Abstract
Yeasts have been used to manage a large number of plant diseases, but little is known about the mechanisms used by these biocontrol agents. The objectives of the present study were to evaluate the antagonistic effect of yeasts against Rhizoctonia solani and possible mechanisms of action in cowpea plants. Seventy yeast isolates were obtained from leaf, root and stem tissues of cowpea and common bean plants. Screening experiments were conducted in a greenhouse at temperatures ranging from 15 to 26 °C in the first and from 22 to 31 °C in the second experiment. Candida saopaulonensis C6A, Cryptococcus laurentii FVC10 and Bullera sinensis FVF10 (R1) reduced disease severity by 57.4%, 48.5% and 66.3%, respectively. Cowpea plants treated with FVF10 (R1) showed the highest peroxidase and catalase activities. The mechanisms of action were based on competition and induction of enzymes such as peroxidase, catalase and ascorbate peroxidase in cowpea. Candida saopaulonensis C6A, C. laurentii FVC10 and B. sinensis FVF10 (R1) are potential biocontrol agents of damping-off and stem rot caused by R. solani on cowpea plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Damping-off and stem rot caused by Rhizoctonia solani are important soil-borne diseases of cowpea plants (Vigna unguiculata) (González-Garcia et al. 2006). These diseases are responsible for severe economic losses, mainly because they reduce seed germination and emergence (Sartorato et al. 2006; Sikora 2004). Rhizoctonia solani is difficult to control because of its broad host range. This pathogen can persist in the soil due to its overwintering structures, its strong saprophytic capacity and high virulence (González et al. 2011). Therefore, taking actions to decrease inoculum density, promote rapid seedling development and prevent favorable environmental conditions for the pathogen to infect susceptible hosts is necessary (Sneh et al. 1996).
The adverse effects caused by fungicides have encouraged researchers to develop alternatives to synthetic chemicals such as biological control (Pal and Gardener 2006). Yeasts have been successfully used to suppress soil-borne plant pathogens and help plant growth (Botha 2011; El-Tarabily and Sivasithamparam 2006). El-Tarabily (2004) used Candida valida, Trichosporon asahii and Rhodotorula glutinis to control diseases caused by Rhizoctonia solani in sugar beet seedlings. The use of Hansenula arabitolgenes, Candida incommunis and Candida steatolytica was effective to inhibit Rhizoctonia solani, and these yeasts helped to reduce damping-off incidence in cotton plants (El-Mehalawy et al. 2006). El-Mehalawy et al. (2007) and Mohammed et al. (2008) showed the effectiveness of yeasts to reduce damping-off severity on potato plants and stem rot on cotton plants attacked by Rhizoctonia solani. According to El-Tarabily and Sivasithamparam (2006), yeasts promoted plant growth and induced resistance by increasing phenylalanine ammonia lyase, phytoalexins, peroxidase and ethylene levels in plant tissues.
There are many studies about yeasts controlling plant diseases, however, little is known about the mechanisms used by these biocontrol agents. The objectives of this study were (i) to isolate yeasts from the leaf, root and stem tissues of cowpea and common bean plants; (ii) to select the most effective yeast isolates against damping-off and stem rot caused by Rhizoctonia solani on cowpea plants; (iii) to investigate their possible mechanisms of action; and (iv) to assess antioxidative enzyme activity in cowpea plants.
Materials and methods
Pathogen and antagonistic yeasts
Rhizoctonia solani (CMM-3643) was obtained from the culture collection “Profa. Maria Menezes” (CMM) at the Federal Rural University of Pernambuco. It was isolated from the root of a bean plant and preserved by the Castellani method. This fungus was grown in potato-dextrose agar (PDA) medium and incubated for 3 days at 25 ± 2 °C. The Rhizoctonia solani inoculum was produced in Erlenmeyer flasks containing 150 g of sterile parboiled rice and 150 mL of water, which were colonized by the pathogen for 10 days at room temperature, according to the methodology described by Barbosa et al. (1995).
The yeasts were isolated from the leaf, root and stem tissue fragments of healthy cowpea and common bean plants. These fragments were separately immersed in test tubes (five pieces per tube) containing 10 mL of sterile water and chloramphenicol (50 mg/L). Subsequently, the tubes were stirred in an ultrasonic bath for 10 min, vortexed and the content was serially diluted. Aliquots of 0.1 mL of these dilutions were spread on Petri plates containing Sabouraud dextrose agar (SDA) composed of 40 g dextrose, 10 g neopeptone and 20 g agar per litre; the agar solution was supplemented with yeast extract (1.5 g/L). The plates were incubated for 72 h at 25 ± 2 °C and, thereafter, the yeast colonies were transferred to tubes containing SDA medium. The isolates were preserved in mineral oil at room temperature (25 ± 2 °C).
Screening the yeasts showing biocontrol potential
Seventy yeast isolates from cowpea and common bean plants were collected in production areas of Pernambuco state, Brazil. The yeasts were cultivated on SDA medium supplemented with yeast extract and chloramphenicol for 48 h at 28 ± 2 °C under a 12 h photoperiod. The suspensions were adjusted to 107 CFU m/L using a Neubauer chamber. Cowpea seeds (cv. IPA-207) were immersed in yeast suspensions with 0.02% tween 80 for 10 min. Treated seeds were kept at 28 °C for 12 h before sowing. The experimental design was completely randomized with 71 treatments (70 yeast isolates plus the control) and five replications, using two seeds in each replication.
The Rhizoctonia solani inoculum prepared as described above was incorporated in the soil (50 mg of colonized rice per kg of soil) 48 h before sowing. The treated seeds were sown in expanded polystyrene trays containing humic dystrophic cohesive yellow latosol with a clayey texture. Two experiments with five replications containing two seeds each were conducted at different temperatures, 22–31 °C and 15–26 °C, under 80% humidity in a greenhouse. The control treatment was the soil infested with the pathogen and common bean seeds without treatment with yeasts.
After 15 days, damping-off and stem rot severity were assessed with a descriptive scale (0–4), where: 0 = no symptoms, 1 = hypocotyl with small injuries, 2 = hypocotyl with large lesions and no constrictions, 3 = completely constricted hypocotyl showing damping-off, and 4 = non-germinated seeds and/or non-emerged seedlings (Noronha et al. 1995). The disease index (DI) was calculated using data from the descriptive scale according to McKinney’s formula (McKinney 1923). The DI was subjected to variance analysis (ANOVA) and the means were compared with the Scott-Knott test at 5% probability.
Three isolates [C6A, FVC10 and FVF10 (R1)] were tested in a second experiment as they were among the ones that consistently reduced disease severity in the first two screening experiments. The same methodology was used in this experiment. The experiment was installed in a completely randomized design with four treatments (three yeast isolates plus the control) and five replications. Each replication contained nine plants. The control treatment consisted of soil infested with Rhizoctonia solani and cowpea seeds without yeasts. Two experiments were conducted at different temperatures (14–26 °C and 19–30 °C) under 80% humidity. The DI and statistical analyses were calculated as described above.
The in vitro antagonism of Candida saopaulonensis C6A, Cryptococcus laurentii FVC10 and Bullera sinensis FVF10 (R1) against Rhizoctonia solani was assessed through the method of dual cultures. The yeasts were cultivated in SDA agar as described above. Cell suspensions containing 107 CFU m/L were prepared in sterile saline solution. Sterilized glass funnels (7 cm diameter) were immersed in the yeast suspensions and used to stamp the surface of the PDA culture medium in each Petri dish (9 cm). A Rhizoctonia solani mycelial disk (5 mm) containing three-day-old colonies was placed at the center of each plate. The plates were kept at 25 ± 2 °C under a 12 h photoperiod. Control treatment contained Rhizoctonia solani, and the yeast suspension was replaced with sterile saline solution. Plates were kept in BOD for five days at 25 ± 2 °C under a 12 h photoperiod. Mycelial growth inhibition was calculated in relation to the control treatment without any yeast isolate.
Identification of the selected yeast isolates
Three yeast isolates in the biocontrol assays were identified according to classical (Kurtzman et al. 2011) and molecular taxonomies (Negri et al. 2014). The ITS and a fragment of the 26S rDNA genomic regions were amplified by PCR and sequenced using primers ITS1 and ITS4 (White et al. 1990). The DNA sequences generated from the isolates were compared to the sequences deposited in GenBank using Blastn (Altschul et al. 1997).
Induction of systemic resistance
The enzymatic activity was assessed in cowpea plants inoculated with the three selected yeast isolates. The inoculum of R. solani was prepared and incorporated in the soil as described above. Seeds were treated and seeded in plastic trays kept in a greenhouse. The design was completely randomized with four treatments (three yeasts plus the control) and five replications. Cowpea leaves were collected 15 days after the beginning of the experiment and processed as follows: 0.1 g of cowpea leaves were immersed in liquid N2 added with 4 mL of 50 mM potassium phosphate buffer (pH 7.0) and 0.05 g of polyvinylpyrrolidone (Silva et al. 2016). This extract was placed in microtubes and centrifuged for 10 min at 10.000×g at 4 °C (Silva et al. 2017). The supernatant was kept in microtubes and stored at −20 °C. Catalase activity (CAT, EC 1.11.1.6) was determined according to Havir and Mchale (1987). Ascorbate peroxidase activity (APX, EC 160 1.11.1.11) was determined according to the method of Nakano and Asada (1981), which was modified by Koshiba (1993). Peroxidase activity (POX, EC 1.11.1) was determined according to Urbanek et al. (1991), using guaiacol and H2O2 as substrates. Polyphenol oxidase activity (PPO, EC 1.10.3.1) was determined through pyrogallol oxidation (Kar and Mishra 1976). All enzyme activities were expressed as U/min/mg of protein.
Results
Screening potential biocontrol yeast isolates
In the first assay (Fig. 1a), performed at 22–31 °C, 62 isolates showed disease indices significantly lower than that of the control (P < 0.05) and eight isolates did not differ from the control (P < 0.05). In the second experiment (Fig. 1b), conducted at 15–26 °C, 36 isolates differed from the control, whereas 34 isolates showed disease indexes similar to the control. Isolates C6A, FVF10 (R1) and FVC10 were randomly chosen among the ones that significantly reduced disease severity in both assays for molecular identification and further experimentation. These isolates were able to decrease severity by 55–72% in the first and by 58–75% in the second experiment.
Screening of potential biocontrol yeast isolates in bioassays performed with cowpea. Treatments were 70 yeast isolates inoculated on cowpea seeds, and the control was non-inoculated with yeasts. Disease severity was determined 15 days after sowing. a Assay 1, temperature ranging from 22 to 31 °C. b Assay 2, temperature ranging from 15 to 26 °C. Means followed by the same letters are not significantly different according to Scott-Knott test (P = 0.05). Disease index (DI) was calculated according to McKinney (1923)
Yeast identification
The ITS and 26S rDNA sequences identified isolate C6A as Candida saopaulonensis, as its sequence was 99% identical to that of isolate JYC1092 (KM555179). The sequence of isolate FVC10 was identical to that of Cryptococcus laurentii isolate IMUFRJ 51996 (FN428921). Isolate FVF10 (R1) was identified as Bullera sinensis as its sequence was 97% identical to an isolate of this species recovered from a landfill site (DQ297411). The sequences varied from 644 bp to 837 bp and were deposited in GenBank under accession numbers KX781276, KX781277 and KX781278, respectively.
Biocontrol activity in vitro and in vivo of selected biocontrol isolates
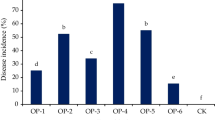
A dual plate assay showed that isolate C6A of Candida saopaulonensis inhibited 52% of Rhizoctonia solani mycelial growth. This was the only isolate that significantly inhibited Rhizoctonia solani compared to the control (data not shown). Two experiments were performed to test the biocontrol activity of the three selected isolates against Rhizoctonia solani. All isolates significantly reduced disease severity in both experiments. This reduction varied from 49 to 66% in the first experiment and from 31 to 65% in the second experiment (Fig. 2).
Control of damping-off and stem rot caused by Rhizoctonia solani with three selected yeast isolates in assays with the temperature ranging from 14 to 26 °C (Assay 1) and from 19 to 30 °C (Assay 2). The data are representative of two independent experiments. Means with the same letters are not significantly different according to the Scott-Knott test (P = 0.05). Disease index (DI) was calculated according to McKinney (1923)
Induction of systemic resistance
Cowpea plants exposed to B. sinensis FVF10 (R1) showed peroxidase and catalase activities significantly higher than other treatments, but a low activity of ascorbate peroxidase (Fig. 3). Plants treated with Candida saopaulonensis C6A showed low ascorbate peroxidase activity whereas the activities of the other enzymes were higher than the activities of their controls. The plants treated with Cryptococcus laurentii FVC10 showed peroxidase activity lower than that of the other treatments, but the activities of the other enzymes were higher than that of their respective controls. The application of Bullera sinensis FVF10 (R1) led to maximum activity of the enzymes in the ROS group (peroxidase and catalase) in cowpea plants.
Enzymatic activities in cowpea leaves inoculated with Rhizoctonia solani and treated with Bullera sinensis, Cryptococcus laurentii and Candida saopaulonensis suspensions at 107 CFU m/L or sterile water (control). a Peroxidase; b Catalase; c Polyphenol oxidase; d Ascorbate peroxidase. Means with the same letters are not significantly different based on the Scott-Knott test (P = 0.05)
Discussion
Damping-off and stem rot diseases caused by Rhizoctonia solani are the most severe diseases of cowpea worldwide. There are no efficient control strategies against these diseases (González et al. 2011). In this study, 70 yeasts were isolated from cowpea and common bean and screened against Rhizoctonia solani. Three yeast isolates, Candida saopaulonensis C6A, Cryptococcus laurentii FVC10 and Bullera sinensis FVF10 (R1), were selected as potential biocontrol agents of Rhizoctonia solani.
Candida saopaulonensis C6A, Cryptococcus laurentii FVC10 and Bullera sinensis (FVF10 (R1) decreased the severity of damping-off and stem rot caused by Rhizoctonia solani in cowpea plants. A higher reduction in disease severity was observed at higher temperatures (19–30 °C). Teixidó et al. (1998) demonstrated that yeasts were more efficient as biological control agents at approximately 30 °C. The temperature is one of the main environmental stresses that affect yeasts (Sui et al. 2015). When applied before harvesting in the field, antagonistic yeasts need to tolerate environmental stresses. Beneficial microorganisms need to overcome these challenges in order to effectively establish themselves and control plant diseases (Conway et al. 1999; Deacon 1991; Sui et al. 2015). In the present study, most yeasts significantly reduced the disease index (DI) at different temperatures. This relatively good response at different temperatures may be explained by the fact that the temperature could have influenced the virulence of Rhizoctonia solani (Goulart 2002; Santos et al. 2005; Tanaka 1994).
Candida species may produce antifungal compounds as observed in our study with Candida saopaulonensis C6A and by other authors (El-Tarabily and Sivasithamparam 2006). The activity of a biocontrol agent by antibiosis does not rule out other mechanisms (Tuzun and Kloepper 1995).
The significant increase in the concentrations of peroxidase and catalase in plants treated with yeast Bullera sinensis FVF10 (R1) and the absence of in vitro antibiosis are evidences that the mechanism of disease control in question is related to induced resistance. Khalid (2014) found that Saccharomyces cerevisiae reduced the incidence of S. rolfsii disease in common bean plants, resulting in increased peroxidase, polyphenoloxidase and chitinase activity.
Cryptococcus laurentii FVC10 and Candida saopaulonensis C6A increased polyphenol oxidase levels in plant tissues as observed for other yeast species (El-Tarabily and Sivasithamparam 2006; Zhao et al. 2008; Khalid 2014). Furthermore, there are reports that ascorbate peroxidase and catalase act in the modulation and removal of the excess of reactive oxygen species (ROS) that result from stresses. Thus, helping the plant’s defense mechanism by preventing ROS accumulation and the consequent cell death (Mittler 2002; Tománková et al. 2006).
Candida saopaulonensis C6A, Cryptococcus laurentii FVC10 and Bullera sinensis FVF10 (R1) were effective against Rhizoctonia solani and are promising control agents against the pathogen in cowpea plants. The current study is the first to report the isolation of Candida saopaulonensis and Bullera sinensis from bean plants, as well as their efficacy in biological control of R. solani. Understanding the role of yeasts in the control of soil pathogens will contribute to future sustainable agricultural practices.
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25:3389–3402
Barbosa MAG, Michereff SJ, Mariano RLR, Maranhão E (1995) Biocontrole de Rhizoctonia solani em caupi pelo tratamento de sementes com Pseudomonas spp. fluorescentes. Summa Phytopathologica 21:151–157
Botha A (2011) The importance and ecology of yeasts in soil. Soil Biology and Biochemistry 43:1–8
Conway WS, Janisiewicz WJ, Klein JD, Sams CE (1999) Strategy for combining heat treatment, calcium infiltration, and biological control to reduce postharvest decay of ‘gala’ apples. HortScience 34:700–704
Deacon JW (1991) Significance of ecology in the development of biocontrol agent against soil-borne plant pathogens. Postharvest Biology and Technology 1:5–20
El-Mehalawy AA, Hassanein SM, Hassanein NM, Abd-Allah S (2006) Induction of resistance and biocontrol of Rhizoctonia in cotton against damping-off disease by rhizosphere yeast and fungi. Applied Ecological and Environmental Research 3:1–12
El-Mehalawy AA, Hassanim SM, Hassanim NM, Zakis SA (2007) Induction on resistence and biocontrol of Rhizoctonia in cotton against damping-off disease by rhizosphere microorganisms. New Egyptian Journal of Microbiology 17:148–168
El-Tarabily KA (2004) Supression of Rhizoctonia solani disease of sugar beet by antagonists and plant growth-promoting yeasts. Journal of Applied Microbiology 96:69–75
El-Tarabily KA, Sivasithamparam K (2006) Potential of yeasts as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Mycoscience 47:25–35
González M, Pujol M, Metraux J, González-Garcia V, Bolton MD, Borrás-Hidalgo O (2011) Tobacco leaf spot and root rot caused by Rhizoctonia solani Kühn. Molecular Plant Pathology 12:209–216
González-Garcia V, Portal OMA, Rubio SV (2006) Biology and systematics of the form genus Rhizoctonia. Spanish Journal of Agricultural Research 4:55–79
Goulart ACP (2002) Efeito do tratamento de sementes de algodão com fungicidas no controle do tombamento de plântulas causado por R. solani. Fitopatologia Brasileira 27:399–402
Havir EA, Mchale NA (1987) Biochemical and development characterization of multiple forms of catalase in tobacco leaves. Plant Physiology 84:450–455
Kar M, Mishra D (1976) Catalase, peroxidase and polyphenoloxidase activities during rice leaf senescence. Plant Physiology 57:315–319
Khalid EE (2014) Biological control of bean damping-off caused by Sclerotium rolfsii. Egyptian Journal of Phytopathology 42:1–12
Koshiba T (1993) Cytosolic ascorbate peroxidase in seedlings and leaves of maize (Zea mays). Plant & Cell Physiology 34:713–721
Kurtzman CP, Fell JW, Boekhout T (2011) The yeasts - a taxonomic study, 5th edn. Elsevier Science Publieshers, Amsterdam
McKinney RH (1923) Influence of soil temperature and moisture on infection of wheat seedlings by Helminthosporium sativum. Journal of Agricultural Research 6:195–218
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7:405–410
Mohammed AS, El Hassan SM, El Balla MMA, El Sheik EAE (2008) The role of Trichoderma, VA mycorriza and dry yeasts in the control of Rhizoctonia disease of potato (Solanum tuberosum L.). The University of Khartoum. Journal of Agricultural Science 16:285–301
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-especific peroxidase en spinach chloroplasts. Plant & Cell Physiology 22:867–880
Negri CE, Gonçalves SS, Xafranski H, Bergamasco MD, Aquino VR, Castro PTO, Colombo AL (2014) Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. Journal of Clinical Microbiology 52:3633–3640
Noronha MA, Michereff SJ, Mariano RLR (1995) Efeito do tratamento de sementes de caupi com Bacillus subtilis no controle de Rhizoctonia solani. Fitopatologia Brasileira 20:174–178
Pal KK, Gardener BM (2006) Biological control of plant pathogens. Plant Health Instructor 1:1–25
Santos FS, Souza PE, Oliveira CA, Magalhães FHL, Laurenti MA (2005) Ajuste do inóculo de R. solani AG-4 em substrato para estudo de rhizoctoniose em algodoeiro e feijoeiro. Summa Phytopathologica 31:374–376
Sartorato A, Nechet KL, Halfeld-Vieira BA (2006) Diversidade genética de isolados de Rhizoctonia solani coletados em feijão-caupi no estado de Roraima. Fitopatologia Brasileira 31:297–301
Sikora EJ (2004) Rhizoctonia root rot on garden beans. Alabama cooperative extension system. Available at: www.aces.edu/pubs/docs/A/ANR-1006/. Accessed on June 12, 2014
Silva JAT, Medeiros EV, Silva JM, Tenório DA, Moreira KA, Nascimento TCES, Souza-Motta C (2016) Trichoderma aureoviride URM 5158 and Trichoderma hamatum URM 6656 are biocontrol agents that act against cassava root rot through different mechanisms. Journal of Phytopathology 164:1003–1011
Silva JAT, Medeiros EV, Silva JM, Tenório DA, Moreira KA, Nascimento TCES, Souza-Motta C (2017) Antagonistic activity of Trichoderma spp. against Scytalidium lignicola CMM 1098 and antioxidant enzymatic activity in cassava. Phytoparasitica 45:219–225
Sneh B, Jabaji-Hare S, Neate S, Dijst G (1996) Rhizoctonia species: taxonomy, molecular biology, ecology, pathology and disease control. 1st Ed. Kluwer Academic Publishers, Dordrecht
Sui Y, Wisniewski M, Droby S, Liu J, Müller V (2015) Responses of yeast biocontrol agents to environmental stress. Applied and Environmental Microbiology 81:2968–2975
Tanaka M (1994) Patógeno causadores de tombamento do algodoeiro e seus efeitos sobre a germinação das sementes em diferentes temperaturas. Fitopatologia Brasileira 19:29–33
Teixidó N, Vinas I, Usall J, Sanchis V, Magan N (1998) Ecophysiological responses of the biocontrol yeast Candida sake to water, temperature and pH stress. Journal of Applied Microbiology 84:192–200
Tománková K, Luhová L, Petrivalský M, Pec P, Lebeda A (2006) Biochemical aspects of reactive oxygen species formation in the interaction between Lycopersicon spp. and Oidium neolycopersici. Physiological and Molecular Plant Pathology 68:22–32
Tuzun S, Kloepper JW (1995) Potential applications of plant growth-promotig rhizobacteria to induced systemic disease resistence. In: Romeiro RS (ed) Controle biológico de enfermidades de plantas. Editora UFV, Viçosa. pp. 39–56
Urbanek H, Kuzniak-Gebarowska E, Herka K (1991) Elicitation of defense responses in bean leaves by Botrytis cinerea polygalacturonase. Acta Physiologiae Plantarum 13:43–50
White TJ, Bruns T, Lee S, Taylor RJ (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Shinsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, London, pp 315–322
Zhao Y, TU K, Shao X, Jing W, Su Z (2008) Effects of the yeast Pichia guilliermondii against Rhizopus nigricans on tomato fruit. Postharvest Biology and Technology 49:113–120
Acknowledgements
We thank CAPES for granting a scholarship to the first author, as well as CNPq for the research scholarship granted to DL, CSL, and EVM (306401/2015-0). We thank Dr. Antônio Félix da Costa (IPA) for his contribution concerning the plant material used in the present study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Section Editor: Jorge T. de Souza
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure S1
The antagonist effect of yeast isolates against Rhizoctonia solani mycelial growth in vitro, calculated in relation to the control treatment without any yeast isolate. (JPG 95 kb)
Rights and permissions
About this article
Cite this article
de Tenório, D.A., de Medeiros, E.V., Lima, C.S. et al. Biological control of Rhizoctonia solani in cowpea plants using yeast. Trop. plant pathol. 44, 113–119 (2019). https://doi.org/10.1007/s40858-019-00275-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-019-00275-2