Abstract
In this study, we evaluate a method for the KPC enzyme detection, using MALDI-TOF MS, for Enterobacterales. A total of 300 clinical Enterobacterales isolates were selected. The collection included 259 carbapenemase-producing (157 KPC and 102 non-KPC) and 41 carbapenemase non-producing isolates. Bacterial proteins were extracted from Mueller–Hinton agar plates using formic acid, isopropyl alcohol, and water (17:33:50). Samples were prepared with a double layer of synapinic acid. Analyses were performed using a Microflex LT mass spectrometer (Bruker Daltonics) and flexAnalysis 4.0 software (Bruker Daltonics). Statistical analyses were performed using SPSS Software. A distinctive peak at m/z 28,643–28,731 was found in all 157 KPC-producing isolates, and it was consistently absent in the 143 KPC non-producing group. KPC-producing peak intensities ranged from 77 to 3893. Considering an intensity cutoff value ≥ 120 for the presence of KPC, this methodology presented 98.09% and 97.90% of sensitivity and specificity, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Infections caused by carbapenem-resistant Enterobacterales (CRE) have become a public health problem in recent decades due to, among other reasons, their remarkable dissemination capacity. Besides, infections caused by CRE are often related to therapeutic failures, being associated with high morbidity and mortality rates [1, 2].
Production of class A carbapenemase is the most important mechanism of carbapenem resistance in many regions of the world. The most relevant enzyme of this class is KPC (Klebsiella pneumoniae Carbapenemase), with KPC-2 standing out [3,4,5]. As carbapenemase genes can spread clonally or by horizontal gene transfer, rapid and accurate detection of CRE is critical not only for therapeutic strategies, but also for infection control purposes [6,7,8].
Resistance to carbapenems is identified through standard susceptibility testing but additional testing is required to identify carbapenemase production. Currently recommended methods for detecting carbapenemases in Enterobacterales are the following: (i) combination disk testing, (ii) biochemical (colorimetric) tests, (iii) carbapenem inactivation method (CIM), and (iv) the detection of carbapenemase activity by MALDI-TOF MS [9]. Unfortunately, these methods are time-consuming or are not able to define the carbapenemase type.
The incorporation of the MALDI-TOF MS technology in the clinical microbiology laboratories has been a revolutionary breakthrough for the bacterial identification. Another promising use of the MALDI-TOF technology is the direct detection of a specific enzyme, such as a carbapenemase, through the analysis of the profile of proteins [10,11,12]. Therefore, in this study, we evaluated a methodology using MALDI-TOF MS to detect the KPC enzyme among Enterobacterales grown in solid culture medium.
Materials and methods
Bacterial isolates and β-lactamases characterization
A total of 300 clinical Enterobacterales isolates (Table 1) collected between 2018 and 2021 from patients attending at a tertiary hospital in Porto Alegre city, southern Brazil, were selected for this study. The collection included 41 carbapenemase non-producing isolates, as well as 259 CRE from which 151 were KPC producers, 84 NDM, 16 OXA-48-like, 6 KPC/NDM co-producers, and 2 NDM/OXA-48-like co-producers. The enzyme detection was performed by high-resolution melting-PCR, as previously described [13], for the presence of blaKPC, blaNDM-1, blaGES, blaOXA-48, blaIMP, and blaVIM genes. Bacteria species were identified by MALDI-TOF Vitek MS (bioMérieux).
Sample preparation for MALDI-TOF MS
Protein extraction was performed according to Figueroa-Espinosa [11] with modifications. Colonies were obtained from Mueller–Hinton (Sigma-Aldrich and Newprov) agar plates, without antibiotics, previously incubated overnight at 37 °C. First, a loopful of bacterial colonies (4 to 5) was suspended in 300 μL of distilled water and vortexed for 30 s at room temperature, and 900 μL absolute ethanol was added. The suspension was vortexed for 30 s and centrifuged at 13,000 rpm (16,060 × g) for 2 min at room temperature. The supernatant was discarted and the pellet was re-suspended in 100 µL of extraction solvent (formic acid–isopropyl alcohol–water, 17:33:50 v/v). Extraction solvent was prepared and used within 1 week. The suspension was vortexed for 30 s and centrifuged for 2 min at 13,000 rpm (16,060 × g) at room temperature. The clean supernatant extract was reserved for MALDI-TOF analysis.
Target spot loading
The sample was prepared with the double layer sinapinic acid (SA) method [11]. The first layer was composed of 0.7 μL of SA saturated solution, which was placed onto MALDI target spots and dried at room temperature. For the second layer, the protein extracts previously obtained were mixed 1:1 with SA 10 mg/mL solution in acetonitrile (30:70 v/v) and 0.1% trifluoroacetic acid in water. One microliter of this sample/matrix mixture was deposited onto the spot containing the first layer. Samples were dried at room temperature and then analyzed by MALDI-TOF MS. Each sample was analyzed in triplicate, loaded once in three different spots (3 spectra per sample). Both SA solutions were prepared and used within 48 h.
Spectra acquisition
The mass spectrum was obtained using the Microflex LT mass spectrometer, with flexControl 3.4 software (Bruker Daltonics). The parameters were configured as follows: linear positive ion mode within the mass range of 17,000 Da to 50,000 Da; spectrometer ion source 1, 20.08 kV; ion source 2, 18.16 kV; lens 6.03 kV; pulsed ion extraction 550 ns; detection gain, 2803 V; sample rate and electronic setting 0.50 GS/s. It was used a laser frequency of 60 Hz and laser power range between 60 and 85%. Each spectrum was obtained after 100 shots per spot.
Data were automatically acquired using autoXecute mode and the spectrum was analyzed using flexAnalysis 3.4 software [11]. Before each run, the spectrometer was externally calibrated using Protein Standard II Calibration Mix (Bruker Daltonics).
Data analysis
KPC peak interpretation was performed searching for a peak closer to the value expected for the enzyme, as determined by Yoon [12] (28,707 to 28,730 Da). Two parameters were taken into consideration: the presence of the KPC peak and its relative intensity of the ions [arbitrary units (a.u.)] after the spectra were baseline subtracted and smoothed. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated considering the results of HRM-PCR [13] as reference. Mann–Whitney test and the area under the curve (AUC) of the ROC curve were also performed in order to confirm that the peak found is KPC. Statistical analyses were performed using SPSS Statistics (PASW Statistics, version 18.0.3).
Results and discussion
Protocol modifications
Initially, we follow the protocol established by Figueroa-Espinosa [11]. After preliminary tests, some modifications became necessary, both in protein extraction and MALDI-TOF parameters, in order to obtain clearer KPC peaks and closer to KPC mass values described elsewhere [11, 12].
For protein extraction, different centrifugation speeds were tested. Figueroa-Espinosa [11] used 17,000 × g; however, we observed that this speed resulted in a very compacted pellet in the microtube bottom which made difficult the extraction, resulting in poor visualization of the peak. A lower speed, 13,000 rpm (16,060 × g), was tested which proved to present better results of peak visualization.
The main parameters adjusted in MALDI-TOF were pulsed ion extraction and laser power. The pulsed ion extraction used by Figueroa-Espinosa [11] was 260 ns., while, more recently, Yoon [12] used 1200 ns. We evaluated the parameters above and an intermediate value of 550 ns. When using a pulsed ion extraction of 260 ns, the peaks were close to 28,400 Da. With 1200 ns, the peaks were close to 28,600 Da and presented some background noise in the mass spectra. Best results were obtained with pulsed ion extraction of 550 ns, from which the KPC-producing isolates showed clear peaks close to 28,700 Da and less background noise.
Regarding laser power, Figueroa-Espinosa [11] used a range of 80–100%. Since a high laser power tends to decrease lifetime of the laser, we tested lower values to improve cost-effectiveness and found a satisfactory laser power of 60–85%. Also, to improve cost-effectiveness of the method and reduce spectra acquisition time in our study, the spectra were obtained after only 100 shots per spot, which proved to be sufficient for clear peak results. Figueroa-Espinosa [11] used 800 shots/spot and Yoon [12] used 400 shots/spot.
The m/z range and intensities of KPC peaks
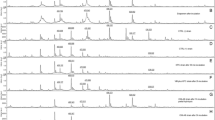
Based on Yoon [12], we searched for KPC peak around 28,718 Da. For analysis, after spectra were baseline subtracted and smoothed, by visual observation, all 157 KPC-producing isolates presented a peak at 28,643–28,731 m/z, with arithmetic mean ± SD of 28,688 ± 20 (95% CI = 28,685–28,691).
Statistical results showed a non-normal distribution by Shapiro–Wilk normality test (statistical significance considered when p ≤ 0.050; p-value < 0.001). Then, Mann–Whitney U test was applied, confirming a significant difference (p ≤ 0.050; p-value < 0.001) for the KPC peak selected in each spectrum when both groups (KPC producers and non-producers) were compared. The area under the curve (AUC) of the ROC curve (Fig. 1) for this specific peak was 0.996. Moreover, the boxplot (Fig. 2) showed the difference of intensities related to the KPC peak of KPC producers and non-producers.
KPC peak intensities ranged from 77 to 3893, with a median value of 380. KPC non-producing isolates, despite not having any peak by visual observation, had intensity ranging from 13 to 217 over the same m/z range, with a median value of 56. The presence and absence of KPC peak can be observed in Fig. 3. Since intensities varied among KPC isolates, we aimed to set a cutoff value to determine whether the isolate is positive or negative for KPC production using ROC curve analysis (Fig. 1).
Table 2 shows different cutoff values evaluated. For our isolates, the intensity cutoff value ≥ 120 for the KPC peak presented high sensitivity (98.09%), specificity (97.90%), and accuracy (98%) results, with 3 false positives and 3 false negatives. Therefore, isolates with intensity ≥ 120 were defined as KPC producers. Yoon [12] decided for an intensity cutoff of 110, having a sensitivity and specificity of 98.1% and 99.5%, respectively. Figueroa-Espinosa [11] did not use the intensity cutoff criteria.
Considering the intensity cutoff ≥ 120, among 157 KPC-producing isolates, we identified 3 false-negative results, all K. pneumoniae, which presented intensities of 77, 111, and 118. The false-positive results included Klebsiella oxytoca (HRM negative; n = 1), Escherichia coli (NDM; n = 1), and Serratia marcescens (HRM negative; n = 1), with intensities of 142, 158, and 217, respectively. All false-positive and false-negative results were repeated and confirmed. Noteworthy, it is important to highlight that even with the KPC-positive isolates presenting the characteristic peak of KPC and with KPC-negative isolates not presenting, as visual interpretation may be subjective, we believe it is important to define a criterion of analysis, such as the intensity of the peak.
Figueroa-Espinosa [11] had no false-positive or negative results, with 100% sensitivity and specificity; however, the intensity cutoff was not applied. Yoon [12] used an intensity cutoff (> 110 for KPC-positive) and detected 2 false-positives (1 Providencia stuartii and 1 Proteus mirabilis) and no false-negative results.
The methodology we tested to detect the KPC presence by MALDI-TOF MS directly from bacterial colonies had better results when compared to other methodologies used for the same purpose. The boronic acid synergy test presents only 72% and 88% of sensitivity and specificity, respectively, for the detection of KPC-producing isolates in addition to other disadvantages such as time-consuming and variability in sensitivity/specificity among different species [9, 14, 15]. Carba-NP and other similar methods presented 100% of specificity but only 84% of sensitivity and it is not able to identify the carbapenemase type [9, 15,16,17]. Another alternative is the carbapenem inactivation method, which presented sensitivity of 97% and specificity of 99%, but it takes around 18 h to get the results after bacterial growth and has variable performance in different studies [9, 15, 16, 18, 19].
More recently, MALDI-TOF MS has been used for the indirect detection of carbapenemases by a method of carbapenem hydrolysis. The latter needs a turn-around-time of up to 4 h after bacterial growth and presents a sensitivity of 77–100% and specificity of 94–100% [9, 14, 20, 21]. Nevertheless, difficult interpretation and doubtful results for enzymes that weakly hydrolyze carbapenems or are weakly expressed limit the use of the carbapenem hydrolysis methodology [15, 16].
In addition to the high sensitivity and specificity, the methodology tested in this study also has a low cost of consumables per sample (< $1.00) [14]. The method is rapid and easy to perform as after the growth of the isolate on solid culture medium, the extraction, target spot loading and spectra acquisition can be performed in less than 20 min per sample, leading to a turnaround time of approximately 1 h for the KPC detection.
Conclusions
The method we evaluated is able to detect KPC-type carbapenemase from colony quickly, efficiently and with low cost, in addition to being easy to be implemented in the routine of clinical microbiology laboratories. Rapid detection of KPC can reduce morbidity and mortality rates, also decreasing clonal and horizontal transfer of carbapenem resistance genes, contributing to decreased antimicrobial resistance.
As other methodologies have been described for direct detection of KPC enzyme [11, 12], our study contributes on stablishing a protocol with high sensitivity and specificity, demonstrating that it must generally be adapted for each equipment and/or laboratory.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not applicable.
References
Villegas MV, Jiménez A, Esparza G, Appel TM (2019) Carbapenemase-producing Enterobacteriaceae: a diagnostic, epidemiological and therapeutic challenge. Infectio 23:388–397. https://doi.org/10.22354/in.v23i4.808
Perez F, Bonomo RA (2019) Carbapenem-resistant Enterobacteriaceae: global action required. Lancet Infect Dis 19:561–562. https://doi.org/10.1016/S1473-3099(19)30210-5
Logan LK, Weinstein RA (2017) The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. https://doi.org/10.1093/infdis/jiw282
Bush K, Bradford PA (2020) Epidemiology of β-lactamase-producing pathogens. Clin Microbiol Rev 33:e00047-e119. https://doi.org/10.1128/CMR.00047-19
Sawa T, Kooguchi K, Moriyama K (2020) Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care 8:13. https://doi.org/10.1186/s40560-020-0429-6
Goff DA, Jankowski C, Tenover FC (2012) Using rapid diagnostic tests to optimize antimicrobial selection in antimicrobial stewardship programs. Pharmacotherapy 32:677–687. https://doi.org/10.1002/j.1875-9114.2012.01137.x
Porreca AM, Sullivan KV, Gallagher JC (2018) The epidemiology, evolution, and treatment of KPC-producing organisms. Curr Infect Dis Rep 20:13. https://doi.org/10.1007/s11908-018-0617-x
Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH (2016) Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. https://doi.org/10.3389/fmicb.2016.00895
European Committee on Antimicrobial Susceptibility Testing (2017) Guidelines for detection of resistance mechanisms and specific resistances o clinical and/or epidemiological importance. Version 2.01. https://eucast.org/. Accessed 23 March 2021
Espinosa RF, Rumi V, Marchisio M, Cejas D, Radice M, Vay C, Barrios R, Gutkind G, Di Conza J (2018) Fast and easy detection of CMY-2 in Escherichia coli by direct MALDI-TOF mass spectrometry. J Microbiol Methods 148:22–28. https://doi.org/10.1016/j.mimet.2018.04.001
Figueroa-Espinosa R, Costa A, Cejas D, Barrios R, Vay C, Radice M, Gutkind G, Di Conza J (2019) MALDI-TOF MS based procedure to detect KPC-2 directly from positive blood culture bottles and colonies. J Microbiol Methods 159:120–127. https://doi.org/10.1016/j.mimet.2019.02.020
Yoon E-J, Lee EH, Hwang DH, Lee H, Baek J-H, Jeong SH (2020) Direct detection of intact Klebsiella pneumoniae carbapenemases produced by Enterobacterales using MALDI-TOF MS. J Antimicrob Chemother 75:1174–1181. https://doi.org/10.1093/jac/dkaa007
Monteiro J, Widen RH, Pignatari ACC, Kubasek C, Silbert S (2012) Rapid detection of carbapenemase genes by multiplex real-time PCR. J Antimicrob Chemother 67:906–909. https://doi.org/10.1093/jac/dkr563
AlTamimi M, AlSalamah A, AlKhulaifi M, AlAjlan H (2017) Comparison of phenotypic and PCR methods for detection of carbapenemases production by Enterobacteriaceae. Saudi J Biol Sci 24:155–161. https://doi.org/10.1016/j.sjbs.2016.07.004
Tamma PD, Opene BNA, Gluck A, Chambers KK, Carroll KC, Simner PJ (2017) Comparison of 11 phenotypic assays for accurate detection of carbapenemase-producing Enterobacteriaceae. J Clin Microbiol 55:1046–1055. https://doi.org/10.1128/JCM.02338-16
Tamma PD, Simner PJ (2018) Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol 56:e01140-e1218. https://doi.org/10.1128/JCM.01140-18
Nordmann P, Poirel L, Dortet L (2012) Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. https://doi.org/10.3201/eid1809.120355
Yamada K, Kashiwa M, Arai K, Nagano N, Saito R (2016) Comparison of the Modified-Hodge test, carba NP test, and carbapenem inactivation method as screening methods for carbapenemase-producing Enterobacteriaceae. J Microbiol Methods 128:48–51. https://doi.org/10.1016/j.mimet.2016.06.019
Tijet N, Patel SN, Melano RG (2016) Detection of carbapenemase activity in Enterobacteriaceae: comparison of the carbapenem inactivation method versus the Carba NP test: Table 1. J Antimicrob Chemother 71:274–276. https://doi.org/10.1093/jac/dkv283
Hrabák J (2015) Detection of Carbapenemases Using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) meropenem hydrolysis assay. Sepsis 1237:91–96. https://doi.org/10.1007/978-1-4939-1776-1_9
Papagiannitsis CC, Študentová V, Izdebski R, Oikonomou O, Pfeifer Y, Petinaki E, Hrabák J (2015) Matrix-assisted laser desorption ionization–time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J Clin Microbiol 53:1731–1735. https://doi.org/10.1128/JCM.03094-14
Acknowledgements
The authors thank the collaboration of the funding from Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) and Fundo de Incentivo a Pesquisa e Eventos from Hospital de Clínicas de Porto Alegre (FIPE/HCPA).
Funding
This work was supported by Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul, Fundo de Incentivo à Pesquisa e Eventos do Hospital de Clínicas de Porto Alegre and a master’s scholarship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Natália Kehl Moreira, Camila Mörshbächer Wilhelm, and Priscila Lamb Wink. The first draft of the manuscript was written by Natália Kehl Moreira and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This study was approved by Hospital de Clínicas de Porto Alegre and Universidade Federal do Rio Grande do Sul (HCPA/UFRGS) ethical committee, under CAAE number 31638920800005327.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Mara Correa Lelles Nogueira
Rights and permissions
About this article
Cite this article
Moreira, N.K., Wilhelm, C.M., Wink, P.L. et al. MALDI-TOF mass spectrometry for direct KPC detection among Enterobacterales. Braz J Microbiol 53, 1907–1913 (2022). https://doi.org/10.1007/s42770-022-00798-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-022-00798-y