Abstract
Epidermal growth factor receptor (EGFR) signaling regulates cell growth and survival, and its overactivation drives cancer development. One important branch of EGFR signaling is through activation of GTPase Rac1, which further promotes cell proliferation, survival and cancer metastasis. Here, we show that EGFR activates Rac1 via inducing the accumulation of its specific guanine nucleotide exchange factor, T-cell lymphoma invasion and metastasis 1 (Tiam1) in non-small-cell lung cancer and colon cancer cells. Conversely, elevated Tiam1 is required for EGFR-induced tumorigenesis. In human lung adenocarcinoma and colon cancer specimens, Tiam1 expression strongly correlates with EGFR expression. We further reveal that AKT, a key downstream protein kinase of EGFR, phosphorylates Tiam1 at several consensus sites, facilitates the interaction of Tiam1 with scaffold proteins 14-3-3 and leads to an increase of Tiam1 stability. Subsequently, Tiam1 is dephosporylated and destabilized by PP2A. Together, our study identifies a bidirectional (phosphorylation and dephosphorylation) regulatory mechanism controlling Tiam1 stability and provides new insights on how EGFR signaling triggers Rac1 activation and cancer development.
Similar content being viewed by others
Introduction
Overexpression or mutational activation of the epidermal growth factor receptor (EGFR) contributes to the initiation and progression of multiple types of human cancer, including non-small-cell lung cancer (NSCLC) and colon cancer.1 Mechanistically, EGF binding to receptor induces receptor dimerization and autophosphorylation, which leads to activation of the kinase domain of EGFR. Activated EGFR in turn stimulates oncogenic signaling pathways, mainly including phosphatidylinositol 3 kinase (PI3K)/AKT and Ras/mitogen-activated protein kinase (MAPK), to promote cell survival and proliferation.2 EGFR could also trigger the activation of small GTPase Rac1, which further contributes to cancer development, via the Rac1/p21-activated kinase/rapidly accelerated fibrosarcoma/MAP kinase kinase/extracellular signal-regulated kinase (Rac1/PAK/RAF/MEK/ERK)3 and the Rac1/c-Jun kinase4 cascades.
Rac1, a Ras-related small GTPase, has important roles in diverse cellular pathways, such as nicotinamide adenine dinucleotide phosphate oxidase activation, the formation of membrane ruffles and lamellipodia, transcriptional activation, cell proliferation and apoptosis.5 Its activity is subtly regulated through functional interaction with guanine nucleotide exchange factors (GEFs) that facilitate GDP to GTP exchange or GTPase-activating proteins that perform GTP hydrolysis.6 Evidence is mounting that abnormally activated Rac1 is heavily involved in the progress of tumorigenesis.7, 8 As upstream regulators of Rac1, GEFs are also implicated in many cancers.9 It is therefore of great importance to decipher the mechanisms by which the Rac1-GEF activity is modulated by growth factor receptor, like EGFR.
T-cell lymphoma invasion and metastasis 1 (Tiam1) is a specific GEF for Rac1.10 Mice deficient in Tiam1, which are viable and with no obvious abnormality, are largely resistant to Ras-induced skin tumors.11 Besides, Tiam1, as a Wnt-responsive gene, is essential for intestinal tumor development and overexpressed in colon cancer samples.12, 13 In breast cancer, its expression level is correlated with advanced tumor grade.14 It has also been demonstrated that Tiam1 is essential for Neu-induced mammary tumor formation and progression but not for c-Myc-induced tumors.15 Therefore, it is important to elucidate the mechanisms by which Tiam1 is implicated in tumor progression in the context of different oncogenic signaling pathways or cancer cell types.15 Also, in NSCLC, the oncogenic function of Tiam1 remains to be elucidated. Moreover, the mechanisms for Tiam1 upregulation in cancer samples are ambiguous. In this study, we show that EGFR/PI3K/AKT-induced Tiam1 upregulation is required for Rac1 activation and cancer development.
Results
EGFR signaling maintains Tiam1 protein level in NSCLC and CRC cells
In cancer cells, inhibition of EGFR signaling pathway could reduce Rac1-GTP level,16, 17 but the molecular mechanisms was not fully understood. Tiam1, as a specific GEF for Rac1, was required for EGF-induced Rac1 activation (Supplementary Figure 1a), which is consistent with the previous studies.18 We therefore examine the relationship between EGFR signaling and the Tiam1-Rac1 cascade. The therapeutic EGFR antibody cetuximab, which antagonizes EGFR signaling via blocking ligand binding, was applied to treat NSCLC cell line HCC827 bearing EGFR mutation (E746_A750 deletion). Consistent with previous studies, the phosphorylation of EGFR and AKT was ablated and the expression of cyclin D1 and c-Myc was both downregulated (Figure 1a). Rac1-GTP was also obviously reduced when treated with high concentration of cetuximab. Unexpectedly, Tiam1 protein was almost vanished upon the treatment of cetuximab (Figure 1a). Gefitinib, a tyrosine kinase inhibitor, used to treat NSCLC patients with EGFR mutations,19 was also able to drastically reduce Tiam1 protein level in HCC827 cells, together with the downregulation of P-AKT, Rac1-GTP, cyclin D1 and c-Myc (Figure 1b). As Rac1 activation contributes to cell proliferation through enhancing oncogenic signaling,5 blocking of EGFR signaling significantly reduced the expression of cyclin D1 and c-Myc (Figures 1a and b), at least in part, through the reduction of Tiam1 protein level and the subsequent downregulation of Rac1 signaling.
EGFR signaling controls Tiam1 protein level in cancer cells. (a) HCC827 cells were treated with cetuximab for 72 h at different concentrations. Total cell lysates were subjected to immunoblotting with the indicated antibodies. (b) HCC827 cells were treated with or without 5 μM gefitinib for 48 h. Total cell lysates were subjected to immunoblotting with the indicated antibodies. (c, d) A549 (c) and H1299 (d) cells were treated with AG1478 for 24 h with increasing concentrations. Total cell lysates were subjected to immunoblotting with the indicated antibodies. (e, f) A549 (e) and H1299 (f) cells were transfected with EGFR siRNA and 48-h post-transfection, total cell lysates were subjected to immunoblotting with the indicated antibodies. (g) BEAS-2B stable cell lines expressing EGFR mutants were subjected to detection of activated Rac1. (h) Total cell lysates derived from BEAS-2B stable cell lines expressing EGFR mutants were subjected to immunoblotting with the indicated antibodies.
The greatest inhibition of cancer cell proliferation with treatment of cetuximab and gefitinib was achieved in cells with EGFR mutations.20, 21 Hence, tyrphostin AG1478, a potent and specific inhibitor of EGFR tyrosine kinase, was applied to treat A549 and H1299, two NSCLC cell lines that harbor wild-type EGFR, rather than mutated HCC827 cells. Similarly, Tiam1 protein level was reduced in a dose-dependent manner in both A549 and H1299 cells treated with AG1478 (Figures 1c and d).
To further verify whether EGFR contributes to maintaining Tiam1 protein level, we knocked down EGFR by small interfering RNAs (siRNAs) in A549 and H1299 cells. Consistently, Tiam1 protein level was reduced in the EGFR knockdown cells (Figures 1e and f). As EGFR is also overexpressed or overactivated in colon cancer cells,22 we examined whether Tiam1 protein level could be affected when EGFR tyrosine kinase was inactivated in colon cancer cells. Inhibition of EGFR activity by AG1478 or EGFR knockdown by siRNAs both markedly downregulated Tiam1 protein level in DLD-1 cells (Supplementary Figures 1b and c), further strengthened the importance of EGFR signaling in maintaining Tiam1 protein level. Remarkably, EGFR knockdown had no effect on the mRNA level of Tiam1 (Supplementary Figure 1d), suggesting that EGFR signaling maintained Tiam1 protein level via posttranscriptional regulation.
Conversely, to test whether EGFR activation could elevate Tiam1 level, human lung bronchial epithelial cell BEAS-2B expressing EGFR harboring exon 19 deletion or L858R were generated (Figures 1g and h). As anticipated, overexpression of mutant EGFR significantly upregulated Tiam1 protein level, accompanied with elevated Rac1-GTP and c-Myc (Figures 1g and h), suggesting that overexpression of EGFR effectively enhanced Rac1 activity through increasing Tiam1 protein level. Together, EGFR signaling is essential for Tiam1 expression in cancer cells.
Positive correlation of Tiam1 and EGFR expression in lung adenocarcinoma and colon cancer samples
Having established that EGFR signaling contributed to Tiam1 expression in lung or colon cancer cell lines, we performed immunohistochemistry analyses to examine the expression level of EGFR and Tiam1 in 60 human lung cancer specimens (Supplementary Table 1). As shown in Figure 2a, the immunostaining intensity of Tiam1 and EGFR was significantly stronger in lung cancer than those in matched adjacent tissues. Quantification analyses based on the staining of EGFR and Tiam1 further exhibited that both proteins were remarkably elevated (Figures 2b and c, P=1.34 × 10−43 and P=4.02 × 10−33, respectively). Importantly, Pearson’s correlation analysis on the scores from immunostaining sections indicated that EGFR expression level was strongly correlated with that of Tiam1 (Figures 2d, r=0.69). These results suggest that EGFR may regulate Tiam1 expression during tumorigenesis.
Tiam1 upregulation is correlated with EGFR overexpression in lung and colon cancer samples. (a) Immunohistochemical staining with anti-EGFR and anti-Tiam1 antibodies was performed on 60 human lung adenocarcinoma and adjacent tissues. Representative images of cancer samples (right panels) and adjacent tissues (left panels) were shown. Scale bar represents 100 μm. (b, c) EGFR expression scores (b) and Tiam1 expression scores (c) were shown as dot blots. Lung adenocarcinoma and matched adjacent tissues were compared using Student’s t-test. n=60. (d) Semiquantitative scores of EGFR and Tiam1 expression in lung adenocarcinoma were analyzed using Pearson’s correlation test. r=0.69. (e) Total cell lysates originated from 12 human colon cancer samples (T) and matched adjacent tissues (N) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blots with the indicated antibodies.
EGFR is also frequently elevated in human colorectal cancer samples and this is correlated with metastasis and poor prognosis.23 To confirm the correlation between Tiam1 and EGFR in colon tissues, we analyzed 12 colon cancer samples with their matched normal tissues by immunoblotting. The results showed that both EGFR and Tiam1 have a similar expression pattern between normal and paired tumor tissues (Figure 2e). Together, these results indicated that Tiam1 upregulation is correlated with elevated expression of EGFR in tumor tissues.
Tiam1 is required for non-small lung cancer growth
Although it has been demonstrated that Tiam1/Rac1 signaling has important roles in cell growth in multiple cancers,24 whether EGFR signaling induced upregulation of Tiam1 is required for tumor growth in a xenograft model bearing EGFR-driving lung cancer cells remained unverified. To resolve this question, we first generated A549 and H1299 cells expressing Tiam1 short hairpin RNA (shRNA), which presented lower levels of Rac1-GTP, cyclin D1 and c-Myc in comparison with control cells (Figure 3a and Supplementary Figure 2a). Subsequently, these A549 cells were subcutaneous injected into athymic nude mice and the results revealed that knockdown of Tiam1 inhibited in vivo tumorigenesis when compared with cells expressing control shRNA (Figure 3b). Similar experiments were carried out with HCC827 cells bearing a mutant EGFR and silencing of Tiam1 significantly downregulated the levels of Rac1-GTP, cyclin D1 and c-Myc, and inhibited tumor growth in a xenograft model (Figures 3c and d). Consequently, the final tumor size and tumor weight were markedly smaller and lighter in the mice injected with Tiam1 knockdown cells than those of control cells (Figures 3e and f). These results suggested that Tiam1 contributes to lung tumor growth.
Tiam1 is required for tumor growth in xenografted mice. (a) Total cell lysates derived from A549 stable cells expressing Control (shCtrl) or Tiam1 shRNA (shTiam1) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blots with the indicated antibodies. (b) A549 stable cells expressing Control (shCtrl) or Tiam1 shRNA (shTiam1) were injected into nude mice (n=6). Representative images of xenografted mice (shCtrl, right; shTiam1, left) were shown in the graph. * indicates P<0.05. (c) Total cell lysates derived from HCC827 stable cells expressing Control (shCtrl) or Tiam1 shRNA (shTiam1) were subjected to SDS–PAGE and western blots with the indicated antibodies. (d, e) HCC827 stable cells expressing Control (shCtrl) or Tiam1 shRNA (shTiam1) were injected into nude mice (n=5) and then the tumor volumes were measured (d). Images of all tumors were shown (e). ** indicates P<0.005. (f) The weight of tumors (e) was measured. Two groups of data were compared using Student’s t-test. (g) Total cell lysates derived from BEAS-2B stable cell lines expressing Control (shCtrl) or Tiam1 shRNA (shTiam1) were subjected to SDS–PAGE and western blots with the indicated antibodies. (h) BEAS-2B stable cell lines expressing Control (shCtrl) or Tiam1 shRNA (shTiam1) were injected into nude mice (n=5) and then the tumor volumes were measured.
To further elucidate the role of EGFR-induced Tiam1 upregulation, control or Tiam1 shRNA were introduced into BEAS-2B-EGFR L858R stable cells (Figure 3g). Expression of EGFR L858R mutant in BEAS-2B epithelial cells upregulated Tiam1 (Figure 3g) and induced anchorage-independent growth in soft agar (Supplementary Figure 2b).25 As expected, these cells were strongly tumorigenic in xenografted mice, whereas the parental BEAS-2B cells formed hardly any tumor (Figure 3h). Knockdown of Tiam1 in BEAS-2B-EGFR L858R cells markedly inhibited tumor growth (Figure 3h), suggesting that the tumorigenic role of EGFR is through Tiam1. Taken together, these results indicated that upregulation of Tiam1 by overactivated EGFR signaling contributed significantly to EGFR-induced tumorigenesis.
AKT1 interacts with and phosphorylates Tiam1 in vitro and in vivo
Next obvious question is how EGFR signaling upregulates Tiam1 expression. It is well known that EGFR activates PI3K/AKT and Ras/MAPK signaling pathways.26 We then applied specific inhibitors of these two signaling branches to dissect their roles on Tiam1 expression. To our surprise, treatment with U0216, an Erk inhibitor, had no effect on Tiam1 protein level in DLD-1 cells (Supplementary Figure 3a), suggesting that Ras/MAPK signaling was not involved in the regulation of Tiam1 expression. In contrast, treatment with PI3K/AKT signaling inhibitor LY294002 significantly reduced the level of Tiam1 protein in a dose-dependent manner in three cell lines (Figures 4a and b; Supplementary Figure 3b), implying that the PI3K/AKT pathway is critical for the regulation of the Tiam1 protein level. Moreover, knockdown of AKT1 by a verified siRNA27 also reduced the Tiam1 protein level in all the three cell lines (Figure 4c; Supplementary Figures 3c and d). Importantly, AKT1 silence did not result in obvious changes of Tiam1 mRNA level (Supplementary Figure 3c).
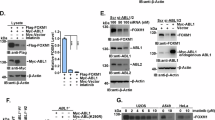
AKT interacts with Tiam1 and phosphorylates it at several sites. (a, b) H1299 (a) or DLD-1 (b) cells were treated with LY294002 for 24 h with increasing concentrations. Total cell lysates were subjected to immunoblotting with the indicated antibodies. (c) Total cell lysates derived from H1299 (left panel) or DLD-1 (right panel) cells transfected with Control (Ctrl) or AKT1 siRNA for 48 h were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and western blots with the indicated antibodies. (d) Cell lysates from HEK293T cells transfected with Myc-Tiam1, HA-AKT1 and Myr-HA-AKT1 plasmids were subjected to immmunoprecipitation (IP) with anti-Myc beads. WCL stands for whole-cell lysate. (e) Cell lysates from H1299 cells were subjected to IP with IgG or anti-Tiam1 antibodies. (f) The amino-acid sequences of Tiam1 from different species were analyzed and four conserved AKT recognizing sequences were shown. (g) Cell lysates from HEK293T cells transfected with Myc-Tiam1 and Myr-HA-AKT1 plasmids were subjected to IP with anti-Myc beads. (h) Western blots of protein samples from in vitro kinase assay. Myr-HA-AKT1 purified from transfected HEK293T cells was incubated with GST-Tiam1-A fragment containing S60, S172 and S231sites purified from E. coli. GST-Tiam1-A was shown as Coomassie blue staining. (i) Cell lysates from HEK293T cells transfected with Myc-Tiam1 wild type (WT) or its single point mutants were subjected to IP with anti-Myc beads and then detected with indicated antibodies after SDS–PAGE. (j) Cell lysates from HEK293T cells transfected with the indicated plasmids were subjected to IP with anti-Myc beads. 2A, 3A and 4A indicate S172/231A, S60/172/231A and S60/172/231/695A, respectively. L and S stand for long and short exposure, respectively. (k) H1299 cells were serum-starved for 24 h and then treated with EGF (100 ng/ml) for 4 h. Total cell lysates were directly subjected to western blotting (WCLs) to show the upregulation of Tiam1 or subjected to IP with anti-Tiam1 antibody. The immunoprecipitates were then subjected to western blots with the indicated antibodies.
To test whether AKT directly regulate the Tiam1 protein stability, their interaction was verified in transiently transfected HEK293T cells. The interaction between Tiam1 and wild-type AKT1 was evidently detected, while a stronger binding was observed with Myr-AKT1, a constitutively active form (Figure 4d). Furthermore, the association between endogenous Tiam1 and AKT1 was confirmed in H1299 cells (Figure 4e). These results promoted us to think that AKT is able to phosphorylate Tiam1. According to previous reports, the conserved substrate recognition sequence for AKT is RXRXXS/T or RXXS/T (X indicates any residue).28 We therefore analyzed Tiam1 protein sequences from several species and recognized four potential AKT phosphorylation motifs (S60, S172, S231 and S695) (Figure 4f; www.phosphosite.org). To confirm that these sites were indeed phosphorylated in cells, immunopurified Myc-Tiam1 from transfected HEK293T cells was analyzed by mass spectrometry. The results indicated that the four conserved AKT sites were all indeed phosphorylated (Supplementary Figures 4a and b).
Next, we used a commercial anti-phospho-AKT substrate antibody and a self-made rabbit polyclonal antibody against phospho-Tiam1 S231 (the specificity of the p-Tiam1 S231 was shown in Supplementary Figure 4c) to testify whether AKT could phosphorylate Tiam1 in vivo and in vitro. As shown in Figure 4g, co-transfection of Myr-AKT1 and Tiam1 in HEK293T cells obviously led to Tiam1 phosphorylation at S231 and increased phosphorylation of AKT substrate of Tiam1, indicating that AKT indeed was capable of phosphorylating Tiam1 in vivo. To further demonstrate a direct phosphorylation event, we purified GST-Tiam1-A (a fragment containing amino-acid 1-392 of Tiam1, see Supplementary Figure 5c) from Escherichia coli (E. coli) and Myr-AKT1 from transfected HEK293T cells and then performed in vitro kinase assay. As shown in Figure 4h, GST-Tiam1-A was remarkably phosphorylated by AKT protein kinase as detected by both anti-P-AKT substrate and anti-P-Tiam1 S231 antibodies. Notably, Myr-AKT1 also phosphorylated GST-Tiam1-B (393-750aa) (see Supplementary Figure 5c) in vitro, which was detected by anti-P-AKT substrate antibody (Supplementary Figure 4d), implying that S695 of Tiam1 was likely phosphorylated, although we failed to produce a specific antibody against Tiam1 S695.
To determine the possibility that Tiam1 could be phosphorylated by AKT at other three sites S60, S172 and S695, we generated a series of single or multiple point mutations on full-length Tiam1. As shown in Figures 4i and j, mutation of all four sites but not single point mutations reduced the recognition by anti-P-AKT substrate antibody to a great extent, suggesting that AKT1 may phosphorylate multiple sites on Tiam1. Finally, we show that, in H1299 cells, EGF treatment induced Tiam1 accumulation and this was correlated with elevated Tiam1 phosphorylation detected by both (P-Tiam1 S231 and P-AKT substrate) antibodies (Figure 4k). Taken together, these results suggested that EGFR signaling may stimulate AKT to phosphorylate Tiam1.
Tiam1 interacts with 14-3-3 via its AKT phosphorylation sites
On substrates, AKT phosphorylation often generates binding motifs for scaffold protein 14-3-3 family members, which affects stabilization or the subcellular localization of the substrates.29, 30, 31 A previous report has identified 14-3-3 as a Tiam1-binding partner.32 Our co-immunoprecipitation results confirmed the interactions between endogenous Tiam1 and 14-3-3 (Figure 5a). Their interaction was also proved using GST pull-down assays, in which GST-14-3-3β purified from E. coli strongly bound to exogenous or endogenous Tiam1 (Supplementary Figures 5a and b). Furthermore, Tiam1 bound to 14-3-3β in a phosphorylation-dependent manner as γPPase treatment significantly reduced the interaction (Figure 5b), consistent with the understanding that 14-3-3 binding is generally phospho dependent.31
Tiam1 interacts with 14-3-3 via its phosphorylation sites. (a) Cell lysates from H1299 cells were subjected to IP with IgG or pan 14-3-3 antibodies. IgG LC stands for IgG light chain. (b) GST or GST-14-3-3β purified from E. coli was incubated with total cell lysates derived from H1299 cells. Where noted, λPPase were added. The inputs and GST pull-down samples were subjected to western blots with the indicated antibodies. (c) GST or GST-14-3-3β purified from E. coli was incubated with total cell lysates derived from HEK293T cells transfected with Myc-Tiam1 five fragments. The inputs and GST pull-down samples were subjected to western blots with the indicated antibodies. GST and GST-14-3-3β were shown as Coomassie blue staining. (d) GST or GST-14-3-3β purified from E. coli was incubated with total cell lysates derived from HEK293T cells transfected with Myc-Tiam1 wild type (WT) or its single point mutations. The inputs and GST pull-down samples were subjected to Western blots with the indicated antibodies. GST and GST-14-3-3β were shown as Coomassie blue staining. (e) GST or GST-14-3-3β purified from E. coli was incubated with total cell lysates derived from HEK293T cells transfected with Myc-Tiam1 wildtype (WT) or its multiple point mutations. The inputs and GST pull-down samples were subjected to western blots with the indicated antibodies. GST and GST-14-3-3β were shown as Coomassie blue staining. (f) Cell lysates derived from HEK293T cells transfected with increasing amount of Flag-14-3-3β were subjected to immunoblotting with the indicated antibodies. (g) Cell lysates from serum-starved H1299 cells were subjected to IP with IgG or anti-pan 14-3-3 antibodies. The whole-cell lysates (WCLs) and immunoprecipitates were subjected to western blots with the indicated antibodies. Where noted, cells were treated with EGF (100 ng/ml) for 4 h.
To further dissect their interaction, domain mapping analysis was carried out. As shown in Supplementary Figure 5c, fragments of Tiam1 were prepared and their interaction with 14-3-3β was monitored in vivo as well as in vitro. In transfected HEK293T cells, 14-3-3β mainly interacted with Tiam1-A, whereas its interaction with Tiam1-B and -E was much weaker and with Tiam1-C and -D undetectable (Supplementary Figure 5d). GST pull-down assay further revealed that 14-3-3β predominantly bound to Tiam1-A, and much weakly to Tiam1-E (Figure 5c). These results suggested that Tiam1 interacted with 14-3-3β largely through its N-terminus, which contains three conserved AKT phosphorylation sites (S60, S172 and S231).
Next, we testified whether those AKT phosphorylation sites (S60, S172, S231 and S695) were required for Tiam1’s association with 14-3-3β. Serine to alanine mutation of each site was introduced into Tiam1 and their interaction with 14-3-3β were verified in transfected HEK293T cells. As shown in Supplementary Figure 5e, the interaction between these single point mutations and 14-3-3β was weakened to certain extents in comparison with the wild type. This was further confirmed in in vitro GST pull-down analysis (Figure 5d). What’s more, multiple point mutations (2A, S172/231A; 3A, S60/172/231A; 4A, S60/172/231/695A) gradually reduced their interaction with 14-3-3β, whereas the 4A mutation almost abolished it (Figure 5e and Supplementary Figure 5f). These results showed that the four AKT phosphorylation sites are largely responsible for Tiam1/14-3-3β association. Consistently, 14-3-3β overexpression caused a gradual increase of endogenous Tiam1 in HEK293T cells (Figure 5f), suggesting that 14-3-3β is indeed able to stabilize Tiam1. More importantly, in response to EGF treatment, the interaction between 14-3-3 and Tiam1 was obviously enhanced and consequently Tiam1 level was elevated (Figure 5g), suggesting that 14-3-3 binding is of importance for EGFR-induced Tiam1 accumulation. Overall, EGFR stimulates AKT, which in turn phosphorylates Tiam1 and facilitates its 14-3-3 binding, leading to Tiam1 stabilization.
PP2A interacts with and dephosphorylates Tiam1
PP2A, an abundant phosphatase, negatively regulate EGFR signaling largely through dephosphorylating AKT.33 PP2A was also identified as a potential interacting protein for Tiam1 by mass spectrometry.32 We thus determined whether PP2A could directly dephosphorylate Tiam1. PP2A consists of three subunits, including structural subunit (A), regulatory subunit (B) and catalytic subunit (C).34 Co-immunoprecipitation results indicated that Tiam1 clearly associated with PP2A-A, as well as PP2A-B subunits (Figure 6a). Moreover, endogenous Tiam1 was also co-immunoprecipitated with PP2A-B subunit (Figure 6b), which is responsible for defining the substrate specificity.35 Consistently and interestingly, PP2A-B subunit selectively interacted with Tiam1-A and -E fragments (Figure 6c), which were also recognized by 14-3-3. In vitro GST pull-down analysis further confirmed that GST-Tiam1-A directly bound to PP2A-B (Figure 6d). Finally, purified Tiam1 from Myr-AKT1 co-transfected HEK293T cells was completely dephosphorylated by a commercial PP2A core enzyme, PP2A-A/C, in an in vitro dephosphorylation assay (Figure 6e).
PP2A is involved in regulating Tiam1 stability through dephosphorylating Tiam1. (a) Cell lysates from HEK293T cells transfected with Myc-Tiam1 and Flag-PP2A-structural (A), regulatory (B) and catalytic (C) subunits were subjected to IP with anti-Myc beads. WCL stands for whole-cell lysate. (b) Cell lysates from HEK293 cells were subjected to IP with anti-HA (as a control) or anti-PP2A-B antibodies. The input and immunoprecipitates were subjected to western blots with the indicated antibodies. (c) Cell lysates from HEK293T cells transfected with Flag-PP2A-B and Myc-Tiam1 five fragments were subjected to IP with anti-Flag beads. (d) GST or GST-Tiam1-A purified from E. coli was incubated with total cell lysates derived from HEK293T cells transfected with Flag-PP2A-B. The inputs and GST pull-down samples were subjected to western blots with the indicated antibodies. GST and GST-Tiam1-A were shown as Coomassie blue staining. (e) Cell lysates from Myc-Tiam1 and Myr-HA-AKT1 co-transfected HEK293T cells were directly subjected to immunoblotting (WCL) or subjected to IP with Myc beads and then in vitro dephosphorylation assay using PP2A core enzyme (PP2A-A/C) was performed. The samples were then subjected to immunoblotting with the indicated antibodies. (f) Total cell lysates derived from HEK293T cells transfected with Control (Ctrl), PP2A-A or PP2A-B siRNA were subjected to western blots with the indicated antibodies. (g) HEK293T and DLD-1 cells were treated with okadaic acid (OA) for 24 h with indicated concentrations. Total cell lysates were subjected to western blots with the indicated antibodies. (h) Cell lysates derived from H1299 cells treated with Vehicle or 5 nM OA for 24 h were subjected directly to immunoblotting (WCL) or to IP with anti-Tiam1 antibody and then detected with indicated antibodies after sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Next, we detected the effect on Tiam1 protein level when PP2A was downregulated. As anticipated, knockdown of PP2A-A or -B subunit using siRNAs led to significant Tiam1 upregulation (Figure 6f). In addition, the treatment with okadaic acid, an inhibitor of PP2A, also resulted in significant accumulation of Tiam1 in HEK293T and DLD-1 cells (Figure 6g). Moreover, okadaic acid-induced Tiam1 accumulation was accompanied with hyper-phosphorylation (Figure 6h). Taken together, PP2A directly interacts with Tiam1 and negatively regulates its stability through dephosphorylation.
Discussion
In this study, we show that EGFR, via PI3K/AKT, promotes the phosphorylation and subsequent 14-3-3 binding of Tiam1, a specific GEF for Rac1. Binding to 14-3-3 causes Tiam1 stabilization and Rac1 activation, and further facilitates lung tumor initiation and progression by upregulating the expression of downstream oncogenes, such as c-Myc and cyclin D1 (Figure 7). Rac1 signaling has well-established oncogenic activity to promote tumorigenesis. Overexpression of Rac1-GEFs, like Vav1, is able to increase Rac1-GTP levels and promote pancreatic cancer tumorigenesis.36 A number of proteins are identified as Rac1 downstream targets, like p21-activated kinases, p70S6K (p70 ribosomal S6 kinase) and c-Jun kinase, which are all implicated in tumor progression.4, 37, 38 As a result of these critical functions of Rac1 signaling in tumorigenesis, chemical inhibitors of Rac1 have been developed to halt cancer cell growth.39, 40
A model. Mutated EGFR or ligand bound wild-type EGFR leads to PI3K/AKT activation. AKT phosphorylates Tiam1 at several serine/threonine sites and then facilitates its binding to 14-3-3. Tiam1 bound to 14-3-3 become stable and accumulates to promote high Rac1-GTP, which contributes to cell growth and proliferation.
GEFs, including Tiam1, are recruited to the plasma membrane to facilitate the exchange of GDP to GTP and achieve Rac1 activation.41, 42 Their PH domains are required for cell membrane localization.6, 10 EGFR kinase or its downstream kinase has key roles in recruiting GEFs to cell membrane.18, 36 EGFR-stimulated translocation of the GEFs to the cell membrane may rely on its downstream signaling molecules, such as PIP36. A recent study indicated that the lipid messenger PtdIns5P can also bind Tiam1 DH-PH domains to activate Tiam1 and Rac1.43 In this study, we suggest that EGFR/PI3K/AKT axis signaling triggers Tiam1 phosphorylation and 14-3-3 binding, which stabilizes Tiam1 protein. Moreover, given that 14-3-3 is able to regulate the subcellular localization of its binding partners,29 it is likely that 14-3-3 binding not only functions in Tiam1 stabilization, but also promotes its cell membrane localization, both of which contribute to Rac1 activation. This is consistent with the previous reports showing that 14-3-3 binding to Tiam1 is required for Rac1 activation.44, 45 A previous study found S60A mutation was stabilized but with reduced interaction with 14-3-3, while other single mutations (S172 or S231) have no effect on the half-life of Tiam1.32 It was hypothesized that 14-3-3 binding to different combination of phosphorylation sites resulted in distinct conformational changes, thereby regulating Tiam1 stability.32 Here, we proposed that phosphorylation of these residues, including S60, may generate binding motifs for an unknown E3 ligase responsible for Tiam1 degradation, whereas 14-3-3 binding could protect Tiam1 from proteasome-mediated degradation.
Abnormal overexpression of the Rac1-GEFs is involved in promoting cancer progression and metastasis in diverse cancer types.36, 46 For example, P-Rex1 is overexpressed in breast cancers and the expression level of P-Rex1 is correlated with the sensitivity to PI3K inhibitors. Therefore, P-Rex1 is suggested a biomarker to predict response to cancer treatment by PI3K inhibitors.47 Tiam1, another important Rac1-GEF, also contributes to cancer development and metastasis.12 In support of this, it has been established that Tiam1 is overexpressed in various tumor types, such as colon cancer and breast cancer.12, 14 We show here that Tiam1 is also highly overexpressed in NSCLC. As TIAM1 gene amplification in cancer cells were not reported, the upregulation of Tiam1 in tumors is probably due to the alteration of transcriptional, translational or posttranslational regulation. Moreover, although mutations in Tiam1 was identified in human renal cell carcinomas,48 whether the mutations have effect on Tiam1 stability remain obscure. In our work, we identified the EGFR/PI3K/AKT axis to directly regulate Tiam1 phosphorylation and stability. Furthermore, we showed that Tiam1 expression was correlated with EGFR level in both NSCLC and CRC cells. AKT activation could be due to many reasons, including but not limited to EGFR mutation or overexpression, PI3K overactivation, loss of phosphatase and tensin homolog deleted on chromosome 10 activity and AKT itself overactivation. Therefore, Tiam1 upregulation may serve as a biomarker to indicate the activation of the signaling cascade and may also predict response to reagents, like cetuximab or PI3K inhibitors, targeting the EGFR/PI3K/AKT axis.
The accumulation of multiple genetic and epigenetic alterations is a hallmark of cancer.49 Hence, it is not always an effective way to treat cancer by targeting one molecule. In addition, drug resistance occurs via bypassing EGFR by obtained enhancement of another oncogenic signaling pathway.50, 51 Therefore, blocking oncogenic signaling from more than one target appears an effective way to treat cancer or restrict drug resistance. The Tiam1/Rac1 signaling can be considered as an oncogenic hub, as it is activated by multiple upstream stimulus and then initiates several downstream oncogenic signaling pathways.52 There may be an option to halt lung cancer cell proliferation by combinatory therapy with the inhibitors for EGFR and Tiam1/Rac1. Supportively, a recent study demonstrated that the Tiam1/Rac1 signaling is also required for proliferation of chronic lymphocytic leukemia (CLL) cells, and that Rac1 inhibition may be of clinical use by antagonizing the chemoresistance of chronic lymphocytic leukemia cells toward fludarabine.53 Inhibitors that suppress Rac1 activation are likely promising therapeutic partners for EGFR-targeted agents, like Erlotinib, in a combined treatment of glioblastoma.54 Hence, as a specific GEF for Rac1, Tiam1 could be a potential therapeutic target in clinical cancer treatment.
Materials and methods
Cell culture
HEK293, HEK293T and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum. DLD-1, A549, H1299 and BEAS-2B cells were cultured in RPMI-1640 containing 10% fetal bovine serum. HCC827 cells were cultured in Dulbecco’s modified Eagle’s medium with 20% fetal bovine serum. All culture medium were supplemented with penicillin and streptomycin.
Plasmids
Myc-Tiam1, Myc-Tiam1−A, −B, −C, −D and −E have been descried previously.55 Myc-14-3-3β and GST-14-3-3β were kindly provided by Dr Yeguang Chen and Myc-14-3-3β was served as a template for cloning 14-3-3β complementary DNA into pcDNA3.1-Flag expression vector. Tiam1-A and -B were subcloned into pPGH vector containing N-terminal GST tag. HA-AKT1 and myr-HA-AKT1 were a kind gift from Dr Joseph R Testa. Complementary DNAs of PP2A-A, PP2A-B and PP2A-C (kindly provided by Dr Yigong Shi and Dr Chuanmao Zhang) were subcloning into pcDNA3.1-Flag expression vector. Mutations in full-length Tiam1 were generated by site-directed mutagenesis and confirmed by DNA sequencing.
Antibodies and reagents
A polyclonal antibody against phosphor-serine 231 of Tiam1 was generated by Abmart (www.ab-mart.com.cn) through immunizing rabbits with two peptides (CQRANSLGDLY and CQRAN(pS)LGDLY) and phosphor-specific antibody was affinity purified. Anti-Myc tag (sc-40), anti-Tiam1 (sc-872), pan 14-3-3 (sc-629) and protein A/G PLUS-Agarose beads were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-Flag M2, anti-AKT1 (P2482) and anti-Flag M2-Agarose were purchased from Sigma-Aldrich (St Louis, MO, USA). Anti-Rac1 (610651) was purchased from BD Transduction Laboratories (Lexington, KY, USA). Anti-cyclin D1 (K0062-3) was from MBL (Nagoya, Japan). Anti-PP2A-A (2041), anti-PP2A-B (PPP2R2A, 5689), anti-PP2A-C (2038), c-Myc (5605), phospho-AKT S473 (4060), phospho-AKT substrate (RXXS/T) (9614), EGFR (4267), P-EGFR Y1068 (3777), rabbit IgG (2729) and tyrphostin AG1478 (9842) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti c-myc (mouse) Agarose was from Abmart. LY294002 (S1737), U0126 (S1901) and okadaic acid (S1786) were purchased from Beyotime (Nantong, Jiangsu, China). Gefitinib (S1025) and cetuximab (Erbitux) were from Selleck Chemicals(Houston, TX, USA) and Merck Serono (Darmstadt, Germany), respectively.
Transfection and RNA interference
Plasmid DNA and siRNA transfections were performed using VigoFect transfection reagent (Vigorous Biotech, Beijing, China) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), respectively, according to the manufacturer’s instructions. siRNA sequences were as follows:
EGFR-1#: 5′-GCAGTGACTTTCTCAGCAA-3′;
EGFR-2#: 5′-GCCCAAAGTTCCGTGAGTT-3′;
AKT1: 5′-CUCACAGCCCUGAAGUACU-3′;27
PP2A-A-1#: 5′-GACCAGGATGTGGACGTCAAA-3′;
PP2A-A-2#: 5′-GGACCCGAAGUGAGCUUCUTT-3′;
PP2A-C-1#: 5′-ATGGAACTTGACGATACTCTA-3′;
PP2A-C-2#: 5′-UCUGUGGAGAUGUGCAUGG-3′.
Tiam1: 5′-CCGACTCCTGAAATCAGAGATCAAA-3′.56
Generation of stable cell lines
BEAS-2B cells stably expressing EGFR-L858R or EGFR-exon 19 deletions were established. Briefly, linearized pCAG-iresNeo vector57 harboring mutant EGFR (L858R or exon 19 deletion) were delivered into BEAS-2B cells with Lipofectamine 2000. After 2 weeks selection with G418 (600 μg/ml), clones were picked and characterized for EGFR expression. BEAS-2B stable cells were then infected with lentivirus vectors LV2-negative control shRNA (5′-TTCTCCGAACGTGTCACGT-3′) or LV2-Tiam1 shRNA (5′-GCGAAGGAGCAGGTTTTCT-3′), which were purchased from GenePharma (Shanghai, China). Similarly, A549, H1299 and HCC827 cells were also infected with lentivirus vector expressing control or Tiam1 shRNA. Forty-eight hours post-infection, cells were cultured in the indicated culture medium containing puromycin (1 μg/ml) and the medium was changed every 2 days. After about 1 week, stable cell lines were obtained and verified by western blots.
Western blot, co-immunoprecipitation assay and GST pull-down assay
These methods were performed as described previously.58
Rac1 activation assay
Rac1 activation assays were performed as described previously59. Briefly, GST-PBD (a p21-activated kinase-1-binding domain) was used to enrich active GTP-bound Rac1. Cells were washed with pre-cold phosphate-buffered saline and lysed in Rac1 lysis buffer (50 mM Tris, pH 7.5, 200 mM NaCl, 2% NP40, 10% glycerol, 10 mM MgCl2 and 1 × protease inhibitor cocktail) on ice for 10 min. Cell lysates were centrifuged by 14 000 r.p.m. for 15 min at 4 °C and the total protein concentration was measured through BCA protein assays (Pierce, Rockford, IL, USA). Equal amounts of cell lysates were incubated with 10 μg GST-PBD agarose beads for 2 h at 4 °C. The precipitated complexes were washed three times with Rac1 wash buffer (25 mM Tris, pH 7.5, 40 mM NaCl, 1% NP40, 30 mM MgCl2 and 1 × protease inhibitor cocktail). Finally, the beads were boiled with 2 × sodium dodecyl sulfate-loading buffer and samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Mass spectrometric analysis
The assay was performed as described previously.60
Immunohistochemistry
Immunohistochemistry was performed by Shanghai Superchip Company (Shanghai, China) with human lung adenocarcinoma tissue array. A semiquantitative analysis of Tiam1 and EGFR expression in stained sections was assessed by an independent pathologist. The German semiquantitative scoring system in considering the staining intensity and area extent was applied. Immunohistochemical staining intensity scores were indicated as: negative (0), weak staining (1), moderate staining (2) and strong staining (3) and the extent of stained cells were indicated as: 0%=0, 1–24%=1, 25–49%=2, 50–74%=3, 75–100%=4. The final scores were defined by multiplying the intensity scores with the scores of the extent of stained cells (0–12).
Human tumor specimens
Cancer tissues analyzed in Figure 2e were collected in the Chinese PLA General Hospital in China. The tissue collection procedure with informed consent was approved by the Clinical Ethic Committee of the Chinese PLA General Hospital.
In vitro kinase assay and in vitro dephosphorylation assay
In vitro kinase assays were performed as described previously.61 Briefly, recombinant GST-Tiam1-A and GST-Tiam1-B purified from E. coli were incubated with Myr-HA-AKT1 purified from transfected HEK293T cells in the presence of 1 mM ATP and kinase buffer (50 mM Hepes, pH 7.5, 50 mM NaCl, 1 mM DTT, 10 mM MgCl2) for 30 min with gentle shaking. For in vitro dephosphorylation assay, purified Myc-Tiam1 from transfected HEK293T cells were incubated with 0.5 units total of PP2A (Millipore, 14–111, Darmstadt, Germany) per reaction according to the manufacturer’s instructions.
Soft agar assay
The assay was performed as described previously.61 For BEAS-2B stable cell lines, 2000 cells per well was seeded in six-well plates.
Tumor xenografts
Animal experiments were approved by Experimental Animal Management and Ethics Committee of Tsinghua University. Six-week old athymic BALB/c nude female mice were injected with different numbers of cells. For detailed, H1299 cells (6 × 105 cells), HCC827 cells (1 × 106 cells) and BEAS-2B stable cell lines (3 × 106 cells) infected with control shRNA or Tiam1 shRNA lentivirus particles in 100 ul Dulbecco’s modified Eagle’s medium/Matrigel (BD Biosciences, Heidelberg, Germany) (1:1) solution was subcutaneously implanted into mice. The tumor size was measured by digital caliper and tumor volume was calculated by the formula: volume=length x width2/2.
Statistical analysis
Data were presented as mean±s.d. A two-tailed Student’s t-test was used to evaluate the statistical significance in the mean value between two populations (P<0.05).
References
Hynes NE, Lane HA . ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer 2005; 5: 341–354.
Avraham R, Yarden Y . Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol 2011; 12: 104–117.
Arai A, Jin A, Yan W, Mizuchi D, Yamamoto K, Nanki T et al. SDF-1 synergistically enhances IL-3-induced activation of the Raf-1/MEK/Erk signaling pathway through activation of Rac and its effector Pak kinases to promote hematopoiesis and chemotaxis. Cell Signal 2005; 17: 497–506.
Davis RJ . Signal transduction by the JNK group of MAP kinases. Cell 2000; 103: 239–252.
Jaffe AB, Hall A . Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol 2005; 21: 247–269.
Schmidt A, Hall A . Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev 2002; 16: 1587–1609.
Vega FM, Ridley AJ . Rho GTPases in cancer cell biology. FEBS Lett 2008; 582: 2093–2101.
Ellenbroek SI, Collard JG . Rho GTPases: functions and association with cancer. Clin Exp Metastasis 2007; 24: 657–672.
Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, Kazanietz MG . Rac signaling in breast cancer: a tale of GEFs and GAPs. Cell Signal 2012; 24: 353–362.
Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A et al. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell 1994; 77: 537–549.
Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG . Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature 2002; 417: 867–871.
Minard ME, Ellis LM, Gallick GE . Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin Exp Metastasis 2006; 23: 301–313.
Malliri A, Rygiel TP, van der Kammen RA, Song JY, Engers R, Hurlstone AF et al. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J Biol Chem 2006; 281: 543–548.
Adam L, Vadlamudi RK, McCrea P, Kumar R . Tiam1 overexpression potentiates heregulin-induced lymphoid enhancer factor-1/beta -catenin nuclear signaling in breast cancer cells by modulating the intercellular stability. J Biol Chem 2001; 276: 28443–28450.
Strumane K, Rygiel T, van der Valk M, Collard JG . Tiam1-deficiency impairs mammary tumor formation in MMTV-c-neu but not in MMTV-c-myc mice. J Cancer Res Clin Oncol 2009; 135: 69–80.
Ramis G, Thomas-Moya E, Fernandez de Mattos S, Rodriguez J, Villalonga P . EGFR inhibition in glioma cells modulates Rho signaling to inhibit cell motility and invasion and cooperates with temozolomide to reduce cell growth. PloS One 2012; 7: e38770.
Dise RS, Frey MR, Whitehead RH, Polk DB . Epidermal growth factor stimulates Rac activation through Src and phosphatidylinositol 3-kinase to promote colonic epithelial cell migration. Am J Physiol Gastrointest Liver Physiol 2008; 294: G276–G285.
Itoh RE, Kiyokawa E, Aoki K, Nishioka T, Akiyama T, Matsuda M . Phosphorylation and activation of the Rac1 and Cdc42 GEF Asef in A431 cells stimulated by EGF. J Cell Sci 2008; 121: 2635–2642.
Sordella R, Bell DW, Haber DA, Settleman J . Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science 2004; 305: 1163–1167.
Mulloy R, Ferrand A, Kim Y, Sordella R, Bell DW, Haber DA et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res 2007; 67: 2325–2330.
Mukohara T, Engelman JA, Hanna NH, Yeap BY, Kobayashi S, Lindeman N et al. Differential effects of gefitinib and cetuximab on non-small-cell lung cancers bearing epidermal growth factor receptor mutations. J Natl Cancer Inst 2005; 97: 1185–1194.
Krause DS, Van Etten RA . Tyrosine kinases as targets for cancer therapy. N Engl J Med 2005; 353: 172–187.
Nicholson RI, Gee JM, Harper ME . EGFR and cancer prognosis. Eur J Cancer 2001; 37: S9–15.
Minard ME, Kim LS, Price JE, Gallick GE . The role of the guanine nucleotide exchange factor Tiam1 in cellular migration, invasion, adhesion and tumor progression. Breast Cancer Res Treat 2004; 84: 21–32.
Steiner P, Joynes C, Bassi R, Wang S, Tonra JR, Hadari YR et al. Tumor growth inhibition with cetuximab and chemotherapy in non-small cell lung cancer xenografts expressing wild-type and mutated epidermal growth factor receptor. Clin Cancer Res 2007; 13: 1540–1551.
da Cunha Santos G, Shepherd FA, Tsao MS . EGFR mutations and lung cancer. Annu Rev Pathol 2011; 6: 49–69.
Caporali S, Levati L, Starace G, Ragone G, Bonmassar E, Alvino E et al. AKT is activated in an ataxia-telangiectasia and Rad3-related-dependent manner in response to temozolomide and confers protection against drug-induced cell growth inhibition. Mol Pharmacol 2008; 74: 173–183.
Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P . Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett 1996; 399: 333–338.
Hermeking H . The 14-3-3 cancer connection. Nat Rev Cancer 2003; 3: 931–943.
Fu H, Subramanian RR, Masters SC . 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol 2000; 40: 617–647.
Morrison DK . The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 2009; 19: 16–23.
Woodcock SA, Jones RC, Edmondson RD, Malliri A . A modified tandem affinity purification technique identifies that 14-3-3 proteins interact with Tiam1, an interaction which controls Tiam1 stability. J Proteome Res 2009; 8: 5629–5641.
Sato S, Fujita N, Tsuruo T . Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci USA 2000; 97: 10832–10837.
Xu Y, Xing Y, Chen Y, Chao Y, Lin Z, Fan E et al. Structure of the protein phosphatase 2 A holoenzyme. Cell 2006; 127: 1239–1251.
Eichhorn PJ, Creyghton MP, Bernards R . Protein phosphatase 2 A regulatory subunits and cancer. Biochim Biophys Acta 2009; 1795: 1–15.
Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, Savoy DN, Molina JR, Fonseca R et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell 2005; 7: 39–49.
Laplante M, Sabatini DM . mTOR signaling in growth control and disease. Cell 2012; 149: 274–293.
Kumar R, Gururaj AE, Barnes CJ . p21-activated kinases in cancer. Nat Rev Cancer 2006; 6: 459–471.
Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y . Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA 2004; 101: 7618–7623.
Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ . Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem 2007; 282: 35666–35678.
Buchanan FG, Elliot CM, Gibbs M, Exton JH . Translocation of the Rac1 guanine nucleotide exchange factor Tiam1 induced by platelet-derived growth factor and lysophosphatidic acid. J Biol Chem 2000; 275: 9742–9748.
Michiels F, Stam JC, Hordijk PL, van der Kammen RA, Ruuls-Van Stalle L, Feltkamp CA et al. Regulated membrane localization of Tiam1, mediated by the NH2-terminal pleckstrin homology domain, is required for Rac-dependent membrane ruffling and C-Jun NH2-terminal kinase activation. J Cell Biol 1997; 137: 387–398.
Viaud J, Lagarrigue F, Ramel D, Allart S, Chicanne G, Ceccato L et al. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nat Commun 2014; 5: 4080.
O'Toole TE, Bialkowska K, Li X, Fox JE . Tiam1 is recruited to beta1-integrin complexes by 14-3-3zeta where it mediates integrin-induced Rac1 activation and motility. J Cell Physiol 2011; 226: 2965–2978.
Kobayashi H, Ogura Y, Sawada M, Nakayama R, Takano K, Minato Y et al. Involvement of 14-3-3 proteins in the second epidermal growth factor-induced wave of Rac1 activation in the process of cell migration. J Biol Chem 2011; 286: 39259–39268.
Sosa MS, Lopez-Haber C, Yang C, Wang H, Lemmon MA, Busillo JM et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell 2010; 40: 877–892.
Ebi H, Costa C, Faber AC, Nishtala M, Kotani H, Juric D et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc Natl Acad Sci USA 2013; 110: 21124–21129.
Engers R, Zwaka TP, Gohr L, Weber A, Gerharz CD, Gabbert HE . Tiam1 mutations in human renal-cell carcinomas. Int J Cancer 2000; 88: 369–376.
Hanahan D, Weinberg RA . Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674.
Vlacich G, Coffey RJ . Resistance to EGFR-targeted therapy: a family affair. Cancer cell 2011; 20: 423–425.
Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, Souglakos J et al. Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med 2011; 3: 99ra86.
Bid HK, Roberts RD, Manchanda PK, Houghton PJ . RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol Cancer Ther 2013; 12: 1925–1934.
Hofbauer SW, Krenn PW, Ganghammer S, Asslaber D, Pichler U, Oberascher K et al. Tiam1/Rac1 signals contribute to the proliferation and chemoresistance, but not motility, of chronic lymphocytic leukemia cells. Blood 2014; 123: 2181–2188.
Karpel-Massler G, Westhoff MA, Zhou S, Nonnenmacher L, Dwucet A, Kast RE et al. Combined inhibition of HER1/EGFR and RAC1 results in a synergistic antiproliferative effect on established and primary cultured human glioblastoma cells. Mol Cancer Ther 2013; 12: 1783–1795.
Zhu G, Fan Z, Ding M, Mu L, Liang J, Ding Y et al. DNA damage induces the accumulation of Tiam1 by blocking beta-TrCP-dependent degradation. J Biol Chem 2014; 289: 15482–15494.
Palamidessi A, Frittoli E, Garre M, Faretta M, Mione M, Testa I et al. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell 2008; 134: 135–147.
Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M et al. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol 2003; 23: 2699–2708.
Su X, Zhu G, Ding X, Lee SY, Dou Y, Zhu B et al. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev 2014; 28: 622–636.
Habas R, He X . Activation of Rho and Rac by Wnt/frizzled signaling. Methods Enzymol 2006; 406: 500–511.
Wang L, Piao T, Cao M, Qin T, Huang L, Deng H et al. Flagellar regeneration requires cytoplasmic microtubule depolymerization and kinesin-13. J Cell Sci 2013; 126: 1531–1540.
Zhu G, Wang Y, Huang B, Liang J, Ding Y, Xu A et al. A Rac1/PAK1 cascade controls beta-catenin activation in colon cancer cells. Oncogene 2012; 31: 1001–1012.
Acknowledgements
We thank Drs Ye-Guang Chen, John G. Collard, Chuanmao Zhang, Joseph R Testa and Yigong Shi for providing plasmids and reagents. We are grateful for Dr Haiteng Deng and Jieyuan Liu for mass spectrometry analysis. This work was supported by grants to WW from the National Natural Science Foundation of China (31221064 and 81272235). GZ is a postdoctoral fellow supported by Tsinghua-Peking Center for Life Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhu, G., Fan, Z., Ding, M. et al. An EGFR/PI3K/AKT axis promotes accumulation of the Rac1-GEF Tiam1 that is critical in EGFR-driven tumorigenesis. Oncogene 34, 5971–5982 (2015). https://doi.org/10.1038/onc.2015.45
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.45
- Springer Nature Limited
This article is cited by
-
Nectin-4 regulates cellular senescence-associated enlargement of cell size
Scientific Reports (2023)
-
A20 interacts with mTORC2 to inhibit the mTORC2/Akt/Rac1 signaling axis in hepatocellular carcinoma cells
Cancer Gene Therapy (2022)
-
T-box transcription factor 19 promotes hepatocellular carcinoma metastasis through upregulating EGFR and RAC1
Oncogene (2022)
-
Hypoxia-induced Fascin-1 upregulation is regulated by Akt/Rac1 axis and enhances malignant properties of liver cancer cells via mediating actin cytoskeleton rearrangement and Hippo/YAP activation
Cell Death Discovery (2021)
-
Role of the V1G1 subunit of V-ATPase in breast cancer cell migration
Scientific Reports (2021)