Abstract
The emerging field of 'predictive analytics in mental health' has recently generated tremendous interest with the bold promise to revolutionize clinical practice in psychiatry paralleling similar developments in personalized and precision medicine. Here, we provide an overview of the key questions and challenges in the field, aiming to (1) propose general guidelines for predictive analytics projects in psychiatry, (2) provide a conceptual introduction to core aspects of predictive modeling technology, and (3) foster a broad and informed discussion involving all stakeholders including researchers, clinicians, patients, funding bodies and policymakers.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Main
Mental disorders are among the most debilitating diseases in industrialized nations today.1, 2 The immense economic loss3, 4, 5 mirrors the enormous suffering of patients and their friends and relatives.6, 7, 8, 9, 10, 11, 12 In addition, health-care costs as well as the number of individuals diagnosed with psychiatric disorders are projected to disproportionately rise within the next 20 years.13 With an ever-growing number of patients, the future quality of health care in psychiatry will crucially depend on the timely translation of research findings into more effective and efficient patient care. Despite the certainly impressive contributions of psychiatric research to our understanding of the etiology and pathogenesis of mental disorders, the ways in which we diagnose and treat psychiatric patients have largely remained unchanged for decades.14
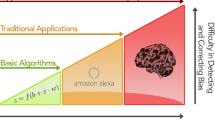
Recognizing this translational roadblock, we currently witness an explosion of interest in the emerging field of predictive analytics in mental health, paralleling similar developments in personalized or precision medicine.15, 16, 17, 18, 19 In contrast to the vast majority of investigations employing group-level statistics, predictive analytics aims to build models which allow for individual (that is, single subject) predictions, thereby moving from the description of patients (hindsight) and the investigation of statistical group differences or associations (insight) toward models capable of predicting current or future characteristics for individual patients (‘foresight’), thus allowing for a direct assessment of a model’s clinical utility (Figure 1).
Predictive analytics in mental health is moving from the description of patients (hindsight) and the investigation of statistical group differences or associations (insight) toward models capable of predicting current or future characteristics for individual patients (foresight), thereby allowing for a direct assessment of a model’s clinical utility.
Within this framework, we can differentiate three main areas of clinical application of predictive analytics models in mental health:
-
1
The prediction of therapeutic response can support the selection of optimized interventions, through comparative effectiveness research, thereby improving the trial-and-error-based approach common in psychiatry. For example, genetic variants have been linked to the outcome of psychotherapy as well as to therapeutic response to pharmacological interventions.20, 21 These individualized treatment optimization might maximizes adherence and minimizes undesired side-effects. Importantly, it also allows clinicians to focus resources on patients who will most likely benefit from the first-line treatment and allocate other resources to those who will require second-line or other treatments. Finally, identifying treatment-resistant individuals with high accuracy would also simplify the development and evaluation of novel drugs and interventions as research efforts could be more focused.
-
2
Supporting differential diagnoses is crucial, whenever the clinical picture alone is ambiguous. Providing additional model-based information to clinicians thus enables a timely administration of disease-specific interventions. Similar to the prediction of therapeutic response, this increases adherence and minimizes undesired side-effects. The differentiation of patients suffering major depression from patients with bipolar disorder before the first manic episode is but one example illustrating clinical utility in this area.
-
3
Models predicting individual risks are important in two respects. On the one hand, short-term predictions of risk can greatly improve outpatient management—for example with regard to prodrome detection in schizophrenia. On the other hand, long-term risk prediction would allow for a targeted application of preventive measures in early stages of a disorder or even before disease onset. Equally important, individual risk prediction could greatly increase the efficiency of the development and evaluation of preventive interventions as research efforts could be focused specifically on at-risk individuals.
In summary, valid models in this area would be instrumental, both, for minimizing patient suffering and for maximizing the efficient allocation of resources for research. For example, children and adults are diagnosed with attention deficit hyperactivity disorder every day and prescribed medications with little or no scientific evidence as to which patient will be likely to benefit from one or the other of the two major classes of medications (methylphenidate or amphetamine) or unlikely to benefit from either medication. In the same vein, the STAR*D study—a large evaluation of depression treatment including 4041 outpatients—showed that approximately 50% of patients respond.22 In both cases, patients would greatly benefit from Predictive Analytics models predicting which treatment would be most effective (for a wide range of predictions possible based on neuroimaging data today, see Gabrieli et al.17).
Against this background, predictive analytics in general and its potential applications in (mental) health have simultaneously been met with exuberant enthusiasm as well as with substantial skepticism. On the one hand, some see ‘previously unimaginable opportunities to apply machine learning to the care of individual patients’,15 prompting others to even propose ‘a shift from a search for elusive mechanisms to implementing studies that focus on predictions to help patients now’.23 On the other hand, critics have pointed out problems of an all too care-free view of predictive analytics in general and big data in particular.24 Considering the tremendous investment into big data infrastructure and predictive analytics capabilities in all areas of science and in the private sector,16 most will agree, however, that this technology—to quote a recent New York Times article24—‘is here to stay’, but that we ought to see it as ‘an important resource for anyone analyzing data, not a silver bullet’. From this, the question arises: How can we best steer the development and implementation of predictive analytics technology to effect the clinical innovations demanded by researchers and practitioners alike?
Now that evidence from initial proof-of-concept studies is accumulating in all areas—from genetics to neuroimaging, from blood-based markers to ambulatory assessments—and the approach is gaining momentum (for reviews, see refs 17, 19, 25, 26, 27, 28, 29, 30, 31), this question is particularly pressing. As the field of predictive analytics in mental health is faced with strategic choices which will have formative influence on research and clinical practice for the decades to come, we seek to move beyond the numerous descriptions and reviews of this beginning transformation of psychiatry by (1) proposing general guidelines for predictive analytics projects in psychiatry, (2) providing a conceptual introduction to the core aspects of predictive modeling technology, which distinguish predictive analytics in mental health from other areas of medicine or predictive analytics applications and (3) fostering a broad and informed discussion involving all stakeholders including researchers, clinicians, patients, funding bodies and policymakers. To this end, we will, first, provide an overview of the steps of a predictive analytics project. Secondly, we will consider the challenges that arise from the unique, multivariate and multimodal nature of mental disorders and argue that the combination of expert domain-knowledge and data integration technology is the key for overcoming, both, the conceptual and practical obstacles ahead. Finally, we will briefly discuss perspectives for the field.
Predictive analytics projects in mental health
Every predictive analytics project can be described as a series of steps aimed at ensuring the utility (that is, the validity and applicability) of the resulting model. Although this process is similar for all predictive analytics projects, numerous questions, problems and opportunities are unique to the area of mental health. The guiding questions in Box 1 are intended primarily as a means to support explicit reflection of the essential steps of a project—from defining objectives to deploying the model. Thereby, we hope to foster a broad discussion leading to common standards and procedures in the field.
Predictive analytics efforts in psychiatry parallel developments in other fields of medicine. Generally, we have witnessed a trend towards ever more precise specification of the genetic, molecular and cellular aspects of disease. This so-called precision medicine approach (for an overview, see for example, the US National Academy of Sciences report on the topic32), in many cases, led to the realization that disease entities which appear to be a single disorder actually have distinct genetic precursors and pathophysiology. For example, cancer diagnosis is—for many forms of cancer—defined by analysis of genetic variants based on which the optimal treatment can be predicted.33 While communalities are particularly obvious with regard to technology, researchers in psychiatry are also faced with rather unique challenges.
Apart from the massively multivariate and multimodal nature of mental disorders which we will discuss in detail below, a traditionally much discussed issue arises from the often-times fuzzy and relatively unreliable labels of disease entities in psychiatry. As predictive models learn from examples, training a model aiming to support the differentiation between patients suffering from major depressive disorder and individuals with bipolar disorder before conversion (that is, before any (hypo)manic symptoms have become apparent), for instance, might proof difficult simply because it may be very hard to reliably categorize patients with certainty. In practice, most studies either mitigate this problem by employing resource-intensive, state-of-the-art diagnostic procedures in combination with multiple clinical expert ratings or circumvent it by acquiring data first and then waiting for the quantity of interest to become more easily accessible (for example, until the end of a therapeutic intervention or until a disorder actually manifests in at-risk individuals screened years ago). Complementing these efforts to render labels more accurate, fuzzy and unreliable labels can also be handled directly using machine learning algorithms specifically designed for this purpose (for a straightforward introduction, see ref. 34, 35). Although currently, it seems as if researchers in psychiatry almost exclusively rely on the optimization of data acquisition rather than trying to inherently model label uncertainty, combining the two approaches might be highly beneficial.
In addition, current disease entities as defined by DSM-5 or ICD-10 are very heterogeneous regarding, both, (neuro)physiology as well as clinical endophenotypes.36 On the one hand, this will make the classification of disease entities difficult as each entity is in fact a conglomerate of different (neuro)physiological and behavioral deviations. On the other hand, the underlying causes or correlates of therapeutic response or disease trajectory may qualitatively as well as quantitatively vary for different, more homogeneous sub-samples of the data. Although this makes training predictive models more difficult (that is, either more training data or more prior information will be needed), machine learning algorithms are generally well equipped to handle such cases. In fact, learning multiple rules mapping features to labels are quite common (model averaging, stacking and voting are but three ways outlined in the next section to handle this). That said, homogeneous disease entities would not only make discovering rules easier (especially on small data sets), but definitely lead to more interpretable models which—though not technically the goal of predictive analytics—is still desirable from a scientific point of view. Most importantly, however, discovering homogeneous disease entities would enable us to move beyond merely reproducing the presently established diagnostic classification using considerably more expensive and complicated procedures. While this has thus far been a seemingly unattainable goal not only for DSM-5, the recent success of so-called unsupervised machine learning approaches might reinvigorate this line of research (for an introduction to unsupervised machine learning, see ref. 37).
Challenges arising from the unique multivariate and multimodal nature of mental disorders
Although the guidelines outlined above provide a straightforward framework for predictive analytics projects in psychiatry, the main challenge for the field arises from the unique, multivariate and multimodal nature of mental disorders. In the following, we will outline the conceptual and practical problems in more detail and argue that the combination of expert domain-knowledge and data integration technology is the key when aiming to construct valid predictive model for clinical use.
Modeling massively multivariate data
Overwhelming evidence shows that no single measurement—be it a gene, a psychometric test or a protein—explains substantial variance with regard to any practically relevant aspect of a psychiatric disorder (compare, for example, ref. 38). In contrast, it has been recognized that multiple measures are necessary to gain meaningful information even within a single modality. It is this profoundly multivariate nature of mental disorders that has driven researchers to, for example, conduct genome-wide association studies and acquire whole-brain neuroimaging data.
When aiming to build predictive models, this complexity necessitates the use of methods suitable for high-dimensional data sets, in which the number of variables (that is, measurements) may far exceed the number of samples (that is, patients). Generally, the so-called curse of dimensionality is addressed in three ways (for an excellent review detailing this issue, see ref. 39). First, unsupervised methods for dimensionality reduction—such as principal component analysis—may be used. These algorithms apply more or less straightforward transformations to the input data to yield a lower-dimensional representation. Also, they can extract a wide range of predefined features from raw-data. For example, distance measures can be extracted from raw protein sequences for classification in a fully automated fashion.40 Second, techniques integrating dimensionality reduction and predictive model estimation (for example, regularization, Bayesian model-selection and cross-validation) may be applied. In essence, they use penalties for model complexity, thereby enforcing simpler, often lower-dimensional models. Simply speaking, models containing more parameters must enable proportionally better predictions to be preferred over simpler models. These algorithms are at the heart of predictive analytics projects and include well-known techniques such as Support Vector Machines and Gaussian Process Classifiers as well as the numerous tree algorithms (for details, see refs 37, 41). Third, feature-engineering, that is, all methods aiming to create useful predictors from the input data—can be used. In short, feature-engineering aims to transform the input data (that is, all measures acquired) in a way that optimally represents the underlying problem to the predictive model. An illustrative example comes from a recent study which constructed a model predicting psychosis onset in high-risk youths based on free speech samples. Whereas it would have been near impossible to build a model based on the actual recordings of participants’ speech, the team achieved high accuracy in a cross-validation framework using speech features extracted with a latent semantic analysis measure of semantic coherence and two syntactic markers of speech complexity.42 While these results still await fully independent replication, the approach shows that transforming the input data (speech samples) using domain-knowledge (in this case the knowledge that syntax differs in certain patients) can greatly foster the construction of a predictive model. Demonstrating the problem-dependent nature of feature-engineering, it might have been much easier to decode, for example, participants’ gender from the actual recordings than from latent semantic analysis measures given the difference in pitch between males and females. In that it links data acquisition and model algorithms, feature-engineering is not primarily a preprocessing or dimensionality-reduction technique, but a conceptually decisive step of building a predictive model.
While important for all modalities, feature-engineering often has a particularly crucial role when constructing predictive models based on physiological or biophysical data. On the one hand, these data are often especially high-dimensional (for example, genome, proteome or neuroimaging data with regularly tens of thousands of variables), thus often requiring dimensionality-reduction. On the other hand, alternative transformations of the raw-data can contain fundamentally different, non-redundant information. For example, the same funational magnetic resonance imaging raw-data, that is, measures of changes in regional blood-oxygen levels—can be processed to yield numerous, non-redundant representations (for example, activation maps or functional connectivity matrices). In addition, domain-knowledge regarding the choice of relevant regions-of-interest or atlas parcellations also fundamentally affects the representation of information in neuroimaging data.43 As different parameters can be meaningful in the context of different disorders, these examples powerfully illustrate the fundamental importance of domain-knowledge in feature-engineering. The sources of domain-knowledge needed to decide which data representations might be optimal with regard to the problem at hand may range from large-scale meta-analyses, reviews and other empirical evidence to clinical experience.
Taking the traditionally somewhat subjective ‘art of feature-engineering’ a step further, are automated feature-engineering algorithms. The former are akin to other unsupervised methods for dimensionality reduction, but can learn meaningful transformations from large, unlabeled data sets (for example, using Deep Learning algorithms44). In short, these algorithms form high-level representations of more basic regularities in the data (for a large-scale example, see ref. 45). It is these high-level representations which can then be used to train the model. For example, we might use large data sets of resting-state funational magnetic resonance imaging to automatically uncover regularities (such as network-structure) using unsupervised learning. These newly constructed features might then provide a lower-dimensional, more informative basis for model-building in future funational magnetic resonance imaging projects. Note that domain-knowledge is not provided directly, but learned from independent data sources in this framework. Although these techniques appear highly efficient as no expert involvement is required, discovering high-level features for the massively multivariate measures commonly needed in psychiatry will require extraordinarily large—though possibly unlabeled—data sets as well as computational power beyond the capabilities of most institutions today. Considering the developments in other areas such as speech recognition, we believe, however, that the significance of automated feature-engineering techniques can only grow in the years to come.
In many ways, the theory-driven approach to computational psychiatry is following an at least equally promising—albeit extreme opposite—strategy. This approach builds mechanistic models based on theory and available evidence. After a model is validated, model parameters encapsulate a theoretical, often mechanistic, understanding of the phenomena (for an excellent introduction, see ref. 39). In many ways, the resulting models thus constitute highly formalized (one might say ‘condensed’) representations of domain-knowledge, custom-tailored to the problem at hand. Unlike virtually all other approaches to feature-engineering, computational models allow researchers to test the validity of data representations while simultaneously fully explicating domain-knowledge. While certainly more scientifically satisfying and theoretically superior to feature-engineering, constructing valid models is far from simple. Thus, we believe that this technology will gain in importance to the degree that building valid models proofs feasible, further intertwining theoretical progress and predictive analytics.
Having discussed feature-engineering in greater detail, it is important to point out that model construction algorithms are not limited to the use of one single data-representation. To the contrary, it is a particular strength of this approach—with algorithms usually allowing for massively multivariate data and model integration—that multiple, meaningful data representations can be combined to enable valid predictions (for a more detailed discussion of model integration, see below).
Summarizing, the acquisition of high-dimensional data is regularly required to capture the massively multivariate nature of the processes underlying psychiatric disorders. Even on a single level of observation, we thus need to deal with the curse of dimensionality. To this end, model building commonly includes steps such as simple dimensionality-reduction techniques (for example, principal component analysis) and penalizing model-complexity as part of machine learning algorithms. Most importantly, however, feature-engineering is used to create data representations from the input data which enable machine learning algorithms to build a valid model. Feature-engineering may draw on partially (meta-analyses or clinical experience) or fully formalized domain-knowledge (for example, parameters from previously validated computational models) or a combination thereof. This prominent role of domain-knowledge underlines the interdependence of classic scientific approaches seeking mechanistic insight fostering theoretical development and predictive analytics approaches in mental health. While theoretical progress and meta-analytic evidence aid the construction of optimal features, a predictive analytics approach, in turn, allows for a direct assessment of the clinical utility of group-level evidence and theoretical advances. Thus, it is evident that these two branches of research are not mutually exclusive, but complementary approaches when aiming to benefit patients.
Incorporating (interactions across) multiple levels of observation
Substantially aggravating the problem of dimensionality discussed above, mental disorders are characterized by numerous, possibly interacting biological, intrapsychic, interpersonal and socio-cultural factors.46, 47 Thus, a clinically useful patient representation must probably, in many cases, be massively multimodal, that is, include data from multiple levels of observation—possibly spanning the range from molecules to social interaction. All these modalities might contain non-redundant, possibly interacting sources of information with regard to the clinical question. In fact, it is this peculiarity—distinguishing psychiatry from most other areas of medicine—which has hampered research in general and translational efforts in particular for decades now. As applying a simple predictive modeling pipeline on a multi-level patient representation would increase the already large number of dimensions for a unimodal data set by several orders of magnitude, it might seem that Predictive Analytics endeavors are likely to suffer from similar if not larger problems. Indeed, neither of the dimensionality-reduction, regularization or even feature-engineering approaches outlined above is capable of seamlessly integrating such ultra-high-dimensional data from so profoundly different modalities. Considering the tremendous theoretical problems of understanding phenomena on one level of observation, we also cannot rely on progress regarding the development of a valid theory spanning multiple levels of observation in the near future. Likewise, detailed domain-knowledge across levels of observation is extremely difficult to obtain as empirical evidence as well as expert opinions are usually specific to one modality. Given the extreme amounts of data and the combinatorial explosion due to their potential interactions, fully automated feature-engineering approaches across levels of observation (as opposed to such techniques for single levels of observation) also appear unlikely in the near future. Finally, the often qualitatively different data sources alone—including genetics, proteomics, psychometry, and neuroimaging data as well as ambulatory assessments and information from various, increasingly popular wearable sensors—would make this a herculean task.
A somewhat trivial solution would be to limit the predictive model to a single level of observation. If high-accuracy predictions can be obtained in this way—which might be considered unlikely at least for the most difficult clinical questions—such unimodal models are always preferable due to their comparatively high efficiency. Apart from the inherent multimodal nature of mental disorders which might render unimodal models less accurate, it is, however, exactly these efficiency considerations, which obviate the need for predictive analytics research to consider multiple levels of observation. In order to identify the most efficient combination of data sources in a principled way in the absence of detailed cross-modal expert knowledge and evidence, we have to learn it from the data. To this end, a plethora of machine learning approaches which can be broadly described as model integration techniques—have been developed.
Probably, the most intuitive way to combine information from different high-dimensional sources is by voting. In this framework, a predictive model is trained for each modality and the majority vote is used as the overall model prediction. In a binary classification—if we wish to predict therapeutic response (yes, patient will benefit vs. no, patient will not benefit from the intervention) from five multivariate data sources—we first train a model for each modality. Then, we count the number of models predicting a response (#yes) and the number of models predicting no response (#no). The final prediction of therapeutic response is given by the option receiving more votes across modalities. A slightly more sophisticated approach is stacking or stacked generalization. Here, again, a model is trained for each modality. The predictions are, however, not combined by voting, but used as input to another machine learning algorithm which constructs a final model with the unimodal predictions as features (note that both examples might technically be considered automatic feature-engineering techniques). In addition to these simple approaches, numerous other techniques (for example, (Bayesian) model averaging, bagging, boosting or more sophisticated ensemble algorithms) exist—each with different strengths and weaknesses which affect the computational infrastructure needed and interact with data structure within and across modalities. That said, most predictive analytics practitioners would agree that models—in the field—are most often constructed by evaluating a large number of approaches, that is, by trial-and-error relying on computational power. However, it cannot be emphasized enough that this strategy must rely on the training data only. At no time and in no form, may the test set—that is, the samples later used to evaluate (out-of-bag) model performance—be used in this process. Only in this way, we guarantee a valid estimation of predictive power in practice. Note that the techniques for model combination can generally be used also to construct predictive models from unimodal multivariate data sets as well (see for example refs 43, 48 for an in-depth introduction, see ref. 49). Given the multimodal nature of psychiatric disorders, however, they hold particular value for cross-modal model integration.
Importantly, the construction of models from multimodal data does not mean that the final predictive model used in the clinic must also be multimodal. To the contrary, by training models with multimodal data, we not only guarantee maximum predictive power but also gain empirical evidence regarding the utility of each modality. Analyzing the final model, we can investigate which modalities (and which variables within each modality) contribute substantial, non-redundant information. In an independent sample, we could then train a model based only on those modalities (or variables) most important in the first model. With this iterative process, we can obtain not only the most accurate, but also the most efficient combination of modalities and variables in a principled manner. Thus, final models might only consist of very few modalities and variables fostering their widespread use also from a health economics point of view.
Perspectives
Effective translation of research findings into clinical practice using predictive analytics will not only require the combination of expert domain-knowledge and data integration technology as outlined above. Effective translation will also need to address more general issues regarding the organization and structure of the emerging field. This will require joint efforts from all stakeholders including researchers, clinicians, patients, funding bodies, and policymakers. One such example is the Patient Centered Outcomes Research Network (PCORnet.org) and its associated psychiatric networks, the MoodNetwork, the Interactive Autism Network, and the Community and Patient-Partnered Centers of Excellence which focuses on behavior disorders in underserved communities. 50
Given the often sensitive nature of the data needed to build predictive models—which might for example include electronic health records—an adequate level of security must be maintained at all times. Whether this speaks for decentralized infrastructure or outsourcing to specialized institutions is likely to remain a matter of intensive debate. As an example, PCORnet uses a federated datamart with a common data model infrastructure for multiple health-care systems across USA that includes over 90 million people. Similar discussions will probably arise with regard to the predictive models themselves. Although only easy access to validated, pre-trained models will make them widespread, useful tools in the clinic, predictive models might also enable the prediction of sensitive personal data from the combination of seemingly harmless information an individual might readily provide. Thus, it is in the interest of all stakeholders to reach a public consensus regarding the regulation of access to pre-trained models before practically applicable models become available. While some level of regulation is likely beneficial with regard to industry use, it will be essential for efficient model construction to encourage model sharing (similar to data sharing) for research purposes. Especially for multimodal models, sharing modality-specific, pre-trained models (for example, in dedicate model databases) will save substantial amounts of time and money. Finally, we need experts to consider the legal implications of deploying models (publicly or within the field) which predict health-related information which potentially guides medical decisions.
From a more applied perspective, we believe that technology will continue to simplify data acquisition and improve data quality in the years to come, thus bringing predictive mobile health (mHealth) applications within reach. Although holding great promise, especially mHealth applications raise the question of whether it is generally better to rely on mechanistic predictors or instead on a pragmatic approach.23, 51 Although we firmly believe that the identification of causal relationships provides the most robust and scientifically satisfying features for prediction, we expect a pragmatic approach to prevail in the years ahead for two reasons. First, while causal predictors might be most effective, they will often be inefficient. For example, measuring variables of brain metabolism causally linked to a disorder might enable the construction of highly accurate predictive models. If however, we can use cheaper and more readily obtainable (for example, smartphone-based) measures not causal to the disorder with comparable or even slightly lower predictive power, those would probably be more efficient and thus more useful to clinicians in practice. Secondly, as decades of research have only begun to uncover causal links on single levels of observation, we think it highly unlikely that unified theoretical models across levels of observation will be established even in the mid-term.
To promote the endeavor of creating individualized predictive models to improve patient care and maximize cost efficiency in psychiatry, concrete steps can to be taken by institutions, researchers and practitioners. For example, we have recently seen numerous educational efforts such as organizing workshops and seminars on the various technical topics. Conferences such as the European College of Neuropsychopharmacology Congress or the Resting-State Conference and many others will continue to host sessions and satellite symposia dedicated to predictive analytics. Common in the field of machine learning, but currently scarce in psychiatry, predictive analytics competitions in which teams compete for the best predictive model performance (for example, ADHD-200 global competition) bring together clinicians, researchers, and machine learners and may accelerate the availability of pre-trained, validated models in the mid-term as well as make this research more visible to the public.
Although patients, clinicians, and researchers share a common interest in improving mental health outcomes, there will need to be a thoughtful balancing of issues related to privacy, data security and ethics in relation to the contrasting priorities and roles of various stakeholders. Currently, research and curation of shared data bases arise primarily from publicly funded, academic research groups, where data sharing is viewed as a common good to support greater utilization of large data sets to enhance predictive accuracy. A private business, on the other hand, could have the different role of using predictions to make decisions about reimbursing health-care options or to advise on hiring practices or to identify potential customers for advertisements. Although these contrasting goals could lead to some tensions about the use of predictive analyses, there are examples where a public-private hybrid could be advantageous. For example, because intervention research is costly and complex, it tends to have limited numbers of subjects and relatively short durations (such as evaluation immediately after an intervention). Public–private partnerships could take advantage of the ongoing administration of treatments to very large numbers of subjects over extended time periods.
In summary, we believe that unimodal feature-engineering and model integration across levels of observation will be the key to highly accurate and efficient predictive analytics models in mental health. Successful predictive analytics projects will thus require (1) substantial domain-knowledge-based or technology-driven efforts (e.g. from computational modelling or deep learning) to enable optimal feature-engineering for the often massively multivariate data sets obtained on each level of observation and (2) profound machine learning expertise with a focus on model integration techniques. With technology rapidly simplifying data acquisition and model construction, we urge all stakeholders including researchers, clinicians, patients, funding bodies and policymakers to initiate an open discussion regarding key-issues such as data-sharing and model access-regulations to enable predictive analytics technology to close the gap between bench and bedside.
References
Kessler RC, Aguilar-Gaxiola S, Alonso J, Chatterji S, Lee S, Ormel J et al. The global burden of mental disorders: an update from the WHO World Mental Health (WMH) surveys. Epidemiol Psichiatr Soc 2009; 18: 23–33.
Wittchen HU, Jacobi F, Rehm J, Gustavsson A, Svensson M, Jonsson B et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur Neuropsychopharmacol 2011; 21: 655–679.
Gustavsson A, Svensson M, Jacobi F, Allgulander C, Alonso J, Beghi E et al. Cost of disorders of the brain in Europe 2010. Eur Neuropsychopharmacol 2012; 22: 237–238.
Insel T . Assessing the economic costs of serious mental illness. Am J Psychiatry 2008; 165: 663–665.
Stamm K, Salize H-J Volkswirtschaftliche Konsequenzen. In: Stoppe G, Bramesfeld A, Schwartz F-W (eds). Volkskrankheit Depression? Springer: Berlin Heidelberg, Germany, 2006, pp 109–120.
Dore G, Romans SE . Impact of bipolar affective disorder on family and partners. J Affect Disord 2001; 67: 147–158.
Gianfrancesco FD, Wang R-h YuE . Effects of patients with bipolar, schizophrenic, and major depressive disorders on the mental and other healthcare expenses of family members. Soc Sci Med 2005; 61: 305–311.
Guan B, Deng Y, Cohen P, Chen H . Relative impact of Axis I mental disorders on quality of life among adults in the community. J Affect Disord 2011; 131: 293–298.
Hasui C, Sakamoto S, Sugiura T, Miyata R, Fujii Y, Koshiishi F et al. Burden on family members of the mentally ill: a naturalistic study in Japan. Compr Psychiatry 2002; 43: 219–222.
Olatunji BO, Cisler JM, Tolin DF . Quality of life in the anxiety disorders: a meta-analytic review. Clin Psychol Rev 2007; 27: 572–581.
Saunders JC . Families living with severe mental illness: a literature review. Issues Ment Health Nurs 2003; 24: 175–198.
Wittmund B, Wilms HU, Mory C, Angermeyer MC . Depressive disorders in spouses of mentally ill patients. Soc Psychiatry Psychiatr Epidemiol 2002; 37: 177–182.
Bloom DE, Cafiero ET, Jané-Llopis E, Abrahams-Gesse S, Bloom LR, Fathima S et al The Global Economic Burden of Noncommunicable Diseases. Geneva: World Economic Forum. 2011, available from: http://www3.weforum.org/docs/WEF_Harvard_HE_GlobalEconomicBurdenNonCommunicableDiseases_2011.pdf.
Stephan KE, Bach DR, Fletcher PC, Flint J, Frank MJ, Friston KJ et al. Charting the landscape of priority problems in psychiatry, part 1: classification and diagnosis. Lancet Psychiatry 2016; 3: 77–83.
Darcy AM, Louie AK, Roberts LW . Machine learning and the profession of medicine. JAMA 2016; 315: 551–552.
Eyre HA, Singh AB, Reynolds C 3rd . Tech giants enter mental health. World Psychiatry 2016; 15: 21–22.
Gabrieli JD, Ghosh SS, Whitfield-Gabrieli S . Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron 2015; 85: 11–26.
Jordan MI, Mitchell TM . Machine learning: trends, perspectives, and prospects. Science 2015; 349: 255–260.
Wolfers T, Buitelaar JK, Beckmann CF, Franke B, Marquand AF . From estimating activation locality to predicting disorder: a review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neurosci Biobehav Rev 2015; 57: 328–349.
Eley TC, Hudson JL, Creswell C, Tropeano M, Lester KJ, Cooper P et al. Therapygenetics: the 5HTTLPR and response to psychological therapy. Mol Psychiatry 2012; 17: 236–237.
Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet 2016; 387: 1085–1093.
DeRubeis RJ, Siegle GJ, Hollon SD . Cognitive therapy versus medication for depression: treatment outcomes and neural mechanisms. Nat Rev Neurosci 2008; 9: 788–796.
Paulus MP . Pragmatism instead of mechanism: a call for impactful biological psychiatry. JAMA Psychiatry 2015; 72: 631–632.
Marcus G, Davis E (2014). Eight (No, Nine!) Problems With Big Data. In Sulzberger AOJ (ed). The New York Times: New York, NY, USA, p A23.
Chan MK, Gottschalk MG, Haenisch F, Tomasik J, Ruland T, Rahmoune H et al. Applications of blood-based protein biomarker strategies in the study of psychiatric disorders. Prog Neurobiol 2014; 122: 45–72.
Fu CH, Costafreda SG . Neuroimaging-based biomarkers in psychiatry: clinical opportunities of a paradigm shift. Can J Psychiatry 2013; 58: 499–508.
Hofmann-Apitius M, Ball G, Gebel S, Bagewadi S, de Bono B, Schneider R et al. Bioinformatics mining and modeling methods for the identification of disease mechanisms in neurodegenerative disorders. Int J Mol Sci 2015; 16: 29179–29206.
McMahon FJ . Prediction of treatment outcomes in psychiatry—where do we stand ? Dialogues Clin Neurosci 2014; 16: 455–464.
Savitz JB, Rauch SL, Drevets WC . Clinical application of brain imaging for the diagnosis of mood disorders: the current state of play. Mol Psychiatry 2013; 18: 528–539.
Scarr E, Millan MJ, Bahn S, Bertolino A, Turck CW, Kapur S et al. Biomarkers for psychiatry: the journey from fantasy to fact, a report of the 2013 CINP Think Tank. International Journal of Neuropsychopharmacology 2015; 18: pyv042.
Veronese E, Castellani U, Peruzzo D, Bellani M, Brambilla P . Machine learning approaches: from theory to application in schizophrenia. Comput Math Methods Med 2013; 2013: 867924.
National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. National Academies Press: Washington DC, 2011.
Mirnezami R, Nicholson J, Darzi A . Preparing for precision medicine. N Engl J Med 2012; 366: 489–491.
Lin CF, Wang SD . Fuzzy support vector machines. IEEE Trans Neural Netw 2002; 13: 464–471.
Qin ZC, Lawry J . Decision tree learning with fuzzy labels. Inform Sci 2005; 172: 91–129.
Regier DA, Narrow WE, Kuhl EA, Kupfer DJ . The conceptual development of DSM-V. Am J Psychiatry 2009; 166: 645–650.
Hastie T, Tibshirani R, Friedman J, Franklin J . The elements of statistical learning: data mining, inference and prediction. Math Intell 2005; 27: 83–85.
Ozomaro U, Wahlestedt C, Nemeroff CB . Personalized medicine in psychiatry: problems and promises. BMC Med 2013; 11: 132.
Huys QJM, Maia TV, Frank MJ . Computational psychiatry as a bridge from neuroscience to clinical applications. Nat Neurosci 2016; 19: 404–413.
Ofer D, Linial M . ProFET: feature engineering captures high-level protein functions. Bioinformatics 2015; 31: 3429–3436.
MacKay DJC . Information Theory, Inference, and Learning Algorithms. Cambridge University Press: Cambridge, UK; New York, NY, USA, 2003.
Bedi G, Carrillo F, Cecchi GA, Slezak DF, Sigman M, Mota NB et al. Automated analysis of free speech predicts psychosis onset in high-risk youths. NPJ Schizophr 2015; 1: 15030.
Hahn T, Kircher T, Straube B, Wittchen HU, Konrad C, Strohle A et al. Predicting treatment response to cognitive behavioral therapy in panic disorder with agoraphobia by integrating local neural information. JAMA Psychiatry 2015; 72: 68–74.
LeCun Y, Bengio Y, Hinton G . Deep learning. Nature 2015; 521: 436–444.
Building High-Level Features Using Large Scale Unsupervised Learning. I EEE International Conference on Proceedings of the Acoustics, Speech and Signal Processing (ICASSP): Vancouver, BC, 2013, pp 8595–8598.
Kendler KS . The nature of psychiatric disorders. World Psychiatry 2016; 15: 5–12.
Maj M . The need for a conceptual framework in psychiatry acknowledging complexity while avoiding defeatism. World Psychiatry 2016; 15: 1–2.
Hahn T, Marquand AF, Ehlis AC, Dresler T, Kittel-Schneider S, Jarczok TA et al. Integrating neurobiological markers of depression. Arch Gen Psychiatry 2011; 68: 361–368.
Bishop CM . Pattern Recognition and Machine Learning. Springer: New York, NY, USA, 2006.
Collins FS, Hudson KL, Briggs JP, Lauer MS . PCORnet: turning a dream into reality. J Am Med Inform Assn 2014; 21: 576–577.
Pine DS, Leibenluft E . Biomarkers with a mechanistic focus. JAMA Psychiatry 2015; 72: 633–634.
Pereira F, Mitchell T, Botvinick M . Machine learning classifiers and fMRI: a tutorial overview. Neuroimage 2009; 45: S199–S209.
Yang Z, Fang F, Weng X . Recent developments in multivariate pattern analysis for functional MRI. Neurosci Bull 2012; 28: 399–408.
Lueken U, Hahn T . Functional neuroimaging of psychotherapeutic processes in anxiety and depression: from mechanisms to predictions. Curr Opin Psychiatry 2016; 29: 25–31.
Lazer D, Kennedy R, King G, Vespignani A . Big data. The parable of Google Flu: traps in big data analysis. Science 2014; 343: 1203–1205.
Wolpert DH . The lack of a priori distinctions between learning algorithms. Neural Comput 1996; 8: 1341–1390.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
TH and SW-G declare no conflicts of interest. AAN is a consultant for the Abbott Laboratories, Alkermes, American Psychiatric Association, Appliance Computing Inc. (Mindsite), Basliea, Brain Cells, Inc., Brandeis University, Bristol Myers Squibb, Clintara, Corcept, Dey Pharmaceuticals, Dainippon Sumitomo (now Sunovion), Eli Lilly and Company, EpiQ, L.P./Mylan Inc., Forest, Genaissance, Genentech, GlaxoSmithKline, Hoffman LaRoche, Infomedic, Intra-Cellular Therapies, Lundbeck, Janssen Pharmaceutica, Jazz Pharmaceuticals, Medavante, Merck, Methylation Sciences, Naurex, NeuroRx, Novartis, Otsuka, PamLabs, Parexel, Pfizer, PGx Health, Ridge Diagnostics Shire, Schering-Plough, Somerset, Sunovion, Takeda Pharmaceuticals, Targacept, and Teva; consulted through the MGH Clinical Trials Network and Institute (CTNI) for Astra Zeneca, Brain Cells, Inc, Dianippon Sumitomo/Sepracor, Johnson and Johnson, Labopharm, Merck, Methylation Science, Novartis, PGx Health, Shire, Schering-Plough, Targacept and Takeda/Lundbeck Pharmaceuticals. He receives grant/research support from American Foundation for Suicide Prevention, AHRQ, Brain and Behavior Research Foundation, Bristol-Myers Squibb, Cederroth, Cephalon, Cyberonics, Elan, Eli Lilly, Forest, GlaxoSmithKline, Janssen Pharmaceutica, Intra-Cellular Therapies, Lichtwer Pharma, Marriott Foundation, Mylan, NIMH, PamLabs, PCORI, Pfizer Pharmaceuticals, Shire, Stanley Foundation, Takeda and Wyeth-Ayerst. Honoraria include Belvoir Publishing, University of Texas Southwestern Dallas, Brandeis University, Bristol-Myers Squibb, Hillside Hospital, American Drug Utilization Review, American Society for Clinical Psychopharmacology, Baystate Medical Center, Columbia University, CRICO, Dartmouth Medical School, Health New England, Harold Grinspoon Charitable Foundation, IMEDEX, Israel Society for Biological Psychiatry, Johns Hopkins University, MJ Consulting, New York State, Medscape, MBL Publishing, MGH Psychiatry Academy, National Association of Continuing Education, Physicians Postgraduate Press, SUNY Buffalo, University of Wisconsin, University of Pisa, University of Michigan, University of Miami, University of Wisconsin at Madison, World Congress of Brain Behavior and Emotion, APSARD, ISBD, SciMed, Slack Publishing and Wolters Klower Publishing ASCP, NCDEU, Rush Medical College, Yale University School of Medicine, NNDC, Nova Southeastern University, NAMI, Institute of Medicine, CME Institute, ISCTM. He was currently or formerly on the advisory boards of Appliance Computing, Inc., Brain Cells, Inc., Eli Lilly and Company, Genentech, Johnson and Johnson, Takeda/Lundbeck, Targacept, and InfoMedic. He owns stock options in Appliance Computing, Inc., Brain Cells, Inc, and Medavante; has copyrights to the Clinical Positive Affect Scale and the MGH Structured Clinical Interview for the Montgomery Asberg Depression Scale exclusively licensed to the MGH Clinical Trials Network and Institute (CTNI).
PowerPoint slides
Rights and permissions
About this article
Cite this article
Hahn, T., Nierenberg, A. & Whitfield-Gabrieli, S. Predictive analytics in mental health: applications, guidelines, challenges and perspectives. Mol Psychiatry 22, 37–43 (2017). https://doi.org/10.1038/mp.2016.201
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2016.201
- Springer Nature Limited
This article is cited by
-
Mental Health in Tech: Analysis of Workplace Risk Factors and Impact of COVID-19
SN Computer Science (2023)