Abstract
Glaucoma is the most significant cause of irreversible vision loss and the second biggest cause of blindness globally. The first signs of the disease will only appear in an advanced stage when there is no more recovery. Early diagnosis is of the utmost importance and is currently performed primarily through fundoscopy. This fundus image exam is a tedious and manual process, prone to human errors that can result in false negatives, which could promote vision loss. Deep learning approaches are being used to detect glaucoma directly from eyes fundus images, achieving promising results. However, there is no apparent interest in deploying these systems in medical clinics, which would require lightweight models with minimal false negatives and comprehensive outputs. The present study explores these gaps and proposes an architecture composed of one segmentation network for disc and cup and one classification network for direct glaucoma classification. Our main contribution is optimizing a glaucoma-aiding system by increasing model sensitivity by 3% and simplifying the architecture. Unlike related works, we present a lighter model that shows valuable information to the physician, building their trust in the system.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glaucoma is an ocular disease responsible for most global irreversible vision loss and the second biggest cause of blindness [8]. Early diagnosis is of utmost importance to prevent vision loss. The degeneration of the optic nerve is the primary pathology of glaucoma, usually preceded by an increase in intraocular pressure [9]. This increase causes a common enlargement on the angle of the optic disc cup, erosion of the rim tissue, and, as a result, visual field damage. Evaluation of the optic nerve head through digital fundoscopy is one of the most feasible exams because of its less invasive approach and capability of generating high-resolution images of the back of the eye [4].
However, fundoscopy exam is manual, tedious, complex, and prone to human errors. As a result, this can lead to irreversible vision loss, a consequence of false-negative prognosis done by tired or inexperienced physicians [1]. To address these issues, deep learning models are being explored to classify glaucoma from fundus images, achieving promising results [2, 6, 15]. However, only a few studies discuss the possibility of deploying their systems in the medical clinic. In these cases, expert trust in the system is required and can be increased by showing more than just the final prediction, such as the segmentation of the eye structures and the system’s confidence level on the prognostic. Also, lighter models must be adopted to make this possible while aiming to increase sensitivity more than just accuracy.
The main goal of this study is to optimize the state-of-the-art models used on the diagnosis aiding tool created by Civit-Masot et al. [4]. Their system is an ophthalmic aid tool that uses deep learning and fundus images to classify and segment the image directly. It provides robust information to support the final diagnosis, in addition to using modules light enough to be installed in embedded systems of low storage and processing power. We choose this study as the basis for this research.
The base model architecture consists of a direct classification network based on MobileNetV2 [13], and two modified U-net [11] segmentation networks, for the disc and cup segmentation. This study adds an ensemble of MobileNetV2 and EfficientNet for the direct classification network and a unified segmentation network for the disc and cup features. By adding a second classification network, we increased sensitivity by 3% compared to the base model. We achieved comparable results with the unified segmentation network while simplifying the architecture and reducing processing time by 36.35%. Therefore, our contribution is made by exploring the gaps in the literature left in evidence by the authors, namely:
-
Increasing the sensitivity of the model, further reducing the number of false negatives and thus ensuring even more value for its use in the clinic;
-
Architecture simplification, taking up less storage space and using less processing resources, without losing accuracy, having as an only consequence the need for more initial training time.
The remainder of the paper is organized as follows. In Sect. 2 we give a review of related works. Section 3 introduces the proposed model and the implementation process. Then, Sect. 4 report the experiments made and the results obtained. Lastly, Sect. 5 concludes this work.
2 Related Work
The evaluation of related works was done to understand the state-of-art of glaucoma detection using machine learning. Our study followed the systematic review pipeline proposed by Kitchenham et al. [7] and can be addressed in four main steps: 1) Define research questions; 2) Elaborate a research protocol; 3) List inclusion and exclusion criteria; 4) Review selected papers and answer the research questions. This questions include describing the techniques and technologies commonly used in automated glaucoma classification, the metrics used to measure the efficacy and accuracy of glaucoma classification models, the private and public datasets and, finally, the state-of-the art models.
Research process started by creating a search string and applying it in different journals, followed by inclusion and exclusion filtering and finishing with the acquisition and review of selected papers.
Civit-Masot el al. [4] used a U-shaped network for the segmentation and RANSAC to calculate and improve the expected elliptic shape. Xu et al. [17] also made use of the U-shaped network. However, it has segmented not only the optic disc and cup, but also defects in the retinal nerve fiber layer. Yu et al. [18] used a modified U-net with ResNet-34 as the encoder, achieving top performance in dice values for the cup and the disk.
Sreng et al. [15] used the DeepLabV3+ network, which had not been explored for segmentation yet, testing its performance with five different convolutional neural networks (CNNs), the best combination being DeepLabV3+ with MobileNet. Sallam et al. [12] used several pre-trained CNN architectures and trained them all using transfer learning from the same dataset, thus comparing their performances. Chai et al. [2] used heterogeneous data such as retinal images, medical indicators, and patient complaint texts, integrate them using representation integration, and predict using a Bayesian’s model.
Ahn et al. [1] elaborated on three models to classify glaucoma. The first uses logistic regression, the second InceptionV3 with transfer learning, and the last was an CNN manually architected by the authors. The authors elaborated the one with the better performance among the three. Phasuk et al. [10] combined the results of several integrated networks with the DenseNet-121 for feature extraction and a neural network for the final classification.
Chai et al. [2] created a heterogeneous dataset using retina images, medical charts and text-based pacient complaints, training a bayesian neural network model using representation integration.
Sallam et al. [12] gathered 9 of the most successful models in image classification challenges and, with their pre-trained weights, compared their performance against a single glaucoma classification dataset, the Large-scale Attention based Glaucoma (LAG) dataset. Selected models were AlexNet, VGG16, VGG19, GoogleNet (Inception V1), ResNET-18, ResNET-50, ResNet-101 and ResNet-152. Best results were achieved by ResNet-152, with an accuracy, precision and recall of 86,9.
The work of Serte et al. [14] compared four popular image classification networks using five public glaucoma classification datasets. While a single dataset was used as the testing set, the other four were used to train the models, comparing model performance for each dataset. Selected networks included Xception, ResNet-50, GoogLeNet and ResNET-152. Datasets used in this work were HRF, Drishti-GS1, RIM-ONE, sjchoi86-HRF and ACRIMA. According to author’s results, each network performed better on specific datasets.
As seen, deep learning has shown promising results on classifying fundus images as normal or glaucomatous in an automated way. However, more studies are needed to be focused on the clinic, where issues in addition to accuracy are of paramount importance, such as the explainability and transparency of the results obtained by the models. Moreover, the best-performing models need a large volume of data to extract relevant features and patterns. In the medical field, such information is difficult to obtain, with considerable effort from researchers to model systems capable of working with small amounts of data without losing performance.
Our research focuses on recreating the methodology of Civit-Masot et al. [4] and tackling the open issues of architecture simplification and sensitivity increase, leading to better clinical usage of the model. Section 3 describes the proposed model.
3 Proposed Method
In this paper, we extend the work of Civit-Masot et al. [4] by adding a second classification network and unifying the two segmentation networks. In that way, we aim to increase model sensitivity and simplify its architecture to enable its deployment in medical clinics. Initially, the pipeline of the base model was replicated, followed by the implementation of the ensemble of the classification networks, and lastly, the unification of the segmentation networks. Both architectures and execution flow are presented in Fig. 1. Highlighted colors represent the correlated areas between models.
The network proposed by Civit-Masot et al. [4] employed distinct networks for disc and cup segmentation. Such an improvement simplified development and helped adopting datasets with separated label masks for each structure. On the other hand, this approach leads to an increase in processing and storage loads. In order to enhance user’s trust in the system while keeping it light enough to deploy it in embedded systems, our proposed method joins both disc and cup segmentation networks into a single one, reducing processing time and cost.
The application pipeline is as follows: first, the back of the patient’s eye is captured by a digital fundus camera. Then, the image is loaded into the system and preprocessed, fed afterward to both prediction subsystems, classification, and segmentation. The classification system outputs its prediction, while the segmentation system will extract disc and cup features and compute the cup-to-disc ratio (CDR) as its output prediction.
The diagnosis system then evaluates both outputs, and if one is true for glaucoma, this is the final prognostic printed into the medic screen. In addition to the prognostic, a processed image with disc and cup highlighted by the segmentation network is presented, followed by the CDR value. Even though the system’s internal parts are not self fully explainable, these features aim to increase the medical trust in the final prediction. Since the system can output a processed image and its computed values, it is more evident how the machine came to a conclusion.
3.1 Dataset
RIM-ONE DL [5] was used as the dataset for this study. It consists of 485 fundus images, with its matching binary masks for cup and disc. Of the 485 samples, 313 (65%) were labeled as healthy cases, and 172 (35%) as glaucomatous cases. This dataset is part of a research project, developed as a joint work of three Spanish’s hospitals. The main purpose of the work was to offer a dataset of reference ophthalmology images, specifically developed for glaucoma diagnosis. Labels were made by five experts in the field, with a final segmentation unifying the singular results. An example of images and labels can be seen in Fig. 2 and Fig. 3.
3.2 Preprocessing
Following the details presented in the baseline work, the data were preprocessed as follows. First, the images and their masks were concatenated in a list, followed by the transformation of this list in a tf.dataset object. Thereby, Tensorflow preprocessing functions became available and were applied to the dataset.
Images were of different dimensions and had to be resized to specific input dimensions, this is \(224 \times 224\) px in the classification system and \(128 \times 128\) px in the segmentation system.
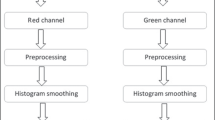
Image quality was also variable, and to help enhance those poor-quality samples, contrast limited adaptive histogram equalization (CLAHE) was applied. By doing so, the morphological structures present in the images were highlighted.
The dataset’s number of samples was insufficient to train robust deep learning models. Data augmentation techniques were utilized to solve that issue, increasing samples from 485 to 9215 images. The process consisted of applying random brightness or random contrast filters, followed by small rotations of less than 15\(^\circ \). It is crucial to notice that glaucoma diagnosis is related to the orientation of the segmented image, so it is crucial to keep rotations to a minimum [4].
To implement the unified segmentation network, it was also necessary to unify the cup and disc masks. In this new mask, each of the structure masks corresponds to a color channel from the RGB system. The disc mask was set to the red channel and the cup mask to green.
Difference between original and preprocessed image samples can be seen at Fig. 4.
3.3 Neural Network Architecture
The baseline system [4] consists of three neural networks, two modified U-nets for disc and cup segmentation, and one direct classification network based on MobileNetV2. On the other hand, in this work, we propose two neural networks, one modified U-net for both disc and cup segmentation, and one direct classification network composed of the ensemble of MobileNetV2 and EfficientNetB1. These networks are detailed below.
Segmentation Using U-Net. For disc and cup segmentation, the baseline work used a generalized U-net architecture [3]. This network has six levels of coding and decoding, with 64 channels in the first coding stage, and a channel growth rate of 1.1. Even though it has one level extra compared to the original U-net, the reduction in the growth rate from 2 to 1.1 reduced the number of parameters from 138M to 2.5M [4].
The unified segmentation network proposed by this study follows the same architecture previously described but changes the optimizer from Adam to RMSprop. Also, due to the increased complexity, the number of training epochs was increased from 100 to 300.
Classification Using MobileNetV2 and EfficientNetB1. For the direct classification subsystem, a MobileNetV2 network was chosen by the baseline authors. The top layers were removed, and an average pooling layer was added. Its output was fed to a 64-node dense layer, followed by a dropout stage and a two-node final layer to distinguish between normal and glaucoma [4].
The proposed classification subsystem consists of an ensemble of the baseline MobileNetV2 model and an EfficientNetB1 model that follows the same structure in its top layers. Each model will output its prediction, passing it to an average layer responsible for the final subsystem output.
3.4 Hyperparameters Optimization
To train the neural networks, some hyperparameters were defined. Those described in the baseline work were chosen. In the case of a lack of details, the selection of hyperparameters was made by the author of this study.
Segmentation Using U-Net. Adam optimizer was used for the segmentation networks, with an adaptive learning rate between 1e−3 (0.001) and 2e−4 (0.0002). The cost function was DICE, and it was trained through 100 epochs, with a batch size of 120. The last layer has a sigmoid activation function, and the metrics for evaluation were IoU and DICE.
The unified segmentation network proposed used the same hyperparameters of the baseline described above, only changing the optimizer from Adam to RMSprop, and increasing training epochs from 100 to 300.
Classification Network: MobileNetV2 and EfficientNetB1. Pre-trained weights from the ImageNet challenge were used for the classification network, with the initial layers frozen. The optimizer was RMSprop with a learning rate of 1e−03. The cost function was Binary Cross-Entropy, and the batch size was 64.
First, the network was trained for 50 epochs, as described in Civit-Masot et al. [4], however it did not achieve the expected metrics. Because of that, the number of epochs was increased to 100. Evaluated metrics were AUC, accuracy, specificity, and sensitivity. The EfficientNetB1 proposed by this study adopted the same hyperparameters.
3.5 Final Output
After passing through both subsystems, the final step in the pipeline is to join their predictions and display them graphically to the physician. If one of the subsystem’s outputs is glaucoma, this is the final prediction. Doing this increases the sensitivity of the model. Not just the prediction but also the semantic segmentation of both disc and cup features are graphically displayed, followed by what each subsystem predicted and the cup-to-disc ratio found. Figure 5 shows the final result of the base model, replicated by the authors of this paper.
4 Experimental Evaluation
This section aims to describe and evaluate the results obtained in each step of the development process. Moreover, validate our hypothesis and contributions based on the results. We start this section from the evaluation of the base model replication, followed by adding the second classification network and its ensemble with the original, then the union of the segmentation networks. Lastly, we present an execution time benchmark for each analog part of the architecture.
Replicating the Base Model. To implement our contributions, the base model was first replicated by following the paper implementation details [4]. The classification system was replicated after collecting the dataset and applying the preprocessing pipeline.
Table 1 compares the original metrics from Civit-Masot et al. [4] classification system to our version. Results are almost identical, which means that the replication was a success. However, specificity went down by two points, mainly because the authors didn’t sufficiently specify the last layers of the base model.
Next, we replicated the segmentation networks, for both disc and cup features. DICE values for the original networks and ours can be seen in Table 2. Our results were far better than the baseline primarily due to the increase in available image samples from the datasets, compared to those obtainable by the time of the baseline paper.
4.1 Contribution 1: Modifying the Classification Network
Aiming to improve the classification network predictions, a second lightweight network was proposed. With that in mind, the chosen network was EfficientNetB1 [16], composed of less than eight million parameters. It is considered a light model capable of providing results as efficient as those of the more robust architectures, like VGG or Inception.
It was trained with the same hyperparameters of the original MobileNetV2, and it resulted in an increase in sensitivity, from 0.9140 to 0.9462. While it did not increase AUC and reduced specificity, it is still an improvement, primarily because of how critical false negatives are to the medical clinic compared to false positives. Table 3 compares the results obtained for the original MobileNetV2, the new network added by this study, EfficientNetB1, and the ensemble of both MobileNetV2 and EfficientNetB1 as the final output of the proposed classification subsystem.
4.2 Contribution 2: Unifying the Segmentation Networks
To simplify the architecture and make it lighter for possible deployment on embedded systems, we developed a single segmentation network capable of identifying both disc and cup features. Due to the increased complexity, training epochs were increased from 100 to 300 to achieve similar results to those of the individual networks. However, it became clear that a single segmentation network can achieve state-of-the-art results with a more straightforward and lighter architecture, increasing the feasibility of an embedded system for the medical clinic. Table 4 compares these results. Comparable results were obtained using only one U-net model. By doing so, we reduced the storage and processing cost of the segmentation subsystem in half. This can prove valuable to those interested in deploying the system in standard desktop hardware.
4.3 Benchmark
To better visualize the impact on the simplification of the system, a benchmark evaluation of the baseline model and the proposed one was made. Both networks were executed from end to end with the same inputs and on the same hardware. Each model ran 50 times, providing high fidelity results. Table 5 shows the obtained results. Each color represents a highlighted region from Fig. 1. As seen in Table 5, the proposed model has a lower average response time for the diagnosis outcome, mostly because of the simplification of the U-net networks. In this manner, the architecture also reduced the times for generating the diagnosis, reducing the patients’ waiting time for the generation of the report.
Unifying the segmentation networks reduced the segmentation subsystem processing time by 24.24%. On the other hand, the ensemble of the two classification systems increased classification subsystem processing time by 33.25%. This is not critical, though, because the pipeline’s bottleneck is the segmentation networks, and by reducing its time, the overall processing time gets reduced. Also, the training process is much slower on the segmentation networks than on the classification networks, increasing the value of this reduction.
5 Conclusion
Glaucoma is a silent disease that only shows signs in the advanced and irreversible stages of the disease. Fundoscopy is the primary diagnostic test, a manual process requiring years of expertise and specialization. It is possible to solve this problem by employing computer vision techniques to generate the diagnosis. However, these techniques often lack transparency and justification, not presenting more than the classification or the why of that result and the algorithm’s reliability.
Our contribution is increasing the model sensitivity, further reducing the number of false negatives and thus ensuring even more value for its use in the clinic. Also, the architecture simplifies, taking up less storage space and using less processing resources without losing accuracy, resulting in the need for more initial training time.
Even though we obtained better results in processing time and sensitivity, our system does not address those situations where the segmented disc or cup does not form a fully connected ellipse. Without this shape enhancement, feature extraction of disc and cup diameter can not be computed, resulting in crashes. Even though CLAHE reduced crash rates by improving image quality, this can only be addressed by changing the logic behind the segmentation feature extraction or optimizing the ellipse fitting function. Because of these issues, our system performed better than the baseline on high-resolution images, and its use is recommended on such occasions. However, this is not the reality in most medical clinics, and fixing it is a priority in future works.
Lastly, we perceive that explainability is crucial for the future implementation of the system in a clinic. Our approach does not entirely address this type of issue. For this, it is necessary to evolve the system into an independent application in a production environment. Learning cycles and direct application are inserted in the process flow from an embedded system that receives the image directly from the capture system. This requires an advance and a future collaboration between researchers and medical institutions, being one of the main future works. Since the study’s main objective is to assist the clinical diagnosis, it is essential to consider the reality of patients. This includes conditions other than glaucoma, varied ethnicities with specific eye characteristics, and images with different resolutions and specifications.
References
Ahn, J.M., Kim, S., Ahn, K.S., Cho, S.H., Lee, K.B., Kim, U.S.: A deep learning model for the detection of both advanced and early glaucoma using fundus photography. PLoS ONE 13 (2018)
Chai, Y., Bian, Y., Liu, H., Li, J., Xu, J.: Glaucoma diagnosis in the Chinese context: an uncertainty information-centric Bayesian deep learning model. Inf. Process. Manag. 58 (2021)
Civit-Masot, J., Billis, A., Dominguez-Morales, M.J., Vicente-Diaz, S., Civit, A.: Multidataset incremental training for optic disc segmentation. In: Iliadis, L., Angelov, P.P., Jayne, C., Pimenidis, E. (eds.) EANN 2020. PINNS, vol. 2, pp. 365–376. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-48791-1_28
Civit-Masot, J., Domínguez-Morales, M.J., Vicente-Díaz, S., Civit, A.: Dual machine-learning system to aid glaucoma diagnosis using disc and cup feature extraction. IEEE Access 8, 127519–127529 (2020)
Fumero, F., Alayón, S., Sanchez, J.L., Sigut, J., Gonzalez-Hernandez, M.: RIM-ONE: an open retinal image database for optic nerve evaluation. In: Proceedings of the IEEE Symposium on Computer-Based Medical Systems (2011)
Hemelings, R., et al.: Accurate prediction of glaucoma from colour fundus images with a convolutional neural network that relies on active and transfer learning. Acta Ophthalmologica 98 (2020)
Kitchenham, B., Charters, S.: Guidelines for performing systematic literature reviews in software engineering (2007)
Mantravadi, A., Vadhar, N.: Glaucoma. Primary Care - Clinics Office Pract. 42(3) (2015)
NICE: Glaucoma: diagnosis and management. NICE Guideline 81(3) (2017)
Phasuk, S., et al.: Automated glaucoma screening from retinal fundus image using deep learning. In: Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Sallam, A., et al.: Early detection of glaucoma using transfer learning from pre-trained CNN models. In: 2021 International Conference of Technology, Science and Administration (ICTSA) (2021)
Sandler, M., Howard, A., Zhu, M., Zhmoginov, A., Chen, L.C.: MobileNetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4510–4520 (2018)
Serte, S., Serener, A.: A generalized deep learning model for glaucoma detection. In: Proceedings of the 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies, ISMSIT 2019 (2019)
Sreng, S., Maneerat, N., Hamamoto, K., Win, K.Y.: Deep learning for optic disc segmentation and glaucoma diagnosis on retinal images. Appl. Sci. (Switzerland) 10, 4916 (2020)
Tan, M., Le, Q.: EfficientNet: rethinking model scaling for convolutional neural networks. In: 36th International Conference on Machine Learning, ICML 2019 (2019)
Xu, Y., et al.: A hierarchical deep learning approach with transparency and interpretability based on small samples for glaucoma diagnosis. NPJ Digit. Med. 4, 1–11 (2021)
Yu, S., Xiao, D., Frost, S., Kanagasingam, Y.: Robust optic disc and cup segmentation with deep learning for glaucoma detection. Comput. Med. Imaging Graph. 74, 61–71 (2019)
Acknowledgements
We thank the anonymous reviewers for their valuable feedback. This research was partially supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001, and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Ceschini, L.M., Policarpo, L.M., Righi, R.d.R., Ramos, G.d.O. (2022). Aiding Glaucoma Diagnosis from the Automated Classification and Segmentation of Fundus Images. In: Xavier-Junior, J.C., Rios, R.A. (eds) Intelligent Systems. BRACIS 2022. Lecture Notes in Computer Science(), vol 13654 . Springer, Cham. https://doi.org/10.1007/978-3-031-21689-3_25
Download citation
DOI: https://doi.org/10.1007/978-3-031-21689-3_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21688-6
Online ISBN: 978-3-031-21689-3
eBook Packages: Computer ScienceComputer Science (R0)