Abstract
Prostate tumours are highly variable in their response to therapies, but clinically available prognostic factors can explain only a fraction of this heterogeneity. Here we analysed 200 whole-genome sequences and 277 additional whole-exome sequences from localized, non-indolent prostate tumours with similar clinical risk profiles, and carried out RNA and methylation analyses in a subset. These tumours had a paucity of clinically actionable single nucleotide variants, unlike those seen in metastatic disease. Rather, a significant proportion of tumours harboured recurrent non-coding aberrations, large-scale genomic rearrangements, and alterations in which an inversion repressed transcription within its boundaries. Local hypermutation events were frequent, and correlated with specific genomic profiles. Numerous molecular aberrations were prognostic for disease recurrence, including several DNA methylation events, and a signature comprised of these aberrations outperformed well-described prognostic biomarkers. We suggest that intensified treatment of genomically aggressive localized prostate cancer may improve cure rates.
Similar content being viewed by others
Main
Prostate cancer is the most commonly diagnosed non-skin malignancy in men, and resulted in 256,000 deaths worldwide in 2010 (ref. 1). Although most men present with localized, potentially curable disease, current clinical prognostic factors explain only a fraction of the heterogeneity of treatment response. These factors therefore do not optimally triage individual patients into risk groupings that can be used to determine how aggressively the cancer should be treated2,3.
Localized prostate cancers exhibit striking inter-tumoural heterogeneity, at both the genomic4,5 and microenvironmental6 levels. In particular, intermediate risk prostate cancers are localized, non-indolent and clinically heterogeneous. Despite management with surgery or radiotherapy, about 30% of men suffer relapses; in 10% of these men (approximately 10,000 per year in North America), rapid biochemical recurrence can portend prostate-cancer-specific death7. Having a rigorous understanding of the genetic factors that drive progression and aggression in the initial pre- and post-treatment settings is essential for both clinicians and genetic researchers, as distinct genomic pathways of progression could define prostate cancer sub-types and lead to novel curative therapies. It is important to identify the genetic drivers of localized, non-indolent prostate cancer, as they cannot be inferred from studies of metastatic castrate-resistant prostate cancer (mCRPC) owing to tumour cell selection and adaption to androgen deprivation therapy8.
Here we describe, to our knowledge, the largest cohort of prostate cancer samples to have been subjected to whole-genome sequencing: 200 non-indolent localized specimens. We provide saturating discovery of recurrent driver single nucleotide variants (SNVs), copy number aberrations (CNAs) and genomic rearrangements in this clinical group, and associate these with epigenomic profiles. Future studies in other clinical settings (for example, early-onset disease) and population-specific contexts (for example, males of African ancestry) will be critical to generalize these findings. We confirm many well characterized recurrent molecular aberrations and identify novel prognostic translocations, inversions and epigenetic events. Together, these data provide insights into the genomic landscape of localized prostate cancer, and highlight molecular aberrations that may help to triage patients for precision prostate cancer medicine.
Saturating genomic interrogations
To address the genetic heterogeneity of non-indolent localized prostate cancer, we first comprehensively profiled CNAs in 284 localized prostate adenocarcinomas (Supplementary Table 1; Supplementary Fig. 1). The profiles recapitulated those previously reported, including recurrent allelic gains of MYC and deletions of PTEN, TP53 and NKX3-1 (Supplementary Results; Supplementary Figs 2–4; Supplementary Tables 2–6). Even in this clinically homogeneous population, we observed large inter-tumoural heterogeneity in the percentage of the genome with a CNA (per cent genome altered (PGA), 0–39.2%)4.
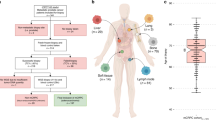
We next performed high-depth whole-genome sequencing (WGS) of 130 of these tumours (and matched blood samples), focusing on localized tumours amenable to surgery (that is, with a Gleason score (GS) of 3 + 3, 3 + 4 or 4 + 3). These were supplemented by 70 pairs of tumour and normal tissue samples with publicly available read-level WGS data9,10,11,12 and 277 read-level exome sequences9,10,12,13, all with similar GSs. WGS data covered 84.2 ± 2.5% (mean ± s.d.) of the non-repetitive genome to at least 17× for tumour samples and 67.1–85.7% to 10× for normal samples, allowing robust analysis of the entire genome. All samples were aligned and profiled for SNVs and genomic rearrangements, using well characterized and validated pipelines14 (Fig. 1a and Supplementary Tables 1, 7–9). Overall, this process yielded 477 prostate tumours with analysis of somatic coding SNVs (Supplementary Data 1, Extended Data Fig. 1, Fig. 1a). These data give 62.9–99.9% power to detect recurrent coding and non-coding SNVs at 0.5–10% recurrence15 (Supplementary Fig. 5a, b). Similarly we had over 99.9% and 44.7% power to detect genomic rearrangements present at 10% and 3% recurrence, respectively (Supplementary Fig. 5c). To supplement these metrics, we performed RNA abundance profiling of 73 tumours, and methylation profiling of 104. We generated methylation subtypes through unsupervised machine learning (Supplementary Table 1).
We analysed genomic profiles of 200 localized, non-indolent prostate tumours. a, Each column represents an individual tumour that underwent WGS, sorted first by GS, then by the number of somatic SNVs identified (top). The middle and bottom panels show the number of genomic rearrangements (GR) and CNAs, respectively. The clinical covariates GS, PSA, T-category, and age are shown, with a colour key for each. Box plots to the right show the association between mutation load and GS, with P values from one-way ANOVAs. b, Correlation between mutation load (PGA, SNV, INV+CTX and CNA) and clinical variables. Background shading indicates Bonferroni-adjusted P values; size and colour of dots show Spearman’s correlation.
We observed a low overall SNV burden, with a median of 0.53 (0.05–6.92) somatic SNVs per million base pairs across all tumours (Fig. 1a). SNV burden was significantly elevated in tumours containing Gleason pattern 4, with a median of 1,063, 1,482 and 1,585 in tumours with GSs of 3 + 3, 3 + 4 and 4 + 3, respectively (P = 1.05 × 10−3; t-test). The number of genomic rearrangements was highly variable across tumours (median 19, 0–499) and those with any GS 4 component (that is, 3 + 4 or 4 + 3) showed elevated rates (median 17 genomic rearrangements in GS 3 + 3 versus 22 in GS 3 + 4 and 4 + 3; P = 5.11 × 10−4; t-test). The number of inversions and translocations was correlated with SNV burden (Fig. 1b, Extended Data Fig. 2a; ρ = 0.56, P = 1.32 × 10−17). We found several other associations between mutational burden and covariates such as serum prostate-specific antigen (PSA) levels, tumour size and ETS gene family fusions (Supplementary Table 9; Extended Data Fig. 2).
Somatic SNV profiles
Individual tumours harboured 0–98 exomic SNVs (Fig. 2). The median number of non-synonymous SNVs increased with GS (GS 3 + 3, 7; 3 + 4, 9; 4 + 3, 10; P = 0.001, one-way ANOVA; Supplementary Data 1, Supplementary Fig. 6). Only six genes were mutated by coding SNVs in more than 2% of tumours: SPOP (8.0%; 38/477), TTN (4.4%, 21/477), TP53 (3.4%; 16/477), MUC16 (2.5%; 12/477), MED12 (2.3%; 11/477) and FOXA1 (2.3%; 11/477). The AR gene was altered by non-synonymous SNVs in only 2 out of 477 tumours (one GS 3 + 3 and one GS 3 + 4), while allelic deletions in AR were observed in 4 out of 284 tumours and amplification in 1 out of 284 tumours. Notably, eight tumours (1.75%) harboured mutations in the DNA damage checkpoint activator gene ATM. Mutations in several genes, most prominently FAT1, were associated with GS (0/78 in GS 3 + 3; 0/261 in GS 3 + 4; 5/133 in GS 4 + 3; P = 0.0048, Fisher’s exact test). Similarly, mutations in multiple genes were associated with increased genomic instability as measured by PGA; these genes included MYO15A (2.7% in wild type versus 6.3% in mutated; false discovery rate (FDR) P = 1.01 × 10−11). Assuming a median background mutation rate of 2.44 × 10−1 mutations per Mbp for transcribed regions (including exons and introns but excluding UTRs), we estimate that there remain no genes to be discovered at the ≥1% rate, but around five undiscovered genes mutated at the 0.5% level. The low frequency of these mutations juxtaposed with the high rate of CNAs confirms the C-class character of localized prostate cancers16.
We created a consistent, standardized set of somatic SNV predictions in the exome from a set of 477 tumours. Tumours are sorted by GS (bottom covariates), then by the total number of coding SNVs identified per sample (top bar plot). The proportion of each type of base change is given in the middle bar plot. The heat map displays the 19 most recurrently mutated genes, each found in at least 6 samples, ranked by the number of somatic SNVs.
We next explored the non-coding regions of the genome in the 200 tumours that underwent WGS. Multiple recurrent noncoding SNVs (ncSNVs) (that is, ones with identical genomic position) were detected: 7 ncSNVs were observed in at least 7 out of 200 patients, and 63 were mutated in 4–6 patients. These SNVs are thus present at a similar mutation rate (about 2–4%) as TP53, MED12 and FOXA1 (Extended Data Fig. 3a). Most tumours harboured at least one recurrent ncSNV (≥2% recurrence; median: 1 per tumour). There was a strong bias in trinucleotide context towards TCT/AGA trinucleotides (from 27/70 SNVs). Validation of these SNVs in further cohorts will be critical to generalize these findings. Several ncSNVs showed trends towards association with GS, PGA and ETS gene fusions, highlighting a potential role in driving mutational phenotypes, and the need for larger cohorts to uncover these effects. Recurrent ncSNVs were not associated with replication time (Supplementary Fig. 7), and encompassed a broad range of variant allele frequencies, from clonal to small subclones (Extended Data Fig. 3b). Recurrent ncSNVs did not generally localize to specific transcription factor binding sites, although genomic rearrangements and CNAs did (Supplementary Results, Extended Data Fig. 3c, Supplementary Fig. 8). We therefore considered the potential impact of SNVs on chromatin structure, across a wide range of marks from multiple cell-types using DeepSEA17 and in a panel of 14 marks characterized in the LNCaP prostate cancer-derived cell line (Extended Data Fig. 3d, Supplementary Table 10). Six out of seventy recurrent ncSNVs showed evidence of perturbing chromatin structure at q < 0.01, but no individual chromatin feature was significantly enriched across ncSNVs.
We next quantified trinucleotide mutational signatures with non-negative matrix factorization18. Three distinct trinucleotide signatures were identified from WGS data (Supplementary Fig. 9a; Supplementary Table 11). Signature 2 reflects the deamination profile previously reported as a hallmark of sequencing false positives14,18. Increased expression of signature 2 showed a marginal positive association with T3 (β = 0.398; q = 0.044; generalized linear model (glm)) and a negative association with age (β = −0.015; q = 0.022; glm); signature 3 showed a weak positive association with age (β = 0.014; q = 0.049; glm). By contrast, signature 1 was characterized by a relatively uniform mutational profile and was not associated with age, GS, PSA, or T category (Supplementary Table 12). These signatures occur in individual patients at different frequencies (Supplementary Fig. 9b, Supplementary Table 13). The fraction of SNVs in a tumour attributed to a given signature (called its ‘exposure’) were correlated with recurrent CNA segments and genomic rearrangement 1-Mbp bins. Supplementary Table 14 shows the significant CNA genes (at the 5% FDR corrected level) for each signature. There were no significant correlations between the exposures of signatures and genomic rearrangements.
Genomic rearrangements in localized prostate cancer have not been extensively studied. As expected, the TMPRSS2:ERG (T2E) fusion on chromosome 21 was the most recurrent genomic rearrangement (38%, 76 out of 200 patients; Extended Data Fig. 4a). Other frequent alterations include translocation of MMS22L (chr6q16.1) and ARHGAP10 (chr4q31.23) in 12 of 200 tumours and translocation of chr17p11.1 and chr1q21.2 in 7 of 200 tumours. These alterations were reflected by several chromosome pairs being involved in more inter-chromosomal genomic rearrangements than expected (Extended Data Fig. 4b), including some without prominent focal genomic rearrangement peaks (for example, chr4–chr6: expected 2 CTXs, observed 14 CTXs; q < 0.001, permutation test). Anticipating that these effects might be induced by inter-chromosomal proximity19,20, we compared pair-wise genomic rearrangement enrichment to Hi-C data measuring inter-chromosomal links in the RWPE1 prostate cancer cell line21. Translocations between a few chromosome pairs co-localized with Hi-C links, but many more were further from Hi-C links than expected by chance (Extended Data Fig. 4c).
To further understand regional genomic rearrangement effects, we divided the genome into 1-Mbp bins and considered the frequency of genomic rearrangements in each (Extended Data Fig. 4a, Supplementary Table 15). Six bins had elevated rates of inversions: chr3:125–126 Mbp and chr3:129–130 Mbp contained inversions in 6% of patients (12 out of 200); chr10:89–90 Mbp contained inversions in 5.5% of patients (11 of 200); and chr3:195–196 Mbp, chr21:39–40 Mbp and chr21:42–43 Mbp all contained inversions in 5% of patients (10 of 200). A recurrent inversion on chr10:89–90 Mbp in 11 of 200 patients was associated with a significant decrease in the mRNA abundance of three genes within it (ATAD1, LOC439994, and PTEN (Extended Data Fig. 5a)), suggesting a novel mode of PTEN repression. Patients with this inversion showed lower PTEN pathway activity than those with deletions of PTEN (Extended Data Fig. 5b). This mode of repression may be more general than just the PTEN locus: inversions in chr3:129–130 Mbp also dysregulate mRNA abundance, with 8 out of 15 genes repressed in tumours harbouring the inversion (P < 0.05, model-based t-test (limma); Extended Data Fig. 5c).
Localized somatic hyper-mutation
Whereas some tumours are initiated or driven by recurrent point mutations, others are driven by focal genomic instability at the level of either DNA double-strand breaks (that is, chromothripsis22) or single-strand breaks (that is, kataegis18). Using ShatterProof23, we detected chromothripsis in 20% of tumours (38 out of 186) with CNA data (Fig. 3a; Supplementary Data 2). Chromothripsis was associated with larger tumour size (Kendall’s τ = 0.23, P = 3.07 × 10−4; Extended Data Fig. 6a), but not with other clinical variables such as age (P = 0.24) or GS (P = 0.35). Chromothripsis was associated with point mutations in FOXA1 (P = 0.008) and CNAs in NKX3-1 (P = 3.5 × 10−4), CHD1 (P = 1.7 × 10−4) and CDKN1B (P = 3.5 × 10−4; Wilcoxon rank-sum test). Chromothriptic tumours were also significantly enriched for deletion of a locus on chr8 q36.32–p11.21 containing ADRA1A, PPP3CC and several genes other whose mRNA abundance was correlated with ShatterProof scores (Extended Data Fig. 7a). Overall CNA burden was modestly increased in chromothriptic tumours, as were essentially all mutation types, but tumour cellularity was not (Extended Data Figs 6b, 7b). Genes within chromothriptic regions largely showed reduced mRNA abundance but not methylation, and were greatly enriched for genes deleted in tumours without chromothriptic events, suggesting that chromothripsis tends to inactivate tumour suppressors (Extended Data Fig. 8). Correlations between methylation probes and mRNA transcripts changed in regions of chromothripsis, suggesting perturbed epigenetic regulation, and genes whose correlation changed in chromothriptic tumours were enriched (FDR < 5%) in pathways associated with development (Extended Data Fig. 9; Supplementary Table 16). The mRNA abundances of 57 genes were strongly correlated with chromothripsis (|R| ≥ 0.35; Supplementary Table 17). The mRNA abundances of several immune genes were negatively correlated with chromothripsis, including the proto-oncogene DBL (also called MCF2; ρ = −0.43, P = 2.0 × 10−4) and CD36 (ρ = −0.39, P = 7.0 × 10−4), suggesting that immune dysregulation might have a role in chromothripsis, although few infiltrating immune cells were identified in primary tumours and their presence was not correlated with chromothripsis (Extended Data Fig. 6d–f).
We assessed the frequency and consequences of chromothripsis and kataegis in tumours with GS 3 + 3, 3 + 4 and 4 + 3. a, For each tumour we quantified the extent of chromothripsis using ShatterProof and ranked samples in descending order of evidence of a chromothriptic event (top barplot). We analysed the association of chromothripsis with measures of mutational burden, known prostate cancer genes with recurrent CNAs, novel chromothripsis-associated genes, T2E fusion status, known prostate cancer genes with recurrent SNVs and clinical variables. Bar plots to the right give the statistical significance of each association, (Mann–Whitney U test for genes, Kendall’s τ for clinical covariates). b, We quantified the presence of kataegic events and visualized them as in a, this time using the tests of proportions. Top bar plot shows the score of the strongest kataegic event.
To quantify kataegis, we developed a sliding-window approach using the binomial test, a test for base change enrichment and an assessment of the expected proportion of variants within a given window. We detected kataegis in 46 out of 200 samples (23%; Fig. 3b, Supplementary Data 3). Kataegic tumours were significantly enriched for CHD1 deletion (15 out of 45 (33%) with kataegis versus 16 out of 141 (11.3%) without kataegis; P = 0.001, prop-test). Additionally, kataegis was preferentially found in tumours with SNVs or CNAs in SPOP (P = 0.05, prop-test) or genomic rearrangements in regions on 4q (129–130 Mbp; FDR q = 0.002, prop-test) or 6q (126–127 Mbp; FDR q = 0.006, prop-test). Furthermore, tumours with kataegic events showed significantly elevated genomic instability (Extended Data Fig. 6c; P = 7.52 × 10−3, t-test). Kataegis was more likely to occur in tumours with elevated Gleason grade (13% of GS 3 + 3 samples had kataegic events versus 19% of GS 3 + 4 and 39% of GS 4 + 3 samples; Kendall’s τ = 0.21, FDR q = 0.004).
Recurrent aberrations predict outcome
To characterize recurrent events better in localized prostate cancer, we evaluated the association of each of these with patient survival. Of the 200 patients whose samples were whole-genome sequenced, 130 had available data on disease relapse, as measured by biochemical recurrence (BCR, see Methods), with a median 7.96-year follow-up. We systematically evaluated the clinical relevance of 40 recurrent genomic alterations in localized prostate cancer: three measures of mutation density, kataegis, chromothripsis, five recurrent coding SNVs, six recurrent non-coding SNVs, six methylation events, six recurrent translocations, four recurrent inversions and eight CNAs. For each, we employed univariate CoxPH modelling (Fig. 4a). Only one SNV was predictive of patient outcome: all patients with point mutations in ATM suffered relapse (Fig. 4b). A recurrent inter-chromosomal translocation breakpoint at the chromosome 7 centromere (chr7:61–62 Mbp) and amplification of MYC were also prognostic for BCR24. By contrast, no measures of mutation intensity (that is, PGA or the number of genomic rearrangements or SNVs) or density (that is, chromothripsis or kataegis) were associated with BCR, although PGA showed a strong trend towards an effect.
a, We defined 40 properties of prostate cancers, including mutation density, presence/absence of chromothripsis and kataegis and a series of recurrent somatic mutations. For each, we calculated the association with BCR using a CoxPH model and show the HR, 95% CI and P value (Wald test). b, Kaplan–Meier plot of biochemical relapse-free survival proportion of patients with and without ATM nonsynonymous SNVs. c, Kaplan–Meier plot of biochemical relapse-free survival proportion of patients with and without hypermethylation of TCERG1L at the 5′ and hypomethylation of TCERG1L at the 3′ probe. d, Receiver operating characteristic (ROC) curves for a multi-modal biomarker predicting biochemical recurrence, tested via cross-validation (yellow) and a PGA marker (green). Blue dots represent the operating point (maximum balanced accuracy). AUC, area under the curve.
Methylation status was much more tightly associated with patient outcome than any other genomic characteristic: of the nine events significantly (P < 0.05; Wald test) associated with disease recurrence, six involved DNA methylation. For example, hyper-methylation of a probe 5′ of a transcriptional elongation regulator (TCERG1L) showed a strong association with poor outcome (hazard ratio (HR) = 2.90; 95% CI, 1.30–6.30; P = 0.007). Another probe on the 3′ end of TCERG1L showed the inverse association, with hypo-methylation associated with good outcome (HR = 0.17; 95% CI, 0.06–0.49; P = 9.45 × 10−4; Fig. 4c). Of the six prognostic methylation events, five were validated in an independent cohort of 100 intermediate-risk patients (Extended Data Fig. 10a–f).
Finally, we evaluated whether these events could be integrated into a multi-modal biomarker to predict disease relapse. We applied multivariate CoxPH modelling using cross-validation to test the outcome of a multi-modal biomarker: T-category, ACTL6B hyper-methylation, TCERGL1 hypo-methylation, the chr7:61 Mbp CTX, ATM SNVs, and MYC CNA. This signature was highly discriminative of patients who would experience disease relapse, with an area under the ROC curve of 0.83 (95% CI: 0.80–0.86), as compared to that of 0.61 for the validated PGA biomarker6,25 (Fig. 4d), and with a concordance index of 0.79. This discriminative ability predicted differences in patient survival (HR = 4.71; 95% CI, 2.17–10.24; P = 9.00 × 10−5; Wald test; Extended Data Fig. 10g, h).
Discussion
We used WGS to identify recurrent mutational events outside the exome in localized, non-indolent prostate cancer. Because of the paucity of driver and prognostic coding aberrations, consideration of the entire prostate cancer genome may be critical in biomarker studies to find driver aberrations that have been missed in smaller studies4,10. For example, we identified several inversions associated with decreases in mRNA abundance, potentially representing a novel mode of tumour-suppressor inactivation. Replication of our newly identified alterations and candidate biomarkers in additional datasets and with additional technologies will be a key next step towards clinical translation. Similarly, functional and mechanistic evaluation of the mutational profiles described here will be important to understand their role in driving aggressive prostate cancer. This study focused solely on index lesions of each tumour, and as such does not directly account for the large spatio-genomic heterogeneity of prostate cancer, except through its large sample size4,5. Understanding of this heterogeneity and the associated evolutionary history of the disease will be an important next step in understanding the aetiology of prostate cancer.
Our data also highlight the differences in mutational profiles between localized intermediate risk cancers and metastatic castrate resistant prostate cancer (mCRPC). Nearly 50% of mCRPCs harbour mutations in AR, ETS genes, TP53 and PTEN and about 20% have aberrations in DNA damage response genes (for example, BRCA1, BRCA2 and ATM, which may portend sensitivity to poly-ADP ribose polymerase (PARP) inhibitors26,27,28). Furthermore, more than 60% of mCRPCs contain clinically actionable mutations that are not related to AR8. By contrast, non-SNV mutations dominate the driver landscape of localized, non-indolent prostate cancer. No single gene was mutated at more than 10% frequency and the only gene in which SNVs were prognostic was ATM.
In the modern era of PSA screening, many patients initially present with aggressive non-indolent prostate cancers with aggressivity. We show that localized disease has a different biology from advanced mCRPCs, which have undergone significant selective pressure, often through multiple courses of treatment29. As recurrent SNV driver aberrations are rare in localized disease, genetically unstable localized tumours requiring intensified therapy may benefit from widespread genotoxic chemotherapy as supported by clinical trials in treatment-naive, metastatic disease30. Similarly, the development of novel therapeutics will be improved by a robust understanding of the non-exomic drivers of aggression in localized prostate cancer.
Methods
Patient cohort
All patients underwent either image-guided radiotherapy (IGRT) or radical prostatectomy (RadP), with curative intent, for pathologically confirmed prostate cancer. All patients were hormone naive at the time of definitive local therapy. In the IGRT cohort, a single ultrasound-guided needle biopsy was obtained before the start of therapy, as previously described6. Fresh-frozen RadP specimens were obtained from the University Health Network (UHN) Pathology BioBank or from the Genito-Urinary BioBank of the Centre Hospitalier Universitaire de Québec (CHUQ). Whole blood was collected and informed consent, consistent with local Research Ethics Board (REB) and International Cancer Genome Consortium (ICGC) guidelines, was obtained at the time of clinical follow-up. Previously collected tumour tissue was used, following University Health Network REB-approved study protocols (UHN 06-0822-CE, UHN 11-0024-CE, CHUQ 2012-913:H12-03-192). To confirm GS and tumour cellularity, all tumour specimens were independently evaluated by two genitourinary pathologists (T.v.d.K., B.T.) on scanned, haematoxylin and eosin (H&E)-stained slides. Serum PSA is reported based on the reading at the time of diagnosis, and is given in ng/ml. Pathological (RadP samples) and clinical (IGRT samples) T category was reported using standard National Comprehensive Cancer Network (NCCN) criteria (http://www.nccn.org/professionals/physician_gls/pdf/prostate.pdf). All patients were N0M0 as an entry criterion for the study. For IGRT patients, BCR was defined as a rise in PSA concentration of more than 2.0 ng/ml above the nadir (after radiotherapy, PSA levels drop and stabilize at the nadir). For RadP patients, BCR was defined as two consecutive post-RadP PSA measurements of more than 0.2 ng/ml (backdated to the date of the first increase). If a patient has successful salvage radiation therapy, this is not BCR. If PSA continues to rise after radiation therapy, BCR is backdated to first PSA > 0.2. If patient gets other salvage treatment (such as hormones or chemotherapy), this is considered BCR.
Sample processing
At UHN, selected samples were cut into 60 × 10-μm sections, with an H&E-stained 4-μm section every 10 cuts. H&E-stained sections were marked by a genitourinary pathologist (T.v.d.K. or B.T.) to indicate areas suitable for macro-dissection (that is, more than 70% tumour cellularity). Manual macro-dissection was performed using sterile scalpel blades, and DNA was obtained by phenol:chloroform extraction, as previously reported4. DNA was extracted from whole blood using an ArchivePure DNA Blood Kit (5 PRIME, Inc.) at the Applied Molecular Profiling Laboratory at the Princess Margaret Cancer Centre.
At CHU de Québec, initial quality control was performed as described above and, if the surface of tumoural glands was considered large enough, 2 cores with 1mm diameter were taken from the tumoural zone using a sterile biopsy punch (Miltex). Tissues were immediately disrupted in ATL buffer using Minilys homogeneizer (Bertin Technologies, Montigny, France). DNA was extracted from the lysate using QIAmp DNA mini kit (Qiagen, Hilden, Germany). The same kit was used to generate DNA extractions on blood samples.
All DNA samples were quantified using a Qubit 2.0 Fluorometer (Life Technologies) and assessed for purity using a Nanodrop ND-1000 spectrophotometer.
SNP microarray data generation
SNP microarrays were performed with 200 ng of DNA on Affymetrix OncoScan FFPE Express 2.0 and 3.0 arrays. Where DNA quantities were limiting (88 samples), we used whole-genome amplification (WGA; WGA2, Sigma-Aldrich), and confirmed that WGA gDNA did not significantly alter CNA profiles4.
Methylation microarray data generation
Illumina Infinium HumanMethylation 450k BeadChip kits were used to assess global methylation, using 500 ng of input genomic DNA at the McGill University and Genome Quebec Innovation Centre (Montreal, QC). All samples were processed from fresh-frozen prostate cancer tissue. In total, there were 104 unique samples from 6 different processing batches in the discovery cohort. The validation cohort comprised 100 methylomes, processed identically.
mRNA microarray data generation
Total RNA was extracted from alternating adjacent sections, using the mirVana miRNA Isolation Kit (Life Technologies), according to the manufacturer’s instructions. In total, three batches were profiled at two locations. For batch 1 samples, 150 ng total RNA was assayed on the Affymetrix Human Gene 2.0 ST array (HuGene 2.0 ST) at The Centre for Applied Genomics (The Hospital for Sick Children, Ontario, Canada). For samples in batches 2 and 3, 100 ng total RNA was assayed on the Affymetrix Human Transcriptome Array 2.0 (HTA 2.0) and HuGene 2.0 ST, respectively, at the London Regional Genomics Centre (Robarts Research Institute, London, Ontario, Canada).
Whole-genome sequencing
Qubit (Life Technologies; Cat #Q32854) quantified gDNA (50 ng) was sheared to 300-bp fragments using the Covaris S2 Ultra-sonicator (Covaris Inc.) followed by 3× volume AMPure XP SPRI bead clean-up (Beckman Coulter Genomics; Cat#A63881). The bead–DNA mixture was transferred to a 96-well PCR plate (Eppendorf; Cat#0030133404) for the remainder of library construction and all subsequent SPRI bead clean-ups. Libraries were constructed using enzymatic reagents from KAPA Library Preparation Kits (KAPA Biosystems; Cat#KK8201) according to protocols as described for end repair, A-tailing, and adaptor ligation31. Adaptor-ligated libraries were enriched using optimized PCR conditions by adding 3 μl Illumina F & R PE enrichment primers (Integrated DNA Technologies), 75 μl 2× KAPA HiFi HotStart ReadyMix (KAPA Biosystems; Cat#KK2602) and 33 μl nuclease-free water (Life Technologies; Cat#AM993) to 36 μl eluted DNA and amplified across three individual PCR reaction tubes. Libraries were incubated in Verti 96-well Thermal Cyclers (Life Technologies) for 45 s at 98 °C and cycled 10 times for 15 s at 98 °C, 30 s at 65 °C, and 30 s at 72 °C. Following a 0.6× SPRI bead clean-up, post-PCR enriched libraries were eluted in 40 μl elution buffer (Qiagen; Cat#19086) and validated using Agilent Bioanalyzer High Sensitivity DNA Kit (Agilent Technologies; Cat#5067-4626). Libraries were quantified on the Illumina Eco Real-Time PCR Instrument (Illumina Inc.) using KAPA Illumina Library Quantification Kits (KAPA Biosciences; Cat#KK4835) according to the standard manufacturer’s protocol. 2 × 101 cycle paired-end sequencing was carried out for all libraries on the Illumina HiSeq 2000 platform (Illumina Inc.), and samples were sequenced to a minimum coverage depth of 50× and 30× for tumour and normal samples, respectively. A subset of the non-tumour reference samples was sequenced using the Illumina FastTrack Sequencing service. Sample preparation is described at www.illumina.com/content/dam/illumina-marketing/documents/services/FastTrackServices_Methods_Tech_Note.pdf.
SNP microarray data analysis
Affymetrix OncoScan FFPE Express 2.0 (n = 4) and 3.0 SNP (n = 280) microarrays were hybridized using 200 ng WGA (n = 88 IGRT biopsies) or genomic DNA (n = 137 RadP samples; n = 59 IGRT biopsies). We compared genomic DNA and WGA DNA from three independent specimens to confirm that WGA did not significantly affect the CNV and SNP profiles. We also evaluated inter-assay variability by analysing duplicate genomic and WGA DNA samples4.
Analysis of Affymetrix OncoScan FFPE Express 2.0 SNP probe assays was performed by Affymetrix using BioDiscovery’s Nexus Copy NumberTM software (http://www.biodiscovery.com/software/nexus-copy-number/). The data from Affymetrix were processed in batches based on version and in some cases LiftOver (http://genome.ucsc.edu/cgi-bin/hgLiftOver) was used to map aberrations from genome reference hg18 to hg19 (http://genome.ucsc.edu/). When the lift-over process deleted a portion of the CNA, the CNA was removed from the analysis.
Analysis of Affymetrix OncoScan FFPE Express 3.0 SNP probe assays was performed using.OSCHP files generated by OncoScan Console 1.1 using a custom reference. A custom reference, which included 119 normal blood samples from male patients with prostate cancer, 2 normal blood samples from females with anaplastic thyroid cancer, and 10 female hapmap cell line samples was created to combat artefacts resulting from differences in sample preparation (FFPE versus Fresh Frozen). BioDiscovery’s Nexus ExpressTM for OncoScan 3 Software was used to call CNAs using the SNP-FASST2 algorithm with default parameters except that the minimum number of probes per segment was changed from 3 to 20. When necessary, samples were re-centred using the Nexus ExpressTM software, choosing regions that showed diploid log2ratio and B allele frequency profiles.
Gene level CNAs for each patient were identified by overlapping CN segments, with RefGene (2014-07-15) annotation, using BEDTools (v2.17.0)32. To account for technical noise, a CNV blacklist was created from matched normal blood samples. Regions were added to the blacklist if they were seen in at least 75% of normal samples and filtered from downstream analyses. PGA was calculated for each sample by dividing the number of base pairs that were involved in a copy number change by the total length of the genome.
Copy number clustering was performed with the BioConductor package ConsensusClusterPlus (v1.8.1)33 using 1,000 iterations of hierarchical clustering with 80% subsampling of the genes for the number of clusters ranging from 2 to 12. Clustering was performed using Ward’s method on Jaccard distances.
We used GISTIC2.0 (v2.0.22) to study the recurrence of gene level CNVs in our sample set34. As input to GISTIC2.0, a profile for each sample was created that segmented each chromosome into regions with neutral, CN loss, and CN gain events. The average copy number intensity for each segment was obtained from the SNP array analysis. GISTIC2.0 was run with the following parameters changed from default (-genegistic 1 -smallmem 1 -broad 1 -brlen 0.5 -conf 0.99 -rx 0).
To test for associations between copy number state and categorical clinical variables, T category and GS, two-sided proportion tests were performed as implemented in R (v3.1.3). Copy number segment data were mapped to the RefGene annotation, classifying each gene’s state as ‘gain’, ‘deletion’ or ‘neutral’. Genes that did not have gains or deletions in 5% of all patients were removed from the analysis. Proportion tests were done separately for gains and deletions. P values were FDR adjusted to account for multiple testing. Similarly, to test for associations between copy number state and the continuous variable PSA, two-sided, unpaired t-tests were performed at the gene. Levene’s test was used to test for equal variance between groups and Welsh’s adjustment was applied if unequal variance was discovered. P values were FDR adjusted to account for multiple testing.
Our 284 samples were assigned to known prostate cancer cluster classifications6,35 by comparing our CNA profiles to their cluster centroids. The cluster centroids were defined as the median copy number of each gene in the patients assigned to that subtype, rounded to the nearest integer copy number. Patients were assigned to the cluster that had the most similar copy number profile based on the Jaccard distance metric.
To estimate the cellularity and purity of our cancer tumour samples we used the qpure (v1.1) and ASCAT (v2.1) algorithms36,37. Both programs require log R ratio (LRR) and B allele frequency (BAF) values obtained from the SNP array probes. These values were computed for the OncoScan 2.0 array platform by using the two intensity values provided for each probe corresponding to the hybridization of each probe using the following equations: LRR = log2(X + Y) and BAF = Y/(X + Y) where Y and X are intensity values corresponding to the minor and major alleles, respectively. For the OncoScan 3.0 array platform, LRR and BAF values were obtained from the.OSCHP files. We used qpure to compute the cellularity of our sample with default parameters and selected the output (tumorpurity.mixture.gam.adjust) as our cellularity estimate. We used ASCAT to compute tumour ploidy and to estimate the aberrant cell fraction for each sample.
The vcflib-tools suite (https://github.com/ekg/vcflib) was used to annotate and compare genotype calls from WGS and the OncoScan FFPE SNP assays. In-house scripts were used to create VCF files for OncoScan data from the OSCHP files. Validation rates (sensitivity) were calculated using TP/TP + FN. A true positive (TP) is identified when both platforms identify a position as AA or AR, where R refers to the reference allele in hg19 and A refers to an alternative allele. A false negative (FN) is identified by the following pairings: AA_AR, AA_RR or AR_RR (Supplementary Table 1).
Whole-genome sequencing data analysis
Each lane of raw sequencing reads was aligned against human reference build hg19 using bwa (v0.5.7)38. Lane-level BAMs from the same library were merged, marking duplicates using picard (v1.92). Library level BAMs from each sample were merged without marking duplicates. The Genome Analysis Toolkit (GATK v2.4.9) was used for local realignment and base quality recalibration, processing tumour/normal pairs together39. Separate tumour and normal sample level BAMs were extracted, headers were corrected using samtools (v0.1.9)40 and files were indexed with picard (v1.107).
Germline SNVs were generated using GATK (v2.4.9). First, UnifiedGenotyper was run on the realigned and recalibrated normal and tumour BAMs together followed by VariantRecalibrator and ApplyRecalibration. In addition, indels, somatic SNVs and ambiguous SNVs that had more than one alternate base separated by comma were removed. We referred to the GATK best practices to develop this pipeline (https://www.broadinstitute.org/gatk/guide/best-practices). The germline SNVs were used to filter somatic SNVs detected by SomaticSniper (v1.0.2)41.
To confirm that there was no cross-individual contamination, ContEst (v1.0.24530) was applied to all 130 normal and tumour sequences42. Both sample and lane-level analyses were performed (Supplementary Fig. 10). Regarding the required input VCFs, genotype information was gained from the germline SNVs generated by GATK (v2.4.9) and the VCF for population allele frequencies for each SNP in HapMap (hg19) was downloaded from https://www.broadinstitute.org/cancer/cga/contest_download.
Positions in read maps were deemed ‘callable’ if they had a minimum coverage of 10× in normal and 17× in tumour samples as calculated using BEDTools (v2.18.2).
Somatic SNVs were predicted using SomaticSniper (v1.0.2). First, somatic SNV candidates were detected using bam-somaticsniper with the default parameters except -q option (mapping quality threshold). The -q was set to 1 instead of 0 as recommended by the developer. To filter the candidate SNVs, a pileup indel file was generated for both normal BAM and tumour BAM file using SAMtools (v0.1.6). SomaticSniper (v1.0.2) package provides a series of Perl scripts to filter out possible false positives (http://gmt.genome.wustl.edu/packages/somatic-sniper/documentation.html). First, standard and LOH filtering were performed using the pileup indel files and then, bam-readcount filter was also performed (bam-readcount downloaded on 10 January 2014) with a mapping quality filter -q 1 (otherwise default settings). In addition, we ran the false positive filter. Subsequently, a high confidence filter was used with the default parameters. The final VCF file that contains high-confidence somatic SNVs was used in the downstream analysis.
After somatic SNV calling using SomaticSniper (v1.0.2), identified SNVs in positions that were not considered ‘callable’ were removed and then were passed through an annotation pipeline. SNVs were functionally annotated by ANNOVAR (v2015-06-17)43, using the RefGene database. Nonsynonymous, stop-loss, stop-gain and splice-site SNVs (based on RefGene annotations) were considered functional. If more than one mutation was found in a sample for a gene, then the mutation of the higher priority functional class was used for visualization. SNVs were filtered using tabixpp (3b299cc0911debadc435fdae60bbb72bd10f6d84), removing SNVs found in any of the following databases: dbSNP141 (modified to remove somatic and clinical variants, with variants with the following flags excluded: SAO = 2/3, PM, CDA, TPA, MUT and OM)44, 1000 Genomes Project (v3), Complete Genomics 69 whole genomes, duplicate gene database (v68)45, ENCODE DAC and Duke Mapability Consensus Excludable databases (comprising poorly mapping reads, repeat regions, and mitochondrial and ribosomal DNA)46, invalidated somatic SNVs from 68 human colorectal cancer exomes (unpublished data) using the AccuSNP platform (Roche NimbleGen), germline SNPs from all 477 samples used in this study and additional 10 WGS samples from prostate cancer patients with higher GS, and the Fuentes database of likely false-positive variants47. SNVs were whitelisted (and retained, independently of their presence in other filters) if they were contained within the Catalogue of Somatic Mutations in Cancer (COSMIC) database (v70)48 (Supplementary Data 1). The mutation rate per megabase of DNA was calculated by dividing the number of somatic point mutations after validation by the count of callable loci × 106 (Supplementary Table 7).
For each patient, aligned tumour and normal BAM files were used to call genomic rearrangements in Delly (v0.5.5)49 at a minimum median mapping quality of 20 and a paired-end cut-off of five. A list of somatic variants were produced by removing germline mutations from the resulting VCF files, which were further filtered using a consolidated list of structural variants from 124 normal samples. To identify genes affected by the genomic rearrangements, bed files were generated for each sample from deleted regions, and breakpoints from inversions, inter-chromosomal translocations, and tandem duplications. The resultant bed files were examined with SnpEff (v3.5)50 and gene names were subsequently extracted for downstream analyses. Recurrent translocation events were visualized using Circos (v0.67-4)51. Input files were bed files containing paired translocation breakpoints and the number of samples in which the event was observed.
Events involving ERG or ETV genes were collectively referred to as ETS events. Genomic rearrangement called using Delly49 were examined in all public data sets and CPC-GENE samples to determine whether breakpoints led to a T2E fusion or were found in both 1-Mbp bins surrounding the following gene pairs: ERG:SLC45A3, ERG:NDRG1, ETV1:TMPRSS2, ETV4:TMPRSS2, ETV1:SLC45A3, ETV4:SLC45A3, ETV1:NDRG1 and ETV4:NDRG1. ETS calls for CPC-GENE samples were further augmented using ERG immunohistochemistry, deletion calls between TMPRSS2 and ERG loci in either aCGH or OncoScan SNP array data, and TMPRSS2:ERG transcript fusion calls in RNA sequencing (RNA-seq). In addition, ETS status from the Berger9, Baca10, TCGA12 and Barbieri13 data sets were retrieved from their corresponding supplementary tables or online documents when applicable and consolidated with Delly breakpoint data.
Methylation microarray data analysis
All methylation analyses were performed in R statistical environment (v3.2.1). The IDAT files were loaded and converted to raw intensity values with the use of wateRmelon package (v1.8.0) from the BioConductor (v3.1) open-source project. Quality control was conducted using the minfi package (v1.14.0) (no outlier samples were detected). Batch effect was also examined across six batches using mclust package (v5.1.0) and no batch effect was found (adjusted Rand index, 0.06). Raw methylation intensity levels were then pre-processed using Dasen52. Probe filtering was conducted after the normalization. For each probe, a detection P value was computed to indicate whether the signal for the corresponding genomic position was distinguishable from the background noise. Probes having 1% of samples with a detection P < 0.05 were removed (1,751 probes). We also filtered probes based on SNPs (65 probes) and non-CpG methylation probes (3,088 probes). Next, we used the DMRcate package (v1.4.2)53 to further filter out 27,309 probes that are known to cross-hybridize to multiple locations in the genome54 and 17,168 probes that contain a SNP with an annotated minor allele frequency of greater than 5% with a maximum distance of two nucleotides to the nearest CpG site. Average intensity levels were taken for technical replicates. Annotation to chromosome location, probe position, and gene symbol was conducted using the IlluminaHumanMethylation450kanno.ilmn12.hg19 package (v0.2.1). Subtype analysis was performed using ConsensusClusterPlus (v1.22.0)33 with k-means and Pearson’s correlation as the similarity metric. Tumour purities were assessed with LUMP55.
For survival analyses, we used 91 samples as our training set, β-values from those were logit-transformed to M-values and median dichotomized to calculate a fold-change per probe. Probes with log2FoldChange >1 were then selected for univariate CoxPH modelling. Six probes associated with prostate cancer progression as well as with high absolute log2HR values (MIR129-2, ACTL6B, TCERG1L-3′, TCERG1L-5′, TUBA3C, SOX14) and P < 0.01 were then selected. These were validated in an independent cohort of 100 prostate tumours processed identically by using the median from our training set to dichotomize values from the validation set followed by univariate CoxPH modelling (Extended Data Fig. 10a–f).
Methylation was obtained from EGA (EGAS00001000682)56 and pre-processed using the same methods as our own Illumina 450k arrays, as described above. As reported in their study, samples 2_TU_1, 2_TU_9, 3_TU_5, 4_LNM_2, 5_TU_10, 5_LNM_2 were removed. Using data from the five remaining patients (n = 80), the coefficient of variance (CV) was calculated across the different samples, per patient in the 20,000 probes used in our survival analysis. Using this distribution of CV, the percentile was calculated for the six probes used in our biomarker. The median CV percentile values were: 26% (cg18360873), 75% (cg03943081), 16% (cg08756887), 60% (cg08073312), 89% (cg26990587) and 66% (cg14944647).
Significance analysis of coding SNVs (SeqSig)
To identify genes recurrently altered by non-synonymous mutations and to gain information from the whole genome, we developed a mathematical model called SeqSig. This model has the following assumptions: A1, only coding regions are considered; A2, only base substitutions are considered, not indels or other structural variants; and A3, for each patient, mutations are independent among nucleotides and homogeneous across all positions on coding regions, that is, there exists a genome-wise transition probability matrix Q = (qxy)x,y∈{T,C,G,A}.
On the basis of the above assumptions, one can compute the non-synonymous mutation probability for each codon and thus the non-synonymous mutation probability for each gene of each patient, if Q is known.
To be able to extract background mutation information from the available patient DNA sequences, we have to assume that only a small amount of mutations are cancer driver mutations. For each patient, we compare the observed DNA sequence to the reference (hg19 is used here), and compute the transition frequency fxy where x,y ∈ {T,C,G,A}. One natural estimate for qxy is:

where

With assumption A3, one can estimate the background mutation rate (probability of random mutation with no natural selection) for each gene. In the paper, we compute the background rate for non-synonymous mutation and refer to it as BMR, which can be calculated by using the transition matrix, reference genome and the codon table.
For a given gene, assume that BMRs p0i of patient i = 1,2,…,n computed as above is known. Assume the true mutation rate is pi, then we have:

Where Y = 1 if patient i has a non-synonymous mutation on given gene, and Y = 0 otherwise.
The hypothesis test is thus:

This is a multi-testing problem, and the null hypothesis may be somehow too strong to be easily rejected as it requires all patients to follow the BMRs. In order to test in an overall sense, we assume the following model:

Where wi is some weight for patient i. When wi = 1, the above is a model with common odds ratio among patients. One may also chose wi = −logp0i or wi = −logp0i/(1 − p0i) (assuming p0i < 0.5, which is almost always true as p0i is usually very small), giving more weights to patients with small BMRs, as one may argue that since mutations on those patients are more ‘difficult’, observing mutations on them should give more evidence. Under this model, the hypothesis test (1) becomes:

It is easy to show by the factorization theorem that:

is a sufficient statistic for β. Obviously, there is a positive relation between β and E(T), the expectation of T.
Many standard tests exist for (3), such as the likelihood ratio test, score test and the Wald-type test, which all require a ‘sufficiently’ large sample size. However, in many practical situations, samples are usually not abundant. We instead use convolution law to find the exact distribution of T under H0. Owing to the positive relation of β and E(T), we can get the P value of (3) by P = P(T > Tobs|H0) where Tobs is the observed T. We can reject H0 under a predefined significance level α if P < α.
In this paper, wi = −logp0i/(1 − p0i) is used, under which T is equivalent to the convolution test statistic used by MuSiC and MutSig (v1.x). However, the general model (2) allows for different choices of wi, and clinical variables can also be incorporated through the values of wi.
Coding SNV power analysis
Based on the median background mutation rate of 2.44 × 10−1 mutations per Mbp for transcribed regions (including exons and introns but excluding UTRs) of the genome (which was obtained by considering only bases that are callable for at least 90% of the whole genome samples—all bases were considered callable for the exome samples), there may be about five SNVs at the 0.5% level that are still to be discovered. With a cohort of 477 samples, there is enough power to find all SNVs altered at the 1% level and above, whereas at the 0.5% level, we have about 76% power to detect aberrations in the coding regions of the genome (that is, the exons) (Supplementary Fig. 5a). BEDTools (v2.21.0) was used to intersect multiple bed files to find callable bases (as defined above) present in at least 90% of samples.
Non-coding SNV power analysis
A similar procedure was done for non-coding SNV power analysis as for coding SNV power analyses. We considered only bases outside the transcribed regions (including exons, introns and UTRs), and calculated the background mutation frequencies for each base (median background mutation rate = 8.89 × 10−1 mutations per Mbp) and subsequently power using SeqSig (Supplementary Fig. 5b).
Transcription factor binding site analysis
To determine whether TFBSs were mutated more than expected by chance, we first downloaded TFBS data from ENCODE (ChIP-seq narrow peaks) and ref. 57. LiftOver (genome.ucsc.edu/cgi-bin/hgLiftOver) was used to convert between the hg18 and hg19 assemblies for the Caco2 and PC3 cell-line data (all options set to default, minimum ratio of bases that must remap = 95, min ratio of alignment blocks or exons that must map = 1; about 0.03–3.2% of bases failed to convert). We then adjusted the TFBS data as well as the aberration data bed files by taking into account callable bases. To obtain genomic rearrangement data, we flanked the genomic rearrangement breakpoints, excluding deletions, by 10 kbp (Extended Data Fig. 3b) and 1 kbp (Supplementary Fig. 8) to show robustness. The adjusted TFBS bed files were then intersected using BEDTools (v2.18.2) with each of the adjusted aberration data bed files, followed by a binomial test. The test was performed for each sample and TFBS combination to see if we observe more aberrations in TFBS than expected by chance; the results were FDR adjusted. Before visualizing, the adjusted P values of replicate TFBS cell lines were averaged, reducing the total TFBS cell line count to 58. Correlations between recurrence of each TFBS and CNAs as well as genomic rearrangement breakpoints flanked with 10 kbp, with FDR adjustment of P values, revealed clinical associations (Supplementary Table 18).
Recurrent non-coding SNV analyses
Non-coding SNVs (ncSNVs) identified in intergenic regions, introns, splicing sites, 1 kbp upstream of transcription start sites or 1 kbp downstream of transcription end sites were extracted from the filtered variant matrix. We used the most recurrent 70 ncSNVs, all of which were found in at least four samples, as our set of recurrent ncSNVs for the following analyses (Supplementary Data 1). First, WebLogo (v3.4) was used for motif discovery in terms of the 10 bp up- and down-stream of ncSNVs58. Second, the variant allele frequency of the ncSNVs was calculated based on the number of reads supporting the alternative base divided by the total number of reads using SomaticSniper VCFs (Extended Data Fig. 3a, b). Third, to see association between the recurrent ncSNVs and replication time, the replication time of genomic regions (bin size: 100 kbp) that harbour a recurrent ncSNV was plotted59 (Supplementary Fig. 7). Fourth, DeepSEA was used to predict the chromatin effects of the recurrent ncSNVs17. The data were generated on the DeepSEA website (http://deepsea.princeton.edu/job/sequence/create/) (version 10/22/2015). Features that have at least one ncSNV with DeepSEA E-value < 0.01 were selected as important features. In addition, we obtained ChIP-Seq data sets generated using the LNCaP cell line and investigated if any of the regulatory regions identified by the ChIP-Seq experiments were overlapped with the ncSNVs. A permutation test was performed (104 − 1 iterations) for each ncSNV to determine whether the mean of the E-values for the important features was less than expected by chance alone. The same analysis was performed for ncSNVs (recurrent ncSNVs versus all ncSNVs) for each feature. Computed P values were then FDR adjusted (Extended Data Fig. 3c, Supplementary Table 10).
Trinucleotide mutation signature analysis
For each SNV in the unfiltered recurrent variant matrix, the 5′ and 3′ bases were extracted from the hg19 reference using BEDTools (v2.17.0) and tabulated into the 96 trinucleotide mutation categories for each patient, and were then input into the NMF (v0.20.6) R package60. Factorizations were generated for ranks 2 to 20. Rank 3 was selected as a balance between the cophenetic and dispersion metrics. We extracted the coefficient (trinucleotide signatures) and basis (signature exposures) matrices from the NMF run. The coefficient matrix was normalized such that each signature could be interpreted as a distribution over the trinucleotide mutational categories18 (Supplementary Table 11). The basis matrix was then scaled by the inverse of the coefficient normalizing matrix. The scaled basis matrix was then normalized per patient (Supplementary Table 13).
Statistical analyses
The survey of PGA, SNV, CNA and genomic rearrangement (including inversions and translocations, separately) data in Fig. 1 and Extended Data Fig. 2b was performed by applying appropriate models for each data type against explanatory variables, including GS, pre-treatment PSA, age at treatment (or age at diagnosis when age at treatment was unavailable), T-category and T2E status. Linear regression was used to find associations between PGA and continuous variables, and a non-parametric approach was taken to find associations between PGA and the categorical variables using the Kruskal–Wallis test. Associations between SNV counts and continuous variables were found using linear regression and one-way ANOVA for categorical variables. CNA, inversion and translocation counts were modelled using a negative binomial generalized linear model for both continuous and categorical clinical variables. For all data types, the mean, median, IQR (25%, 75%), and the overall effect P value for all clinical variables were reported (Supplementary Table 9). Extended Data Fig. 2b displays boxplots of explanatory variables for each data type and reports the overall effect P value corresponding to an appropriate statistic. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Interchromosomal translocation enrichment and spatial proximity
Delly49 outputs two breakpoints for each interchromosomal translocation. The breakpoints were permuted within each sample in a permutation test (106 iterations) to determine whether some chromosome combinations were more abundant in translocations than expected by chance alone. The resulting P values were corrected using FDR. To determine whether breakpoints occurred closer to or further from spatially proximal chromosomal regions than expected by chance alone, a HiC data set of prostate epithelial cells (GSE37752) was retrieved from the NCBI Gene Expression Omnibus21. Because the HiC data set was originally generated using hg18, LiftOver was used to convert the positions to hg19. HiC points missing a coordinate as a result of the conversion were stripped from the data set. Perl (v5.18.2) and R (v3.1.3) scripts were used to calculate the shortest distance between a translocation and its nearest HiC point and the mean of the distances for each chromosome combination was compared to a null distribution of distances (50-bp bins) generated from all possible pairs of positions in each chromosome combination. P values were corrected using FDR.
mRNA abundance analysis
All mRNA analysis was performed using R (v3.2.1). Background correction, normalization algorithms and annotation were implemented in the oligo (v1.32.0) package from the BioConductor (v3.0) open-source project. The Robust multichip average (RMA) algorithm was applied to the raw intensity data60. Annotations were performed using hugene20sttranscriptcluster.db (v2.13.0) and hta20sttranscriptcluster.db (v8.3.1). The sva package (v3.14.0) was used to correct for batch effects between different arrays. Annotated data from HuGene 2.0 ST and HTA 2.0 were combined into one data set based on Entrez Gene IDs. The mRNA expression values were averaged amongst duplicated Entrez Gene IDs. To test the association between mRNA profile and genomic rearrangement (inversion specifically), we used a linear model to compare the mRNA abundance from patients with and without inversions. Genes are selected based on the inversion windows (chr3:129–130 Mbp and chr10:89–90 Mbp). For each gene, mRNA abundances were re-normalized and centred by the median across all patients. Chromothripsis scores were also compared with mRNA abundance levels. For each gene, Spearman’s correlation was calculated between the maximal chromothripsis score per patient and each gene’s respective mRNA abundance levels across all patients; the correlation coefficients and P values were subsequently computed (Supplementary Table 17).
To determine whether PTEN inversions have different effects on the PTEN network from copy losses, the top ten genes most correlated with PTEN mRNA abundances as calculated using Spearman’s ρ were examined. The per sample mean mRNA abundances of the ten genes was used as a proxy for PTEN activity and ultimately overall effects of PTEN inactivation (Extended Data Fig. 5).
To determine the level of infiltrating immune cells in 73 samples with mRNA data, the ‘Estimate of STromal and Immune cells in Malignant Tumours’ (ESTIMATE) method was used, as implemented in the estimate R package (v1.0.11)61. In 23 samples (22 with RNA data), the percent of infiltrating immune cells was measured by a pathologist by screening all available levels of each case for inflammatory cells (which were mostly lymphocytes) located within the tumour areas and scored semiquantitatively the percentage of lymphocytes based on visual estimation. Overall, <1% indicated scattered lymphocytes comprising less than 1% of tumour surface. The presence of aggregates, arbitrarily defined as more than 30 lymphocytes packed together, and the average number of aggregates for each case taking into account the number of them per level were counted.
Chromothripsis and kataegis
Chromothripsis scores were generated using ShatterProof (v0.14) with default settings23. Samples with a max ShatterProof score over 0.517 were defined as having chromothriptic characteristics. Full lists of putative chromothripsis events are shown in the Circos plots (Supplementary Table 7; Supplementary Data 2).
Recurrent somatic variants were used to quantify kataegis in each sample. An overlapping sliding window exact binomial test was conducted to test whether the proportion of variants within given window size was higher than expected. The observed frequency was calculated by dividing the number of variants in a sliding window over the number of bases in that window. The expected frequency was calculated by dividing the number of variants in that chromosome over the number of bases for that same chromosome. The binomial test P values were adjusted for multiple hypothesis testing using FDR and the adjusted P values were converted to a binary variable 0/1 to code for its significance. The R package changepoint was then used to convert those scores into segments. The base change composition for each segment was calculated and segments that are enriched with C/T, C/G, C/A changes (>50% of base change type within a window) were highlighted. Rainfall plots for the whole genome, with SNV position on the x-axis and the log transformed inter-mutational distance plotted on the y-axis, were generated for each sample to visualize kataegic events.
Potential links between chromothripsis and the mutation landscape were explored through various statistical tests on different types of gene mutations. In the R statistical environment (v3.1.3), Mann–Whitney U tests were performed using the maximum ShatterProof scores against genes affected by copy number aberrations, genomic rearrangements, and SNVs separately. In addition, Kendall’s τ was used to determine whether an association existed between the clinical variables and chromothripsis. For the purpose of discovering novel associations, P values were corrected using FDR.
To identify mutations that may be linked to kataegic events, proportion tests were calculated in R (v3.1.3) using kataegis scores against genes affected by copy number aberrations, genomic rearrangements, and SNVs separately. The proportion’s test was also used to explore associations between kataegis and genomic rearrangements at the Mbp bin level, while Kendall’s τ was used to determine whether clinical variables were correlated with kataegic events. P values were corrected using FDR.
For each patient with a chromothriptic or a kataegic event, the transcriptional and methylation profiles of genes within that region were evaluated. The chromosome region that had the maximal chromothripsis or kataegis score for each patient was selected first. A percentile spectrum was then generated by comparing the mRNA abundance levels or methylation β-value of any genes or probes within that region to the same gene or probe in all patients without that particular chromothriptic or kataegic event (Extended Data Fig. 8). The relationship between methylation levels and mRNA abundance was examined in patients with stable genomes (patients with no chromothriptic and kataegic events, n = 46), those with chromothriptic events (n = 14) and those with kataegic events (n = 19).
To evaluate trans effects, Spearman’s correlations were calculated on the top 10,000 probes (for methylation data) and top 10,000 genes (for mRNA), with the highest variance (Extended Data Fig. 9). To understand the association of promoter region methylation and mRNA levels (that is, cis effects), three different approaches were used: 1) the top 10,000 variable mRNA genes were selected and for each gene, all the β-values for that gene were averaged and the Spearman’s correlation was calculated; 2) Same strategy as 1, but instead of taking the average, all β-values for the same gene were used to compute the correlation coefficients. 3) Correlation matrix of methylation and mRNA abundance levels from TCGA was downloaded from https://gdac.broadinstitute.org/. Results from across 28 tumour types (ACC, BLCA, BRCA, CESC, CHOL, COADREAD, DLBC, ESCA, GBMLGG, HNSC, KIPAN, LAML, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, SARC, SKCM, STAD, STES, TGCT, THCA, THYM, UCEC, UCS, UVM) were combined, and the correlation coefficients were scaled based on the sample size of each data set. The mean correlation coefficients were calculated per probe across multiple tumour types. For each gene, the probe showing the highest negative correlation with mRNA abundance levels was kept. Spearman’s correlation coefficients between those selected probes and their corresponding genes were calculated within our data set (Extended Data Fig. 9c). The union of genes (n = 65) that showed differential correlation coefficients between chromothriptic and stable samples (|δ|>0.8) in the above three approaches were used for pathways analysis. Pathways were curated using gene ontology: biological process and REACTOME from g:Profiler. Significantly enriched pathways (q < 0.05; hypergeometric test) were then visualized using Cytoscape (v3.3.0) (Extended Data Fig. 9; Supplementary Table 16).
Prognostic signature generation
The set of 104 patients with whole-genome sequencing, methylation, and survival information was split into four folds. Each fold was balanced by event rate, age, T-category, and GS, in that order. The use of four folds ensured that the young (under 40 years of age) tumours were balanced. In each fold using full, untruncated survival time, each of the 40 features was fit univariately in a CoxPH model without adjustment. From this, a set of candidate features was generated at P < 0.1 from the Wald test. These candidate features were further selected by retaining only the best feature from each molecular class (SNV, ncSNV, CNA, genomic rearrangement) associated with good outcome and the best feature associated with poor outcome. This class-and-direction filtering was not used for mutational density or clinical features, which were considered if they met the P for significance. Features seen in at least two folds were selected, yielding our final six-feature candidate list. A CoxPH model was then fit with these six features separately in each fold, and predictions made on the held-out data. Predictions across the four test-sets were then pooled and performance assessed using the area under the receiver operating characteristics curve (AUROC). For comparison, a CoxPH model was fit using PGA as a continuous variable. Kaplan–Meier plots were generated by binarizing predictions at the event rate thresholds.
Data visualization
Visualizations were generated in the R statistical environment (v3.1.3 or higher) using the lattice (v0.20-31), latticeExtra (v0.6-26), BPG (v5.3.4) and VennDiagram (v1.6.4) packages, along with pdfTeX (v3.1415926-1.40.10). Schematics were created in Inkscape (v0.48) for Ubuntu. Recurrent translocation plots, and overall mutational profiles of each sample, including presence of kataegic or chromothriptic events were produced using Circos (v0.67-4)51.
Data availability
mRNA and methylation data are available in the Gene Expression Omnibus under accession GSE84043. Raw sequencing data are available in the European Genome-phenome Archive under accession EGAS00001000900 (https://www.ebi.ac.uk/ega/studies/EGAS00001000900). Processed variant calls are available through the ICGC Data Portal under the project PRAD-CA (https://dcc.icgc.org/projects/PRAD-CA). Baca and Barbieri WGS/WXS data are available on dbGaP under accession phs000447.v1.p1 (https://www.ncbi.nlm.nih.gov/gap/?term=phs000447.v1.p1). Berger WGS data are available on dbGaP under accession phs000330.v1.p1 (https://www.ncbi.nlm.nih.gov/gap/?term=phs000330.v1.p1). Weischenfeldt WGS data are available on EGA under accession EGAS00001000400 (https://www.ebi.ac.uk/ega/studies/EGAS00001000400). TCGA WGS/WXS data are available at Genomic Data Commons Data Portal (https://gdc-portal.nci.nih.gov/projects/TCGA-PRAD).
Accession codes
References
Lozano, R. et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128 (2012)
Klotz, L. et al. Long-term follow-up of a large active surveillance cohort of patients with prostate cancer. J. Clin. Oncol. 33, 272–277 (2015)
D’Amico, A. V. et al. Cancer-specific mortality after surgery or radiation for patients with clinically localized prostate cancer managed during the prostate-specific antigen era. J. Clin. Oncol . 21, 2163–2172 (2003)
Boutros, P. C. et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat. Genet. 47, 736–745 (2015)
Cooper, C. S. et al. Analysis of the genetic phylogeny of multifocal prostate cancer identifies multiple independent clonal expansions in neoplastic and morphologically normal prostate tissue. Nat. Genet. 47, 367–372 (2015)
Lalonde, E. et al. Tumour genomic and microenvironmental heterogeneity for integrated prediction of 5-year biochemical recurrence of prostate cancer: a retrospective cohort study. Lancet Oncol. 15, 1521–1532 (2014)
Buyyounouski, M. K., Pickles, T., Kestin, L. L., Allison, R. & Williams, S. G. Validating the interval to biochemical failure for the identification of potentially lethal prostate cancer. J. Clin. Oncol. 30, 1857–1863 (2012)
Robinson, D. et al. Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015)
Berger, M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011)
Baca, S. C. et al. Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013)
Weischenfeldt, J. et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell 23, 159–170 (2013)
Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015)
Barbieri, C. E. et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat. Genet. 44, 685–689 (2012)
Ewing, A. D. et al. Combining tumor genome simulation with crowdsourcing to benchmark somatic single-nucleotide-variant detection. Nat. Methods 12, 623–630 (2015)
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014)
Ciriello, G. et al. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 45, 1127–1133 (2013)
Zhou, J. & Troyanskaya, O. G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods 12, 931–934 (2015)
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013)
Gandhi, M., Evdokimova, V. & Nikiforov, Y. E. Frequency of close positioning of chromosomal loci detected by FRET correlates with their participation in carcinogenic rearrangements in human cells. Genes Chromosom. Cancer 51, 1037–1044 (2012)
Nikiforova, M. N. et al. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science 290, 138–141 (2000)
Rickman, D. S. et al. Oncogene-mediated alterations in chromatin conformation. Proc. Natl Acad. Sci. USA 109, 9083–9088 (2012)
Korbel, J. O. & Campbell, P. J. Criteria for inference of chromothripsis in cancer genomes. Cell 152, 1226–1236 (2013)
Govind, S. K. et al. ShatterProof: operational detection and quantification of chromothripsis. BMC Bioinformatics 15, 78 (2014)
Zafarana, G. et al. Copy number alterations of c-MYC and PTEN are prognostic factors for relapse after prostate cancer radiotherapy. Cancer 118, 4053–4062 (2012)
Hieronymus, H. et al. Copy number alteration burden predicts prostate cancer relapse. Proc. Natl Acad. Sci. USA 111, 11139–11144 (2014)
Mateo, J. et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015)
Schiewer, M. J. et al. Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov . 2, 1134–1149 (2012)
Feng, F. Y., de Bono, J. S., Rubin, M. A. & Knudsen, K. E. Chromatin to clinic: the molecular rationale for PARP1 inhibitor function. Mol. Cell 58, 925–934 (2015)
Gundem, G. et al. The evolutionary history of lethal metastatic prostate cancer. Nature 520, 353–357 (2015)
Graff, J. N. & Beer, T. M. Should docetaxel be administered earlier in prostate cancer therapy? Expert Rev. Anticancer Ther. 15, 977–979 (2015)
Fisher, S. et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 12, R1 (2011)
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010)
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26, 1572–1573 (2010)
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011)
Taylor, B. J. et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. eLife 2, e00534 (2013)
Song, S. et al. qpure: A tool to estimate tumor cellularity from genome-wide single-nucleotide polymorphism profiles. PLoS One 7, e45835 (2012)
Van Loo, P. et al. Analyzing cancer samples with SNP arrays. Methods Mol. Biol. 802, 57–72 (2012)
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009)
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010)
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009)
Larson, D. E. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 28, 311–317 (2012)
Cibulskis, K. et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics 27, 2601–2602 (2011)
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res . 38, e164 (2010)
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res . 29, 308–311 (2001)
Ouedraogo, M. et al. The duplicated genes database: identification and functional annotation of co-localised duplicated genes across genomes. PLoS One 7, e50653 (2012)
Gerstein, M. B. et al. Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012)
Fuentes Fajardo, K. V. et al. Detecting false-positive signals in exome sequencing. Hum. Mutat. 33, 609–613 (2012)
Forbes, S. A. et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res . 43, D805–D811 (2015)
Rausch, T. et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics 28, i333–i339 (2012)
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012)
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009)
Pidsley, R. et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics 14, 293 (2013)
Peters, T. J. et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8, 6 (2015)
Chen, Y. A. et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8, 203–209 (2013)
Aran, D., Sirota, M. & Butte, A. J. Systematic pan-cancer analysis of tumour purity. Nat. Commun. 6, 8971 (2015)
Brocks, D. et al. Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Reports 8, 798–806 (2014)
Massie, C. E. et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J . 30, 2719–2733 (2011)
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004)
Lawrence, M. S. et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218 (2013)
Irizarry, R. A. et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res . 31, e15 (2003)
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013)
Acknowledgements
We thank all members of the Boutros and Bristow labs for helpful suggestions, particularly C. M. Lalansingh for technical assistance with recurrent SNV analysis. The results described here are based in part upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. This study was conducted with the support of Movember through Prostate Cancer Canada and with the additional support of the Ontario Institute for Cancer Research, funded by the Government of Ontario, and of the Ontario Institute for Cancer Research to P.C.B. through funding from the Government of Ontario. We thank the Princess Margaret Cancer Centre Foundation and Radiation Medicine Program Academic Enrichment Fund for support (to R.G.B.). R.G.B. is a recipient of a Canadian Cancer Society Research Scientist Award. This work was supported by Prostate Cancer Canada and is proudly funded by the Movember Foundation (grant #RS2014-01 to P.C.B.). P.C.B. was supported by a Terry Fox Research Institute New Investigator Award and a CIHR New Investigator Award. H.H.H. was supported by CIHR operating grant 142246 and CCSRI grant 703800. This project was supported by Genome Canada through a Large-Scale Applied Project contract to P.C.B., R. Morin and S. P. Shah. D.T. was part of the Terry Fox Foundation Strategic Health Research Training Program in Cancer Research at the Canadian Institute of Health Research and Ontario Institute for Cancer Research. E.L. was supported by a CIHR Fellowship. N.S.F. was supported by an NSERC Fellowship.
Author information
Authors and Affiliations
Contributions
Sample preparation and data collection: M.F., A.B., A.M., J.Z., M.C., A.W., T.C., M.S., J.J., L.T., N.B.B., M.O., V.P., H.H., A.B., A.D.P., M.A. and K.K. Pathology analyses: D.T., B.T. and T.v.d.K. Statistical and bioinformatics analyses: V.Y.S., T.N.Y., L.E.H., J.L., V.H., Y.-J.S., F.Y., X.L., A.P.M., N.S.F., M.X., S.D.P., E.L., X.L., T.A.B., A.D., R.E.D., H.K., S.M.G.E., N.J.H., C.P., K.E.H., K.C.C., B.L., F.N., C.H.L., R.X.S., R.d.B., C.I.C., J.F.H., S.K.G., C.F., D.W., J.G., S.H., M.A.C.-S.-Y., E.J., Z.W., M.A., A.M., K.K. and H.H.H. Wrote the first draft of the manuscript: M.F., R.G.B. and P.C.B. Initiated the project: M.F., C.C.C., T.v.d.K., J.D.M., R.G.B. and P.C.B. Supervised research: T.A.B., L.L., C.C.C., C.S., N.E.F., Y.F., B.T., M.L., H.H.H., T.v.d.K., J.D.M., R.G.B. and P.C.B. Approved the manuscript: all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information
Nature thanks S. Chanock, C. Plass and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Figure 1 Study design.
The overall study cohort consisted of 137 patients who underwent radical prostatectomy (surgery) and 147 patients who underwent image-guided radiotherapy for localized prostate cancer (biopsy). For surgery patients, a fresh-frozen tissue specimen from the index lesion was obtained for macro-dissection. For radiotherapy patients, a fresh-frozen needle core ultrasound-guided biopsy to the index lesion was obtained for macro-dissection. All 284 tumour DNA specimens were analysed for CNA by OncoScan SNP arrays. Of these tumour DNA specimens, 130 were selected for further analysis by WGS (as was a matched normal DNA specimen from whole blood). For a subset of analyses, additional data (numbers as indicated) from publicly available whole-genome or whole-exome sequencing data sets were re-aligned and re-analysed and integrated to maximize statistical power.
Extended Data Figure 2 Comparison of molecular aberrations.
a, Pairwise comparison scatter plot of data type as indicated on the x- and y-axes. Spearman correlation and unadjusted P values are provided. b, Scatterplots and box plots of each mutation burden (CNA, CTX, INV, SNV counts and PGA) versus clinical variables (age, GS, T-category, PSA and ETS consensus fusion) is provided along with a model-derived P value, as described in Methods. Grey dots represent values for individual samples.
Extended Data Figure 3 Non-coding SNV profile.
We analysed 70 non-coding recurrent somatic SNVs: defined as at least 2% (4 of 200) of tumours having mutations in the same, non-coding position. a, The central heat map shows the 70 recurrent ncSNVs (rows) and the samples in which they are present (columns), with colour indicating their variant allele frequency (VAF). The top bar plot indicates the total number of ncSNVs mutated in each sample, while the right bar plot gives the total number of samples in which each ncSNV is mutated. b, Box plot showing VAF for recurrent ncSNVs. Each dot indicates the VAF of a recurrent ncSNV for a sample. The recurrent ncSNVs (rows) were sorted by median VAF. c, To determine whether ncSNVs were biased towards specific TFBSs, we tested whether experimentally derived TFBS locations from ENCODE were enriched for aberrations of different types using the binomial test. Heatmap of 58 TFBS cell lines for each sample coloured by the data type or combination of data types (SNV, CNV, and CTX flanked by 10 kbp) if it was aberrant in more samples than expected by chance (binomial test with FDR-adjusted P value). The samples are ordered by the number of significantly aberrant TFBSs (top barplot), the TFBS cell lines are ordered by fraction of samples with significantly mutated TFBSs by cell line (right barplot), covariates of pathological GS, pre-treatment PSA, T-category, and patient age at treatment are displayed at the bottom. d, Predicted chromatin effects of recurrent ncSNVs. The left heat map shows E-values, which measure the expected proportion of SNPs (found in the 1,000 Genomes Project) with a larger predicted effect for a chromatin feature, predicted by DeepSEA. The right heat map shows the overlaps between chromatin elements detected by LNCaP chromatin immunoprecipitation with sequencing (ChIP–seq) experiments and ncSNVs. The FDR adjusted P values (Q values) for the DeepSEA or ChiP–seq experiment features are shown above each plot. The ncSNV Q values for DeepSEA and ncSNV recurrence are shown on the right. Experimental conditions (cell line type, chromatin feature, and treatment) of the ChIP–seq data are represented by the covariates at the bottom. The heatmaps and barplots were sorted by Q values.
Extended Data Figure 4 Genome rearrangements overview.
a, Global overview of somatic structural variants in 180 localized GS 3 + 3, 3 + 4 and 4 + 3 prostate cancers. The central heat map shows per-sample inter-chromosomal translocations (CTXs), inversions and deletions for 1-Mbp bins across the genome (columns) and for each patient (rows). The striking TMPRSS2:ERG peak on chromosome 21 is by far the most frequent aberration, but additional recurrent inversion breakpoints were identified on chromosomes 3 and 10, and CTX breakpoints on chromosome 6. b, Number of CTXs joining each chromosome pair and their occurrences relative to random chance. Dot size represents the number of translocations enriched (number greater than expected) while background colour indicates their significance as calculated using a one-tailed permutation test (1 million replicates) with FDR correction. c, Mean shortest distance between a CTX and the corresponding nearest HiC point in each chromosome pair. Dot size represents the difference between the mean observed CTX–HiC distances and their expected distances, while the background indicates significance as calculated using a one-tailed permutation test (1 million replicates) corrected using the FDR method. Orange dots indicate distances greater than expected by chance alone (top right), while blue dots show distances smaller than expected by chance alone (bottom left).
Extended Data Figure 5 Effects of inversion on mRNA abundance and PTEN.
a, For each gene in the inversion window (chr10:89–90 Mbp), mRNA abundance levels were re-normalized and centred by the median across all patients. Box plot (top) demonstrates the renormalized mRNA abundance levels (y-axis) of patients with no inversion (n = 70, orange) and with inversions (n = 3, green) for each gene. A linear model was used to calculate the P values between the two patient groups. Bar plot (bottom) shows unadjusted P values with genes ordered by chromosome location. b, Spearman’s ρ was used to identify the top ten genes most correlated with PTEN mRNA abundances. The per sample mean mRNA abundances of the ten genes was used to represent the overall effects of various types of PTEN inactivation. PTEN inactivation as a result of CNV loss led to a significantly lower abundance of PTEN-associated proteins when compared to copy number-neutral PTEN (Mann–Whitney U test, P = 2.0 × 10−4) whereas PTEN inversions yielded further reduced abundances (Mann–Whitney U test, P = 0.016). c, For each gene in the inversion window (chr3:129–130 Mbp), mRNA abundance levels were re-normalized and centred by the median across all patients. Box plot (top) shows the renormalized mRNA abundance levels (y-axis) of patients with no inversion (n = 65, orange) or with inversions (n = 8, green) for each gene. A linear model was used to calculate P values between the two patient groups. Bar plot (bottom) shows the P values with genes ordered by chromosome location.
Extended Data Figure 6 Hypermutation associations.
a, Box plot of ShatterProof scores grouped by T-category. Each grey dot represents a single sample. P value is from a one-way ANOVA. b) To assess the association between genome stability (measured as PGA) and the presence of one or more chromothriptic events in a tumour, we compared the mean PGA between tumours with a chromothriptic event (4.28% ± 5.04%) and those without one (7.79% ± 5.3%). This difference of 3.52% was statistically significant (P = 1.10 × 10−3; two-sided t-test). c, To assess the association between genome stability (measured as PGA) and the presence of one or more kataegic events in a tumour, we compared the mean PGA between tumours with a kataegic event (6.87% ± 5.62%) and those without one (4.34% ± 5.13%). This difference of 2.53% was statistically significant (P = 7.52 × 10−3; two-sided t-test). d, Scatter plot of ShatterProof scores against per cent infiltrating immune cells as measured by a pathologist. e, Scatter plot of ShatterProof scores against estimated immune score calculated by the ESTIMATE software. For both these plots, Spearman’s ρ is given, along with its P value. f, Scatterplot showing the correlation between pathologist and ESTIMATE predictions for 22 samples.
Extended Data Figure 7 Chromothripsis associations and mutational burden.
a, Scatter plots of mRNA abundance against ShatterProof scores for four genes found to be associated with chromothripsis. Spearman’s ρ and P values are shown. Box plots of mRNA abundance against copy number status (DEL, deletion; NEU, copy number neutral). P values are from a two-sided t-test. b, Scatterplots of mutation burden (SNV, INV, CNA, CTX counts) and qpure cellularity values against ShatterProof score. Spearman’s ρ and corresponding P values are shown.
Extended Data Figure 8 Characteristics of mRNA genes and methylation probes in chromothripsis region.
a, Histogram of percentiles from mRNA genes (2,197 unique genes) located in a chromothriptic region. Upper left corner indicates Pearson’s correlation between each bin and the frequency of genes that reside in that bin. b, Histogram as in a for the 43,985 unique methylation probes located in chromothriptic regions. c, Box plot of genes that are in chromothriptic regions against genes not in chromothriptic regions and which are deleted in at least one patient. Only non-chromothriptic patients are included, making this analysis conservative. P values were generated by a two-sided Wilcoxon rank-sum test.
Extended Data Figure 9 mRNA–methylation associations in tumours with focal genomic events.
a, Density plot of Spearman correlations between the 10,000 most variable methylation probes and the 10,000 most variable mRNA transcripts in tumours with chromothriptic events, with kataegic events, and with neither focal abnormality. b, Density plot as in a for the 14,778 methylation probes in promoter regions and corresponding mRNA transcripts. c, Scatter plot of methylation (β-values for cg07227024 on chr2q) and mRNA abundance for OR2AK2 (on chr1q), which have the highest difference in correlations between chromothriptic (R = −0.90, P = 9.42 × 10−6) and non-chromothriptic (R = 0.52, P = 2.0 × 10−4) tumours. Dotted lines represent the regression line for each group. d, Enrichment pathway network plot of genes differentially correlated between chromothriptic and stable samples in promoter regions (|δ| > 0.8). Each node represents a gene set, which is defined as a set of genes that underlies a functional profile by g:Profiler. Node size corresponds to the number of genes within the gene set. The colour of the node represents the significance of the enriched gene set (hypergeometric test) ranging from FDR-adjusted P = 1.99 × 10−3 to P = 0.05 (red to pink). Gene sets are connected by a grey line if they share common genes and the thickness of the line corresponds to the size of the overlap. Gene sets with similar functions are grouped together by a purple dotted circle.
Extended Data Figure 10 Methylation survival validation and multi-modal signature survival.
Top, Kaplan–Meier plots of the six prognostic methylation probes in the validation data set (100 prostate tumours). Statistical analysis done using Cox proportional hazards modelling and P values generated by the Wald test, except for a where the log-rank test was performed owing to failure of the proportional-hazards assumption. a, TCERG1L-3′. b, SOX14. c, TUBA3C. d, TCERG1L-5′. e, MIR129-2. f, ACTL6B. g, A Kaplan–Meier plot for a multi-modal biomarker predicting biochemical recurrence, tested via cross-validation. This curve shows prediction of 18-month biochemical relapse-free survival. h, A Kaplan–Meier plot of the same biomarker, showing full biochemical relapse-free survival to the maximum follow-up time. In both plots, P values were generated using the Wald test.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, Legends for Supplementary Tables 1-18 (see separate zipped file), Supplementary Figures 1-10, Legends for Supplementary Data 1-3 (see separate zipped file) and Supplementary References. (PDF 4365 kb)
Supplementary Tables
This file contains Supplementary Tables 1-18 (see pages 3-5 of the Supplementary Information file for details). (ZIP 8162 kb)
Supplementary Data
This file contains Supplementary Data 1-3 (see page 18 of the Supplementary Information file for details). (ZIP 61447 kb)
Source data
Rights and permissions
About this article
Cite this article
Fraser, M., Sabelnykova, V., Yamaguchi, T. et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature 541, 359–364 (2017). https://doi.org/10.1038/nature20788
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature20788
- Springer Nature Limited
This article is cited by
-
Oral pimonidazole unveils clinicopathologic and epigenetic features of hypoxic tumour aggressiveness in localized prostate cancer
BMC Cancer (2024)
-
A reliable transcriptomic risk-score applicable to formalin-fixed paraffin-embedded biopsies improves outcome prediction in localized prostate cancer
Molecular Medicine (2024)
-
TP63–TRIM29 axis regulates enhancer methylation and chromosomal instability in prostate cancer
Epigenetics & Chromatin (2024)
-
Reference-free inferring of transcriptomic events in cancer cells on single-cell data
BMC Cancer (2024)
-
The yin and yang of chromosomal instability in prostate cancer
Nature Reviews Urology (2024)