PSAd below 0.13 ng/mL/cc, especially below 0.09 ng/mL/cc. By establishing these cut-offs, our findings aim to reduce unnecessary biopsies and overdiagnosis of non-clinically significant prostate cancer.
Although opting for an MRI-targeted biopsy may appear to be the primary strategy for handling PI-RADS 3 lesions, it is worth considering a practical and acceptable alternative. This alternative entails monitoring the features of these lesions through subsequent MRI examinations, which can effectively decrease the workload and potential risks linked with additional biopsies. This approach gains significance, especially when other signs like consistent findings in DRE and PSAd are evident. Based on Boschheidgen et al.'s study, individuals who were initially diagnosed with PI-RADS 3 lesions and later confirmed to have PCa consistently experience an elevation in their PI-RADS assessment during the follow-up period when undergoing a subsequent mpMRI within 12 to 24 months. In contrast, individuals with PI-RADS 3 lesions who were negative for PCa tend to experience a downgrade in their PI-RADS assessment after 25 to 36 months [17]. Zhang et al., in their cohort of PI-RADS ≤ 3, individuals who were confirmed to have csPCa exhibited elevated levels of PSA, reduced prostate volume, and higher PSAd. Similarly, in a study involving 1057 men conducted by Venderink et al., it was demonstrated that for PI-RADS 3 lesions, when the PSAd is less than 0.15 ng/ml/cc, the rate of false negatives was only 6%, resulting in a high negative predictive value of 94% for csPCa, which stands in line with our results [18]. Szemplinski et al. retrospectively analyzed 740 men (including 17% of PI-RADS 3) who underwent MRI-targeted biopsy and found a 10% csPCa detection in PI-RADS 3 lesions [19]. A significant association for the detection of csPCa in PI-RADS 3 lesions was demonstrated only for smaller prostate volume. This is in contrast to other studies that demonstrated older age, peripheral zone location, higher PSAd, and positive DRE to be associated with detection of csPCa in PI-RADS 3 lesions [20,21,22,23].
Given the relevant research question and the advancement of artificial intelligence, Aussavavirojekul et al. aimed to address the challenge of the low cancer detection rate in patients with PI-RADS 3 lesions by developing machine learning models to assist in decision-making regarding the need for prostate biopsies or monitoring based on clinical data without biopsy result. The study incorporated clinical factors such as age, PSA, prostate volume, PSAd, and prior biopsy status. Radiological data included the number of lesions, maximum lesion size, lesion location, and zone within the prostate. They showed that machine learning models streamline the process of selecting PI-RADS 3 patients for MRI-targeted biopsy, potentially unlocking the full potential of established clinical risk factors [24].
In addition to PSAd, other biomarkers such as PSA isoforms and the Prostate Health Index (Phi) density have shown promise in enhancing the stratification of patients with PI-RADS 3 lesions. Studies have indicated that incorporating these biomarkers could improve the prediction of csPCa and assist in decision-making regarding prostate biopsies [25, 26]. For instance, the Phi, which combines total PSA, free PSA, and [-2]proPSA, has been demonstrated to provide a higher specificity for csPCa detection. Moreover, the combined use of PHI and mpMRI has shown superior performance in predicting positive biopsy outcomes and identifying high-grade cancers, which helps in minimizing overdiagnosis and overtreatment [25, 26].
The rise of biparametric MRI (bpMRI) has also an impact on the assessment of PI-RADS 3 lesions. The recently published VISIONING study investigates the use of bpMRI as a primary screening tool for PCa without relying on PSA levels [27]. The findings indicate that bpMRI has higher sensitivity for detecting csPCa compared to traditional methods. The protocol adjustment led to a 54.6% reduction in biopsies for PI-RADS 3 lesions, demonstrating that bpMRI can reduce unnecessary procedures while maintaining high detection rates of csPCa [27].
We acknowledge the retrospective nature of the present analysis which confers a potential selection bias. The presented figures likely reflect a pre-existing selection bias towards patients at a higher risk of cancer with a PI-RADS 3 classification. Therefore, despite the excellent results, this must be analyzed in consideration of this potential selection bias. It is probable that there is a limited representation of patients with low PSAd and possibly other risk parameters for prostate cancer, which could have influenced the biopsy outcomes. Moreover, the study was conducted in tertiary referral centers with dedicated physicians for each step of the procedure, which may limit the generalizability of the findings. While all participating centers adhered to prevailing guidelines and terminology, the absence of centralized review resulted in notable heterogeneity in both MRI reporting and biopsy analysis. The variability in mpMRI interpretation and PI-RADS scoring can be attributed to differences in radiologists’ training and experience, since the level of expertise significantly influences the consistency and accuracy of PI-RADS assessments, with expert radiologists more likely to reclassify equivocal lesions accurately. Despite employing the KOELIS system to minimize subjectivity and variability in biopsy core analysis compared to a cognitive approach, we acknowledge variations in prostate biopsy protocols across institutions, potentially introducing sources of variability in csPCa detection and impacting study outcomes. It is worth noting that prostate volume was determined using the ellipsoidal formula based on MRI, and alternative calculations could introduce uncertainty, especially in PSAd estimation. Finally, external validation of the prediction model could not be done due to sample size restriction (i.e., minimum required sample size was 2660 and the number of required events was 426 events using Ryley et al. method).
Conclusion
For individuals with PI-RADS 3 lesions on prostate mpMRI and a PSAd below 0.13, especially below 0.09, prostate biopsy could be omitted, in order to avoid overdiagnosis of non-csPCa. This nuanced strategy may lead to more precise and informed decision-making, potentially sparing certain patients from unnecessary invasive procedures. Prospective and randomized studies are needed to confirm our findings.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Ahmed HU, El-Shater Bosaily A, Brown LC, Gabe R, Kaplan R, Parmar MK, et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study. Lancet. 2017;389:815–22. https://doi.org/10.1016/S0140-6736(16)32401-1
Zhang Y, Zeng N, Zhang F, Huang Y, Tian Y. How to make clinical decisions to avoid unnecessary prostate screening in biopsy-naïve men with PI-RADs v2 score ≤ 3? Int J Clin Oncol. 2020;25:175–86. https://doi.org/10.1007/s10147-019-01524-9
Wysock JS, Mendhiratta N, Zattoni F, Meng X, Bjurlin M, Huang WC, et al. Predictive value of negative 3T multiparametric magnetic resonance imaging of the prostate on 12-core biopsy results. BJU Int. 2016;118:515–20. https://doi.org/10.1111/bju.13427
Haffner J, Lemaitre L, Puech P, Haber G-P, Leroy X, Jones JS, et al. Role of magnetic resonance imaging before initial biopsy: comparison of magnetic resonance imaging-targeted and systematic biopsy for significant prostate cancer detection. BJU Int. 2011;108:E171–8. https://doi.org/10.1111/j.1464-410X.2011.10112.x
Hamoen EHJ, de Rooij M, Witjes JA, Barentsz JO, Rovers MM. Use of the prostate imaging reporting and data system (PI-RADS) for prostate cancer detection with multiparametric magnetic resonance imaging: a diagnostic meta-analysis. Eur Urol. 2015;67:1112–21. https://doi.org/10.1016/j.eururo.2014.10.033
EAU Guidelines on Prostate Cancer - Uroweb. Uroweb - Eur Assoc Urol n.d. https://uroweb.org/guidelines/prostate-cancer#4. Accessed 21 May 2023.
Purysko AS, Rosenkrantz AB, Turkbey IB, Macura KJ. RadioGraphics update: PI-RADS Version 2.1—a pictorial update. RadioGraphics. 2020;40:E33–7. https://doi.org/10.1148/rg.2020190207
Oerther B, Engel H, Bamberg F, Sigle A, Gratzke C, Benndorf M. Cancer detection rates of the PI-RADSv2.1 assessment categories: systematic review and meta-analysis on lesion level and patient level. Prostate Cancer Prostatic Dis. 2022;25:256–63. https://doi.org/10.1038/s41391-021-00417-1
Schoots IG. MRI in early prostate cancer detection: how to manage indeterminate or equivocal PI-RADS 3 lesions? Transl Androl Urol. 2018;7:70–82. https://doi.org/10.21037/tau.2017.12.31
Weinreb JC, Barentsz JO, Choyke PL, Cornud F, Haider MA, Macura KJ, et al. PI-RADS prostate imaging - reporting and data system: 2015, version 2. Eur Urol. 2016;69:16–40. https://doi.org/10.1016/j.eururo.2015.08.052
Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ, et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol. 2019;76:340–51. https://doi.org/10.1016/j.eururo.2019.02.033
Epstein JI, Egevad L, Amin MB, Delahunt B, Srigley JR, Humphrey PA, et al. The 2014 International Society of Urological Pathology (ISUP) consensus conference on gleason grading of prostatic carcinoma: definition of grading patterns and proposal for a new grading system. Am J Surg Pathol. 2016;40:244–52. https://doi.org/10.1097/PAS.0000000000000530
Riley RD, Ensor J, Snell KIE, Harrell FE, Martin GP, Reitsma JB, et al. Calculating the sample size required for developing a clinical prediction model. BMJ. 2020;368:m441. https://doi.org/10.1136/bmj.m441
Zeng J, Cheng Q, Zhang D, Fan M, Shi C, Luo L. Diagnostic ability of dynamic contrast-enhanced magnetic resonance imaging for prostate cancer and clinically significant prostate cancer in equivocal lesions: a systematic review and meta-analysis. Front Oncol. 2021;11:620628.
Hong SK, Song SH, Kim HJ, Lee HS, Nam JH, Lee SB. Temporal changes of PIRADS scoring by radiologists and correlation to radical prostatectomy pathological outcomes. Prostate Int. 2022;10:188–93. https://doi.org/10.1016/j.prnil.2022.07.001
Cornford P, van den Bergh RCN, Briers E, Van den Broeck T, Brunckhorst O, Darraugh J, et al. EAU-EANM-ESTRO-ESUR-ISUP-SIOG guidelines on prostate cancer-2024 update. Part I: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2024:S0302-2838(24)02254-1. https://doi.org/10.1016/j.eururo.2024.03.027
Boschheidgen M, Schimmöller L, Doerfler S, Al-Monajjed R, Morawitz J, Ziayee F, et al. Single center analysis of an advisable control interval for follow-up of patients with PI-RADS category 3 in multiparametric MRI of the prostate. Sci Rep. 2022;12:6746 https://doi.org/10.1038/s41598-022-10859-9
Venderink W, van Luijtelaar A, Bomers JGR, van der Leest M, Hulsbergen-van de Kaa C, Barentsz JO, et al. Results of targeted biopsy in men with magnetic resonance imaging lesions classified equivocal, likely or highly likely to be clinically significant prostate cancer. Eur Urol. 2018;73:353–60. https://doi.org/10.1016/j.eururo.2017.02.021
Szempliński S, Kamecki H, Dębowska M, Zagożdżon B, Mokrzyś M, Zawadzki M, et al. Predictors of clinically significant prostate cancer in patients with PIRADS categories 3–5 undergoing magnetic resonance imaging-ultrasound fusion biopsy of the prostate. J Clin Med. 2023;12:156 https://doi.org/10.3390/jcm12010156
Felker ER, Raman SS, Margolis DJ, Lu DSK, Shaheen N, Natarajan S, et al. Risk stratification among men with prostate imaging reporting and data system version 2 category 3 transition zone lesions: is biopsy always necessary? Am J Roentgenol. 2017;209:1272–7. https://doi.org/10.2214/AJR.17.18008
Sheridan AD, Nath SK, Syed JS, Aneja S, Sprenkle PC, Weinreb JC, et al. Risk of clinically significant prostate cancer associated with prostate imaging reporting and data system category 3 (equivocal) lesions identified on multiparametric prostate MRI. Am J Roentgenol. 2018;210:347–57. https://doi.org/10.2214/AJR.17.18516
Schoots IG, Padhani AR. Risk-adapted biopsy decision based on prostate magnetic resonance imaging and prostate-specific antigen density for enhanced biopsy avoidance in first prostate cancer diagnostic evaluation. BJU Int. 2021;127:175–8. https://doi.org/10.1111/bju.15277
Kim TJ, Lee MS, Hwang SI, Lee HJ, Hong SK. Outcomes of magnetic resonance imaging fusion-targeted biopsy of prostate imaging reporting and data system 3 lesions. World J Urol. 2019;37:1581–6. https://doi.org/10.1007/s00345-018-2565-3
Aussavavirojekul P, Hoonlor A, Srinualnad S. Optimization of clinical risk-factor interpretation and radiological findings with machine learning for PIRADS category 3 patients. Prostate. 2022;82:235–44. https://doi.org/10.1002/pros.24266
Ferro M, Crocetto F, La Civita E, Fiorenza M, Jannuzzi G, Carbone G, et al. Serum (-2)proPSA/freePSAratio, (-2)proPSA/freePSA density, prostate health index, and prostate health index density as clues to reveal postoperative clinically significant prostate cancer in men with prostate-specific antigen 2-10ng/mL. Prostate. 2024. https://doi.org/10.1002/pros.24752
Ferro M, Crocetto F, Bruzzese D, Imbriaco M, Fusco F, Longo N, et al. Prostate health index and multiparametric MRI: partners in crime fighting overdiagnosis and overtreatment in prostate cancer. Cancers. 2021;13:4723. https://doi.org/10.3390/cancers13184723
Wetterauer C, Matthias M, Pueschel H, Deckart A, Bubendorf L, Mortezavi A, et al. Opportunistic prostate cancer screening with biparametric magnetic resonance imaging (VISIONING). Eur Urol Focus. 2024:S2405-4569(24)00023-3. https://doi.org/10.1016/j.euf.2024.02.006
Author information
Authors and Affiliations
Contributions
Conceptualization: GM, LH, TJ, RD. Data collection: TJ, AB, HAB, YL, MF, GS, AF, GF, MO, PG, ABG, AM, JBR, RAZ, GP, GF, AH, KR, CD, GD, JA, NBD, APB, FT, OW, DB, GA, JB, KG, TR, AP, RD. Data analysis: GM, RD. Writing – first draft: GM, LH, TJ. Writing – review and editing: GM, LH, TJ, TJ, AB, HAB, YL, MF, GS, AF, GF, MO, PG, ABG, AM, JBR, RAZ, GP, GF, AH, KR, CD, GD, JA, NBD, APB, FT, OW, DB, GA, JB, KG, TR, AP, RD. Supervision: RD.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
This research is in accordance with the Declaration of Helsinki and was conducted after IRB approval of our institution. A written informed consent was obtained from all subjects.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mjaess, G., Haddad, L., Jabbour, T. et al. Refining clinically relevant cut-offs of prostate specific antigen density for risk stratification in patients with PI-RADS 3 lesions. Prostate Cancer Prostatic Dis (2024). https://doi.org/10.1038/s41391-024-00872-6
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41391-024-00872-6
- Springer Nature Limited