Abstract
Purpose
Glaucoma damages the optic nerve and causes permanent visual impairment. It cannot be recovered, so it is important to distinguish the disease over time. Fortunately, this is usually a state of progress, and if picked early, it is likely to be treated effectively. Early detection is the key to success preventing visual impairment. In this article, we will talk about how to diagnose glaucoma by means of two main steps: segmentation and classification using convolutional neural networks (CNN).
Methods
First, we present different implementations of segmentation and classification on a database that contains a set of fundus data. This article proposes supervised and unsupervised methods for segmentation, detection, and classification of glaucoma in fundus images. A supervised method using U-net and Modified U-net (M-net) is first formed to segment and detect the optical disc and cup disc regions in fundus images. Then classing them in the next step. We are training proposed networks by using a graphics processor and public datasets. The unsupervised method based on K-means algorithm for optic and cup disc segmentation, then calculates CDR for classification to glaucoma or not.
Results
Our proposed M-net achieves an accuracy of 99.89% and unsupervised methods achieve an accuracy of 98.17%.
Conclusion
With the highlights of recent advancements in deep learning-based approaches for glaucoma diagnosis. These techniques have shown promising improvement in detection performance.The results obtained have shown a significant improvement in the diagnostic performance of glaucoma abnormality.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
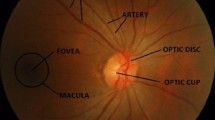
Glaucoma is one of the most serious eye infections. As shown by the number of visual disturbances worldwide. It is the second-most important eye disease (Elmoufidi et al. , 2023). In this sense, the real task of the ophthalmologist is to educate himself early, observe the patient for a long time, and choose the right treatment at the right time (Elmoufidi et al. , 2022a). In short, glaucoma is a never-ending eye disease in which the optic nerve is constantly damaged and gradually begins to cause vision loss. At first, there is no torture, and patients usually have an indication. After a while, glaucoma begins to affect your side/peripheral vision and gradually moves to the cente when not detected. As the World Health Organization (WHO) has said, glaucoma is the cause of poor vision. In total, this has resulted in around 5.2 million cases of visual impairment (accounting for 15% of reported cases of absolute visual impairment) and could affect around 80 million people over the next ten years. So far, there is no cure for glaucoma. Glaucoma is an irreversible common eye disease and is the second clarification of movement after visual impairment. Since there is no qualified early detection facility, it can alert vigilantly in the last days of glaucoma (Almazroa et al. , 2015).In fact, around 80 million individuals have glaucoma worldwide (Tsai et al. , 2019). This number is likely to increase to 111.8 million individuals in 2040 in the world. Unfortunately, at least, half of individuals affected by glaucoma are unaware that they are diseased. In some developing countries, 90% of glaucoma is not detected and this number may even be higher in underdeveloped countries. The difficulty comes from that, glaucoma is an asymptomatic disease and detecting it in its early stages is difficult. If glaucoma is undetected and untreated in early stages it may progress to blindness. Early examination and treatment are essential to prevent vision loss in glaucoma patients. By checking the condition of the retinal fundus image, it can be considered appropriate in time for glaucoma. In this article, we are using convolutional neural networks to assess changes in the retina from a database. When we do image recognition or prediction by deep learning, the most important thing is to have a large database, i.e., to have as many images as possible. Able to provide a maximum of images to the model of a convolutional neural network that one wishes to train, but care must be taken to provide it with a coherent and useful database. When we create a convolutional neural network model or only when we use an existing model, we need to train the model with data so that it can learn and recognize specific objects or things. For this, we use a database that contains all the images. Part of this database will be used to train the CNN model, called the “training database.” The rest of the database is used to evaluate the model and is called the “validation database.” It is important to know that all the images we use must be pre-tagged, i.e., every image we use must belong to a specific class. Training a CNN is to determine and empirically calculate the value of each of its weights. The principle is as follows: CNN processes an image (from the training database) and makes a prediction on the output, that is, to which class it thinks the image belongs. Knowing that we know in advance the class of each of the training images, we can check if the result is correct. Based on the accuracy of this result, all CNN weights are updated using an algorithm called error gradient backpropagation. During the model training phase, the process explained above is repeated several times and with all the images in the training database. The objective is to classify these data as well as possible. When the model has finished updating the weight, it will be evaluated by providing it with a validation database. It classifies all these images (images that the model has never seen before), then calculates its rate of good classification, this is called the model accuracy. At this point, we develop an illustration in which we recognize GLAUCOMA in the starting periods using convolutional neural networks (CNN) using fundus images. These images are being prepared in the perspective of deep learning. A significant learning structure is proposed, recalling the ultimate objective of obtaining a different leveled representation of the images to be isolated among the GLAUCOMA and NON-GLAUCOMA contours. The detection of this disease is based on the segmentation of the cup and the optic disc. The segmentation of retinal images, in particular the fundus, is an important step in the medical follow-up of glaucoma. In this work a segmentation method that is based on unsupervised classification. The K-means are for the cup regions, while, for the optical discs, we have chosen a method of mathematical morphology to measure the ratio between the cup and the disc (CDR) and between the rim and the disc (RDR) as well as, the difference between the upper and lower distances of the rim, and a method based on convolutional neural networks.
In recent years, many studies have been carried out to improve computerized decision-making, such as breast cancer (Elmoufidi et al. , 2014), (Elmoufidi et al. , 2018), (Elmoufidi , 2019), (Elmoufidi et al. , 2022b), brain cancer (Tang et al. , 2019), diabetic retinopathies (Hossi et al. , 2021), and (Skouta et al. , 2022). Algorithms (detection and classification of glaucoma) for extracting optical discs and wells to calculate CDR ratios, reports RDR, the difference between the upper width and the lower and the sum between the latter and the CDR. The first relevant implementations of CAD systems for the detection of glaucoma with fundus images began to appear in 2008 (Hagiwara et al. , 2018).
Majhi and Nayak (2022) proposed a feature modulating two-stream deep convolutional neural network (CNN). The network accepts the full fundus image and the region of interest (ROI) outlined by clinicians as input to each stream to capture more detailed visual features. A feature modulation technique is also proposed as an intrinsic module in the proposed network to further enrich the feature representation by computing the feature-level correlation between two streams. The proposed model is evaluated on a recently introduced large-scale glaucoma dataset, namely G1020 and has achieved state-of-the-art glaucoma detection performance.
Chaudhary and Pachori (2021) are published a paper intituled “Automatic diagnosis of glaucoma using two-dimensional Fourier-Bessel series expansion based empirical wavelet” in this paper, for better segmentation of image spectra in EWT, the Fourier-Bessel series expansion (FBSE)-based spectrum is used instead of Fourier transform (FT)-based spectrum. Because FT-based spectrum segmentation provides poor performance when frequency components are closely spaced. FBSE provides better spectrum representation than FT, so by using FBSE in place of FT in EWT, one can get better boundary segmentation.
Nayak et al. (2021) proposed a novel non-handcrafted feature extraction method termed as evolutionary convolutional network (ECNet) for automated detection of glaucoma from fundus images. The ECNet is trained using a criteria that maximizes the inter-class distance and minimizes the intra-class variance of different classes. The final feature vectors are then subjected to a set of classifiers such as K-nearest neighbor (KNN), backpropagation neural network (BPNN), support vector machine (SVM), extreme learning machine (ELM), and kernel ELM (K-ELM) to select optimum performing model. The experimental results on a dataset of 1426 fundus images (589 normal and 837 glaucoma) demonstrate that the ECNet model with SVM yielded the highest accuracy of 97.20% compared to state-of-the-art techniques.
Ganesh et al. (2021) developed a LayerCAM construction for RoI segmentation based on multiple CAMs. Gradient CAMs (GCAM) are used to analyze the behavior of the binary classification process. This analysis is performed by capturing the GCAM of the convolutional layer in the classification branch of the GD-YNet. The GCAMs are used for the classification of normal and glaucomatous images in the Drishti-gs dataset.
Raghavendra et al. (2019) presented a CAD tool for the precise detection of glaucoma using a machine learning approach. An auto-encoder is trained to determine effective and important features of the code.fundus images. These features are used to develop classes of glaucoma for testing. The method achieved an F-measure value of 0.95 using 1426 digital fundus images (589 control and 837 glaucoma).
In 2016, A. Singh et al. (Singh et al. , 2016) reached a precision of 0.947. They started by identifying the center of the disc, and then proceeded to segment the disc. The intuition that blood vessels represent noisy pixels that affect system performance led to the removal of these vessels from the resulting optical disc images. After this step, the feature extraction was performed using a discrete wavelet decomposition, resulting in a feature vector of 18 features. For the selection of characteristics, two approaches were tried, genetic algorithms and principal component analysis (PCA). The final vectors of each technique were then tested with several classifiers (support vector machine (SVM), K-nearest neighbors (KNN), random forest, Naive Bayes, and artificial neural networks (ANN). Since the fundus images are from a local 63-image dataset, which is rather small, no-indent cross-validation was also performed to account for overfitting issues. The best performing models were the SVM and the KNN, with the selection of PCA features, which resulted in only two components. These two classifiers obtained an accuracy of 0.947. More recently, deep learning approaches have proven capable of outperforming existing techniques and are widely applied.
In 2014, Hafsah Ahmad et al. (Ahmad et al. , 2014) proposed a system for the early detection of glaucoma using CDR and NRR ratio in ISNT quadrants. The strategy is executed in 80 frames and an accuracy of 97.5% in 0.8141 s.
In 2013: Fauzia Khan et al. (Khan et al. , 2013) used morphological techniques to extract two NRR surfaces. The proposed method achieves an average precision of 94%, with an average computational cost of 1.42 s. In 2009: Nayak et al. (Anusorn et al. , 2013) extracted three intrapapillary characteristics for the automated diagnosis of glaucoma, which were the CDR, the size of the optic disc and the ISNT ratio. The red and green planes segmented the optic disc and optic cup, respectively, after the blood vessels by morphological operations. These three characteristics were used to classify normal and glaucomatous images using a neural network classifier, achieving sensitivity and specificity of 100% and 80%, respectively.
These approaches. However, do not target all types of glaucoma and despite the better results recently obtained with deep learning techniques; these solutions have little or no concerns about the interpretation of the decision.
Materiels and methods
Dataset
DRISHTI-GS
In this article, we present a DRISHTI-GS1 database consisting of 50 training images and 51 test images. For each image, manual segmentations for OD and cup region from four different human experts with varying clinical experience (Sivaswamy et al. , 2014). The markings were collected with a dedicated marking tool that provides a fully deformable circle to help capture different shapes of the outside diameter and cup, as well as any localized change in shape, such as the notch. DRISHTI-GS1 is an extension of DRISHTI-GS, a dataset recently released by us for the OD and cup segmentation algorithms. In addition to the structural markings provided in (Sivaswamy et al. , 2014), two other expert opinions are included. These are decisions on:
-
The image representing a normal or glaucomatous eye
-
Presence or absence of notch in the lower and / or upper sectors of the image.
Since the dataset aims to compare different segmentation algorithms, the results of the OD and cup segmentation methods (Joshi et al. , 2011a), (Joshi et al. , 2012), (Chakravarty and Sivaswamy , 2014) tested on this dataset are also provided. A new supervised method of notch detection based on OD and cup segmentations is proposed. The method was evaluated on the dataset and the results are presented.
RIM-ONE
An online database with images of the retinal fundus has been developed to be a benchmark for designing optical nerve head segmentation algorithms. The main differences between all other databases and the presented RIM-ONE are as follows: RIM-ONE focuses exclusively on ONH segmentation; it has a relatively large amount of high-resolution images (Lowell et al. , 2004), (Fumero et al. , 2011a), (Doyle , 2007), and manual reference segmentations of each (Fumero et al. , 2011b). This allows the creation of reliable reference standards, thereby reducing the variability between expert segmentations, and the development of very precise segmentation algorithms. This database is available online at rimone.isaatc.ull.es and can be used for research and teaching purposes, without asking permission from the authors. It is a place of reference for all those who wish to use its images with their own optical disc detection algorithms.
REFUGE
The REFUGE (Orlando et al. , 2020) challenge database consists of 1200 retinal CFPs stored in JPEG format. With 8 bits per color channel, acquired by ophthalmologists or technicians from patients sitting upright and using one of two devices: a Zeiss Visucam 500 fundus camera with a resolution of 2124 x 2056 pixels (400 images) and a Canon CR-2 device with a resolution of 1634 x 1634 pixels (800 images). Images have been centered on the posterior pole, with both the macula and the optic disc visible, to allow the assessment of the ONH and potential defects in the retinal nerve fiber layer (RNFL). These pictures correspond to Chinese patients (52% and 55% female in offline and online test sets, respectively) visiting eye clinics, and were retrieved retrospectively from multiple sources, including several hospitals and clinical studies. Only high-quality images were selected to ensure proper labeling and any personal and/or device information was removed for anonymization.
RIM-ONE DL
The Retinal IMage database for Optic Nerve Evaluation Deep Learning (RIM-ONE DL) (Fumero et al. , 2020) image dataset was captured by cooperation between three Spanish hospitals, which are Hospital Universitario de Canarias (HUC), in Tenerife, Hospital Universitario Miguel Servet (HUMS), in Zaragoza, and Hospital Clínico Universitario San Carlos (HCSC), in Madrid. This database contains 495 images, consisting of 313 retinographics of normal subjects and 172 retinographics from patients with glaucoma. This dataset has been divided into two subset, one for training and the second for test sets, with two variants:
In most cases, the disease remains asymptomatic for several years and can only be diagnosed with an eye exam. In this chapter, we present some methods for the detection of this disease. We are talking here about the unsupervised method (ex: clustering, K-means...), and the supervised method (using convolutional neural networks), concerning the segmentation phase, concerning the classification, we propose a method which depends on CNN to detect glaucoma from fundus images and classify whether these images are glaucomatous or not.
Unsupervised segmentation
The unsupervised context is where the algorithm must operate from unannotated examples; it must automatically display categories to be correlated with the data submitted to it in order to recognize cats such as cats, and cars. The most common unsupervised learning problem is segmentation (or clustering). In this case, we are trying to divide the data into several categories (category, class, class, etc.): group images of cars, cats, etc.
People hope to use anomaly detection for predictive maintenance, network security, and early disease detection.
In this part, the method has five main functions:
-
Extract images and CDR values from each image folder in the Drishti database.
-
Segmentation of the slice and the disc region from each of the fundus images.
-
Calculates CDR values from these segmentations.
-
Main function where the segmented images are passed to these functions and the corresponding CDR values calculated and stored in a csv file.
-
A last function that displays a graph of the images grouped according to the CDR values obtained.
In general, this part presents an image processing technique for optical disc and well segmentation based on adaptive thresholding using image characteristics. The proposed algorithm uses the characteristics obtained from the image, such as the mean and standard deviation, to remove the red and green channel information from a background image and obtain an image that contains only the region of the head of the optic nerve in both channels. The optical disc is segmented from the red channel and the optical cup from the green channel, respectively. The threshold is determined from the smoothed histogram of the preprocessed image. The precision of the algorithm is good and very fast in terms of computation (Fig. 1).
In order to segment the optic nerve area, the fundus image must be preprocessed. In the preprocessing step, local statistical characteristics (such as the mean and standard deviation) are used and they are iteratively subtracted from the image. This pretreatment step only highlights the area of the optic nerve in the fundus image. Pretreatment is applied to the red channel and the green channel to seed the disc and the cup, respectively. The pre-processed images are shown in the figure below (Fig. 2).
The pre-processed histogram is used to find the disc and cup segmentation threshold. The intensity of the grey levels is indicated on the horizontal axis, and the number of pixels with a grey level is indicated on the vertical axis. The different peaks of the histogram show different parts of the inner eye. The first peaks are considered the peaks of the background pixels because of their low grayscale intensity, whereas the last peaks are considered the peaks of the disc and cup because they have pixels at levels of grey with high brightness. The histogram of the preprocessed image is convolved with a Gaussian filter for smoothing, as shown in the following figure (Figs. 3, 4, 5, and 6):
Result: The following figure is an example of the program output, which shows the different CDR values for each frame out of three experiments done on each.
The following graph represents the images.
The following graph represents the images classified according to its CDR values.
Supervised segmentation
In the case of supervised learning, this type depends on artificial intelligence; it functions as a researcher is there to guide the algorithm on the path of learning by providing it with examples that it considers convincing after having them. Previously labeled with expected results.
In our topic, the examination of the optic nerve head involves measuring the cup-to-disc ratio, which is considered one of the most valuable methods for the structural diagnosis of the disease. Estimation of the cup/disc ratio requires segmentation of the optic disc and the optic cup on fundus images. Then can be performed by convolutional neural network algorithms.
This work presents a general method of automatic segmentation of discs and cups based on deep learning, namely U-net and M-net.
M-net
For this deep learning architecture, named M-net, can solve the problem of OD and OC segmentation in multistage systems in one-step? The proposed M-net is mainly composed of a multi-scale input layer, a U-shaped convolutional network, a side output layer and multiple label loss functions (Figs. 7 and 8).
The U-shaped convolutional network is used as the main body network structure to learn the hierarchical representation, while the side output layer acts as an early classifier, which produces an associated local prediction map for different scale layers. Finally, a multi-label loss function is proposed to generate the final segmentation map (Fig. 9).
M-net is an efficient fully convolutional neural network for biomedical image segmentation. Similar to the original U-net architecture, our M-net is an end-to-end multi-label deep network, which consists of three main parts.
-
The first is disc detection for fundus images.
-
Model training
-
Database data test
The model is made with a set of layers, the first is a U-shaped convolutional network, which is used as the main structure for learning rich level representations, which and linked with Max pooling layers which have size filters (2, 2), an element rectified nonlinearity (ReLU) activation function is used.
Then, the large-dimension feature representation at the output of the final decoder layer is applied to a trainable classifier. In our method, the final classifier uses the 1*1 convolutional layer with a sigmoid activation function as a pixel-by-pixel classification to produce the probability map. For multilabel segmentation, the output is a probability map, where K is the class number (K = two for OD and OC in our case). The predicted probability map corresponds to the class with a maximum probability at each pixel. Our M-net is implemented with Python based on Keras with Backend TensorFlow. When the model was tested on RIM-ONE data sets, the model achieved an average value of 98.9% for disc segmentation and cup segmentation (Table 1).
In practice, the source code of M-net works very well, it is divided into three parts:
-
Disc detection for fundus images
-
Model training
-
Database data test
The following table summarizes the results obtained on the database.
U-net
Among the proposed models is an architecture called U-net, which has been widely used in biomedical image segmentation and has obtained good results. The U-net architecture is built using a fully convolutional network and aims to provide better segmentation results in medical imaging competitive with other models based on deep learning.
It was first designed by Olaf Ronneberger, Philipp Fischer, and Thomas Brox in 2015 to process biomedical images. It is a neural network for image segmentation. U-net is presented as a fully convolutional neural network that can train datasets; it accepts images as input and returns a probability map as output. It has a “U” shape. The U-net architecture is symmetrical and its operation is similar to an automatic encoder to some extent. It can be simplified into three main parts: the contraction path (down sampling), the bottleneck, and the expansion path (oversampling).
U-net is presented as a fully convolutional neural network that can train datasets; it accepts images as input and returns a probability map as output. It has a “U” shape. The U-net architecture is symmetrical and its operation is similar to an automatic encoder to some extent. It can be simplified into three main parts: the contraction path (down sampling), bottleneck path, and expansion path (oversampling). This model consists of 10 layers of convolution with an activation function ReLU and a parameter padding = “same” and four layers of Max Pooling which contains a filter of size (2, 2). The model also includes the Adam optimizer, which plays the role of the algorithm corrector so that the precision value is high (Fig. 10 and Table 2).
Result:
When the model was tested on DRISHTI-GS datasets, the model had an average precision value of 94.87% for disc segmentation and cup segmentation (Figs. 11 and 12).
The following table summarizes the rest of the U-net model results:
Evaluation metric
The performance evaluation of a model can be carried out by assessing statistical laws; the objective is to obtain the most accurate estimate possible of the behavior of the classifier under real conditions of use. Additionally, evaluation metrics, such as specificity and sensitivity, provide useful information. Recall sensitivity represents the probability of correctly classifying a class, and specificity is an indirect measure of the probability of a false alarm equal to 1-Recall, referring to the ratio of the number of correctly predicted negative samples to the total number of true negative samples, i.e., how many negative samples can I correctly find from these samples. Also, we have used the metrics for the evaluation of the segmentation model were the intersection over union (IoU) and the dice coefficient (F1 score). The IoU metric measures the accuracy of an object detector applied to a particular database. It measures the common area between the predicted (P) and expected (E) regions, divided by the total area of the two regions, as presented in Eq. 1:
We also note the precision indicator:
Or TP = number of true positives, FP = number of false positives and FN = number of false negative pixels. Then the F score, which is the harmonic average of precision and recall. It is defined as:
This is also known as the F1-score because sensitivity and precision are evenly weighted. The maximum possible value of the F score is 1. Finally, the accuracy indicator (accuracy) refers to the ratio between the number of correctly predicted samples and the total number of predicted samples. It does not consider whether the predicted samples are positive or negative.
Results
In this article, our analysis examined segmentation with different supervised (U-net and M-net) and unsupervised (adaptive thresholding) methods, which present different results. In this part, we present some results obtained by comparing them with old results. 1. Duration: The entire training phase of our M-net and U-net segmentation method takes about 3 h in the Spider environment (100 epochs). However, the training phase could be done offline, in the online phase it may take less time. Regarding the test step to generate the image mask, it only costs 5 s, and 7 s for the U-net method. on the other hand, the super pixel method (Cheng et al. , 2013) takes 10 s, the ASM method (Yin et al. , 2011) takes 4 s, R-Bend Method (Joshi et al. , 2011b) takes 4 s, sequential segmentation of OD and OC using original U-net(Ronneberger et al. , 2015) takes 1 s. 2. Accuracy: The precision value plays the most important role in this work; it refers to the performance of each model, whether it is U-net or M-net. Regarding U-net, this model was able to achieve 97.42% accuracy, and 99.89% for M-net.% in contrast, the sequential segmentation of OD and OC using the original U-net reached 95.3%.
Conclusion
The objective of this article is to present the different steps in the diagnosis of glaucoma disease. First, we present a segmentation method based on a convolutional neural network. For this, we used two models of different architectures (U-net, M-net). We then showed the different results obtained in terms of precision and error. Comparison of results shows that the number of epochs, the size of the bases and the depth of the network are important factors in obtaining better results; similarly, in the classification stage, our CNN-based proposal for glaucoma detection has achieved a maximum precision of 99% that detects glaucomatous and non-glaucomatous retinal images. Seeing the results, we clearly synthesize that the studied system is better compared to other approaches. Although the recommended methods achieve maximum performance, they present certain problems, such as sensitivity to noise, and require large data sets to analyze the detection of glaucoma. The supervised method has high precision, but requires a huge labeled database, which is practically difficult to collect, expensive, and slow to obtain. In addition, they cannot use unlabelled data. Nonetheless, the unsupervised segmentation cited in this review appears to be effective in improving segmentation results despite having expert intervention. At this point, there are many possibilities to extend an idea of the optimal detection method for glaucoma to other segmentation issues that will be incorporated into future research. In this research, there are many possibilities to extend the idea of the optimal detection method for glaucoma to other problems of integrated segmentation.
Data availability
Not applicable.
Code Availability
Not applicable.
References
Elmoufidi A, Skouta A, Jai-Andaloussi S, Ouchetto O. Cnn with multiple inputs for automatic glaucoma assessment using fundus images. Int J Image Graph. 2023;23(01):2350012.
Elmoufidi A, Skouta A, Jai-andaloussi S, Ouchetto O. Deep multiple instance learning for automatic glaucoma prevention and autoannotation using color fundus photography. Prog Artif Intell. 2022;11(4):397–409.
Almazroa A, Burman R, Raahemifar K, Lakshminarayanan V. Optic disc and optic cup segmentation methodologies for glaucoma image detection: a survey. Journal of ophthalmology.2015;2015.
Tsai T, Reinehr S, Maliha AM, Joachim SC. Immune mediated degeneration and possible protection in glaucoma. Frontiers in neuroscience.2019;13:931.
Elmoufidi A, Fahssi KE, Jai-Andaloussi S, Madrane N, Sekkaki A. Detection of regions of interest’s in mammograms by using local binary pattern, dynamic k-means algorithm and gray level cooccurrence matrix. In 2014 International Conference on Next Generation Networks and Services (NGNS), pages 118–123. IEEE, 2014.
Elmoufidi A, Fahssi KE, Jai-Andaloussi S, Sekkaki A, Gwenole Q, Lamard M. Anomaly classification in digital mammography based on multiple-instance learning. IET Image Processing. 2018;12(3):320–8.
Elmoufidi A. "pre-processing algorithms on digital x-ray mammograms". In 2019 IEEE International Smart Cities Conference (ISC2), pages 87–92. IEEE, 2019.
Elmoufidi A, Skouta A, Jai-andaloussi S, Ouchetto O. Deep multiple instance learning for automatic glaucoma prevention and autoannotation using color fundus photography. Progress in Artificial Intelligence, pages 1–13.
Tang W, Fan W, Lau J, Deng L, Shen Z, Chen X. Emerging blood-brain-barrier-crossing nanotechnology for brain cancer theranostics. Chemical Society Reviews. 2019;48(11):2967–3014.
Hossi AE, Skouta A, Elmoufidi A, Nachaoui M. Applied cnn for automatic diabetic retinopathy assessment using fundus images. In International Conference on Business Intelligence, pages 425–433. Springer, 2021.
Skouta A, Elmoufidi A, Jai-Andaloussi S, Ouchetto O. Hemorrhage semantic segmentation in fundus images for the diagnosis of diabetic retinopathy by using a convolutional neural network. J Big Data. 2022;9(1):1–24.
Hagiwara Y, Koh JEW, Tan JH, Bhandary SV, Laude A, Ciaccio EJ, Tong L, Acharya UR. Computer-aided diagnosis of glaucoma using fundus images: a review. Com- puter methods and programs in biomedicine.2018;165:1–12.
Majhi S, Nayak DR. Feature modulating two-stream deep convolutional neural network for glaucoma detection in fundus images. In International Conference on Computer Vision and Image Processing, pages 171–180. Springer, 2022.
Chaudhary PK, Pachori RB. Automatic diagnosis of glaucoma using two-dimensional fourier-bessel series expansion based empirical wavelet transform. Biomed Signal Process Control. 2021;64:102237.
Nayak DR, Das D, Majhi B, Bhandary SV, Acharya UR. Ecnet: an evolutionary convolutional network for automated glaucoma detection using fundus images. Biomedical Signal Processing and Control. 2021;67.
Ganesh SS, Kannayeram G, Karthick A, Muhibbullah M. A novel context aware joint segmentation and classification framework for glaucoma detection. Computational and Mathematical Methods in Medicine. 2021.
Raghavendra U, Gudigar A, Bhandary SV, Rao TN, Ciaccio EJ, Acharya UR. A two layer sparse autoencoder for glaucoma identification with fundus images. Journal of medical systems.2019;43(9):1–9.
Singh A, Dutta MK, ParthaSarathi M, Uher V, Burget R. Image processing based automatic diagnosis of glaucoma using wavelet features of segmented optic disc from fundus image. Computer methods and programs in biomedicine.2016;124:108–120.
Ahmad H, Yamin A, Shakeel A, Gillani SO, Ansari U. Detection of glaucoma using retinal fundus images. In 2014 Interna- tional conference on robotics and emerging allied technologies in engineering (iCREATE), pages 321–324. IEEE.
Khan F, Khan SA, Yasin UU, Haq Iu, Qamar U. Detection of glaucoma using retinal fundus images. In The 6th 2013 Biomedical Engineering International Conference, pages 1–5. IEEE.
Anusorn CB, Kongprawechnon W, Kondo T, Sintuwong S, Tungpimolrut K. Image processing techniques for glaucoma detection using the cup-to-disc ratio. Science & Technology Asia, pages 22–34.
Sivaswamy J, Krishnadas SR, Joshi GD, Jain M, Tabish AUS. Drishti-gs: retinal image dataset for optic nerve head (onh) segmentation. In 2014 IEEE 11th international symposium on biomedical imaging (ISBI), pages 53–56. IEEE.
Joshi GD, Sivaswamy J, Krishnadas SR. Optic disk and cup segmentation from monocular color retinal images for glaucoma assessment. IEEE transactions on medical imaging.2011a;30(6):1192–1205.
Joshi GD, Sivaswamy J, Krishnadas SR. Depth discontinuity-based cup segmentation from multiview color retinal images. IEEE Transactions on Biomedical Engineering. 2012;59(6):1523–31.
Chakravarty A, Sivaswamy J. Coupled sparse dictionary for depth-based cup segmentation from single color fundus image. In Interna- tional Conference on Medical Image Computing and Computer-Assisted In- tervention, pages 747–754. Springer.
Lowell J, Hunter A, Steel D, Basu A, Ryder R, Fletcher E, Kennedy L. Optic nerve head segmentation. IEEE Transactions on medical Imaging. 2004;23(2):256–64.
Fumero F, Alayón S, Sanchez JL, Sigut J, Gonzalez-Hernandez M. Rim-one: an open retinal image database for optic nerve evaluation. In 2011 24th international symposium on computer-based medical systems (CBMS), pages 1–6. IEEE.
Doyle DJ. Health education assets library www.healcentral.org.
Fumero F, Alayón S, Sanchez JL, Sigut J, Gonzalez-Hernandez M. Rim-one: an open retinal image database for optic nerve evaluation. In 2011 24th international symposium on computer-based medical systems (CBMS), pages 1–6. IEEE.
Fumero F, Diaz-Aleman T, Sigut J, Alayón S, Arnay R, Angel-Pereira D. RIM-ONE DL: A Unified Retinal Image Database for Assessing Glaucoma Using Deep Learning. BImage Analysis and Stereology. 2020. p. 39 https://doi.org/10.5566/ias.2346.
Orlando JI, Fu H, Breda JB, Keer Kv, Bathula DR, Diaz-Pinto A, Fang R, Heng PA, Kim J, Lee J, et al. "Refuge challenge: a unified framework for evaluating automated methods for glaucoma assessment from fundus photographs". Medical image analysis, 59:101570.
Cheng J, Liu J, Xu Y, Yin F, Wong DWK, Tan NM, Tao D, Cheng CY, Aung T, Wong TY. Superpixel classification based optic disc and optic cup segmentation for glaucoma screening. IEEE Transactions on Medical Imaging. 2013;32(6):1019–32.
Yin F, Liu J, Ong SH, Sun Y, Wong DWK, Tan NM, Cheung C, Baskaran M, Aung T, Wong TY. Model-based optic nerve head segmentation on retinal fundus images. In 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, pages 2626–2629. IEEE.
Joshi GD, Sivaswamy J, Krishnadas SR. Optic disk and cup segmentation from monocular color retinal images for glaucoma assessment. IEEE transactions on medical imaging.2011b;30(6):1192–1205.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention, pages 234–241. Springer.
Hervella ÁS, Rouco J, Novo J, Ortega M. End-to-end multi-task learning for simultaneous optic disc and cup segmentation and glaucoma classification in eye fundus images. Applied Soft Computing. 2022;116: 108347.
Zhou W, Ji J, Jiang Y, Wang J, Qi Q, Yi Y. Eards: efficientnet and attention-based residual depth-wise separable convolution for joint od and oc segmentation. Frontiers in Neuroscience. 2023;17:1139181.
Almubarak H, Bazi Y, Alajlan N. Two-stage mask-rcnn approach for detecting and segmenting the optic nerve head, optic disc, and optic cup in fundus images. Applied Sciences. 2020;10(11):3833.
Tadisetty S, Chodavarapu R, Jin R, Clements RJ, Yu M. Identifying the edges of the optic cup and the optic disc in glaucoma patients by segmentation. Sensors. 2023;23(10):4668.
Liu B, Pan D, Shuai Z, Song H. Ecsd-net: a joint optic disc and cup segmentation and glaucoma classification network based on unsupervised domain adaptation. Computer Methods and Programs in Biomedicine. 2022;213.
Wang J, Li X, Cheng Y. Towards an extended efficientnetbased u-net framework for joint optic disc and cup segmentation in the fundus image. Biomedical Signal Processing and Control. 2023;85: 104906.
Sun G, Zhang Z, Zhang J, Zhu M, Zhu Xr, Yang JK, Li Yu. Joint optic disc and cup segmentation based on multi-scale feature analysis and attention pyramid architecture for glaucoma screening. Neural Computing and Applications.2021;1–14.
Gómez-Valverde JJ, Antón A, Fatti G, Liefers B, Herranz A, Santos A, Sánchez CI, Ledesma-Carbayo MJ. Automatic glaucoma classification using color fundus images based on convolutional neural networks and transfer learning. Biomedical optics express.2019;10(2):892–913.
Sreng S, Maneerat N, Hamamoto K, Win KY."Deep learning for optic disc segmentation and glaucoma diagnosis on retinal images". Applied Sciences.2020;10(14):4916.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Elmoufidi, A., Hossi, A.E. & Nachaoui, M. Machine learning for glaucoma detection using fundus images. Res. Biomed. Eng. 39, 819–831 (2023). https://doi.org/10.1007/s42600-023-00305-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42600-023-00305-8