Abstract
In order to explore the karyotype and evolutionary trend of Clematis and provide taxonomically useful data, the chromosome number, the number and position of satellites, the karyotype formulae, the karyotype, arm ratio, relative length centromeric index, and the index of the karyotype asymmetry of 11 species of the genus Clematis were studied. All the analyzed species showed the same stable chromosome number (2n = 2x = 16) and basic chromosome number 8. Clematis karyotypes are composed by metacentric, submetacentric, and terminal centromeric chromosomes. Clematis lanuginosa, C. otophora, C. lasiandra, C. shenlungchiaensis, C. florida var. plena, C. fusca var. violacea, C. crispa, and C. viticella belong to Type 2A, a very symmetric and plesiomorphic type. Clematis henryi, C. integrifolia, and C. japonaca belong to Type 2B, which is apomorphic relative to the other group. The 11 species of Clematis are different in karyotype parameters providing data for the taxonomy of the genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Clematis belongs to the family Ranunculaceae and consists of about 230–355 species including 147 native species in China, 93 of which are endemic (Wang and Bartholomew 2001; Wang and Li 2005). Clematis is one of the most popular climbing plants used in landscapes and floriculture as a garden or potted plant (Xie et al. 2011).
Regarding Clematis karyotype, Gurignard firstly reported C. recta with basic chromosome number 8 and then Langlet (1927) confirmed this conclusion. Meurman and Therman (1939) found tetraploid, hexaploid, and hybrid ploidy for Clematis. According to Zhang and He (1990) and Zhang and He (1991), about 106 species/variations with chromosome number and species/varieties with diverse karyotypes were recorded. The evolution of Clematis karyotype showed two trends: change of chromosome number (including polyploidization and aneuploid) and chromosome structure variation (Dennis 1976; Gong et al.1985; Dawson 1993).
The taxonomy of Clematis is mainly based on morphological characters. Clematis plays an important role in the classification of the Ranunculaceae (Scholars had different ideas on the composition of this genus, boundary, interspecific relationship, etc.) (Tamura 1987; Yang 1998; Grey-Wilson 2000; Wang and Li 2005).
In this work, we describe the karyotype of ninr Chinese (C. henryi, C. otophora, C. lasiandra, C. lanuginosa, C. integrifolia, C. japonica, C. shenlungchiaensis, C. florida. var. plena, C. fusca var. violacea) and two Container Nursery species (C. crispa, C. viticella). In particular, C. henryi, C. otophora, C. lanuginosa, and C. shenlungchiaensis are rare and endemic to China (Wang and Bartholomew 2001; Wang and Li 2005). The results are discussed in the context of the current taxonomy, distribution, and evolution of the family.
Materials and methods
Materials
Eleven species of Clematis were used in this study; for each species, five individuals were analyzed totalizing 55 samples. Clematis integrifolia, C. japonica, C. crispa, and C. viticella were from foreign nursery gardens and others were domestic wild resources (See Table 1). Localities of the collections are shown in Fig. 1. Classification and morphological characters of those materials are in Table 2.
Collection localities of the 11 species of Clematis in China. Note The numbers refer to Clematis species are listed in Table 1
Method
Root tips (1 cm in length) were excised and immersed in preconditioning bath (mixture of 2 mM 8-hydroxyquinoline and 0.02 % colchicine) for 6–8 h at 4 °C, fixed in the mixture of ethanol and acetic acid (3:1) for 16–20 h at 4 °C, washed in distilled water, and preserved in 70 % ethanol at 4 °C. Root tips were hydrolyzed in 1 mol.L−1 HCl at 60 °C for 10 min and then put in 2 % cellulose for 30–40 min. Squash preparation was made with carbol fuchsin staining and then slides were observed and photographed (Li and Chen 1985).
Fifty metaphase cells were chosen to count the chromosome number, and five clear metaphase cells were selected for microphotographs. Idiograms were constructed based on the chromosome lengths and relative arm ratios. Index of the karyotypic asymmetry and karyotype type analyses were basically the same as what was respectively described by Levan et al. (1964) and Stebbins (1971). Index of the karyotypic asymmetry was represented by centrometric term inalization value (T.C. for short) as follows: T.C % = (total length of the long arm of chromosome/total length of chromosome) × 100.
The chromosome characteristics of the 11 species of Clematis were analyzed by the SPSS 17.0 software (Statistical Product and Service Solutions) (Liu et al. 2010; Ebrahim et al. 2012). The statistics created by SPSS included data as follows: index of karyotype asymmetry, longest chromosome/shortest chromosome, average arm ratio, percentage of arm ratio >2:1/ %, centromere index, satellite presence (satellite presence was quantified as 1; no satellite presence was quantified as 0), karyotype type (2A was quantified as 0; 2B was quantified as 1), and chromosome number (2x = 16 was quantified as 0). The initial data were converted into standard normal distribution. Then Euclidean distances were calculated, and cluster analysis was carried out by applying the averaging method.
Results
Karyotype and chromosome number of 11 species of Clematis
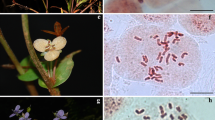
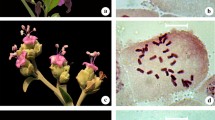
Karyotype descriptions are detailed in Table 3. The morphology of the somatic chromosomes and the karyotype idiograms is shown in Fig. 1. The 11 species showed the same number of chromosomes (2n = 2X = 16) and stable base chromosome number (n = 8). All species showed metacentric (m), submetacentric (st), and terminal centromeric (t) chromosomes.
The arm ratios were variable in the different species. Clematis lanuginose, C. otophora, C. lasiandra, C. shenlungchiaensis, C. florida var. plena, C. fusca var. violacea, C. crispa, and C. viticella belong to Type 2A, a very symmetric and plesiomorphic type. Clematis henryi, C. integrifolia, and C. japonica belong to Type 2B, apomorphic relative to the other type. Each metaphase in all species had one or two pair of satellites which was located on different chromosomes. Clematis lanuginosa had two pair of satellites, while all the others had only one Fig. 2.
Relationship and evolution of the 11 species of Clematis
According to the theory of Levitzky and Stebbins, in angiosperms, the chromosome karyotype changes from symmetry to the asymmetry in biological evolution. Species with symmetrical karyotype correspond to the basal trait, while species with asymmetrical karyotype usually have derived characters (Stebbins 1971; Desal et al. 2012). Therefore, we selected average arm ratio reflecting the karyotypic asymmetry as abscissa and longest/shortest chromosomes as ordinate in the rectangular chart and then compared evolution degree of the 11 species (Fig. 3).
Relative position of the point in the figure reflects the asymmetry, apomorphy degree, and the relative relations of the 11 species of Clematis. The species on the top right of the figure showed relatively high karyotype asymmetry with high apomorphy degree. On the contrary, the species on the bottom left of the figure showed relatively low karyotype asymmetry with low apomorphy degree. According to Fig. 3, C. crispa and C. fusca var. violacea are closer to the bottom left than other species, so they have lower karyotype asymmetry and belong to the plesiomorphic type. Clematis integrifolia, C. lanuginosa, and C. viticella are placed more on the top or the right of the graph, reflecting higher karyotype asymmetry, and belong to the derived type.
Based on the karyotype parameters (Table 3), Euclidian distances were calculated (Table 4), and clustering analysis allowed the stablishment of three groups (Fig. 4). The first group is characterized as karyotype 2A and the lowest index of karyotypic asymmetry, which comprises four climbing species namely C. otophora, C. shenlungchiaensis, C. crispa, and C. fusca var. violacea. The second group is characterized as karyotype 2A and highest index of karyotypic asymmetry. Three climbing species, C. lanuginosa, C. viticella, C. lasiandra, and C. florida var. plena, comprise this group. The third group comprises two climbing species (C. henryi and C. japonica) and one erect species (C. integrifolia), characterized by a normal index of karyotypic asymmetry and karyotype 2B.
Discussion
In this study, karyotype morphology and chromosome number of 11 species were reported for the first time. Our results demonstrate that these species are diploid with a constant number of chromosomes (2x = 16) basic number n = 8, which is in agreement with previous reports (Gong et al. 1985; Zhang and He 1990; Zhang and He 1991).
The average arm ratio ranges from 1.80 to 3.31, and the index of the karyotypic asymmetry ranges from 58.99 to 63.60 %. According to theory of Levitzky and Stebbins, C. lanuginosa, C. otophora, C. lasiandra, C. shenlungchiaensis, C. florida var. plena. D., C. fusca var. violacea, C. crispa, and C. viticella belong to Type 2A, a very symmetrical and basal type. Clematis heneryi, C. integrifolia, and C. japonanca belong to Type 2B, a derived type.
The taxonomy of Clematis has relied mainly on characteristics of morphology in various classification systems. In the past years, many scholars carried out extensive studies on Clematis taxonomy. In this paper, we compared the status of the 11 species in the available taxonomic treatments of Clematis classification systems. There are some incongruences between the results of this paper and the classification system of Wang and Li (2005). Wang and Li divided Clematis into four subgenera, 15 sections, and 355 species. The four subgenera included subgen. Cheiropsis, subgen. Clematis, subgen. Virona, and subgen. Atragene. In this paper, according to the cluster analysis, the 11 species of Clematis are divided into three groups. The first group belongs to subgen. Virona. In the second group, C. lanuginosa, C. viticella, and C. florida var. plena belong to subgen. Clematis. Clematis lasiandra belongs to subgen. Virona. The third group belongs to subgen. Virona. Tamura (1995) classification system divides the genus into four subgenera and 17 sections. Clematis fusca var. violacea and C. crispa are in one of the subgenera, and C. florida var. plena and C. viticella are in another subgenus, which is consistent with the results of the present study. In the Johnson classification system (1997), Clematis was divided into 19 sections and 314 species. Grey-Wilson classification system (2000) recognizes 297 species of Clematis which were divided into nine subgenera and 18 sections. In our study, some results were consistent with the two classification systems. At the same time, some points were not coherent because the morphological classification focused on certain characters and ignored the overall degree of similarity or too many subjective factors in the testing process affected the accuracy of the testing results.
In this paper, we explored the karyotype of 11 Clematis species and compared them with available taxonomic treatments. Although this research provided preliminary cytology evidence for the cytotaxonomy, we need to analyze more karyotypes to establish a reasonable cytology classification system which can reflect the relationship among species of Clematis.
References
Dawson MI (1993) Contributions to a chromosome atlas of the New Zealand flora—31 Clematis (Ranunculaceae). NZ J Bot 31:91–96
Dennis WM (1976) Chromosome morphology of Clematis, subsection Viornae (Ranunculaceae). Can J Bot 54:1135–1139
Desal N, Kawalkar H, Dixit G (2012) Biosystematics and evolutionary studies in Indian Drimia species. J Syst Evol 50:512–518
Ebrahim F, Pakniyat H, Arzani A, Rahimmalek M (2012) Karyotype analysis and new chromosome number reports in Achillea species. Biologia 67:284–288
Gong WZ, Long YY, Li MX (1985) Karyotype studies on clematis from Beijing China. J Wuhan Bot Res 3:371–379
Grey-Wilson C (2000) Clematis the genus. Oregon Timber Press, Portland
Langlet O (1927) Beiträge zur Zytologie der Ranunculaceen. Sv Bot Tids 21:1
Levan A, Fredga K, Sandberg A (1964) Nomenclature for centromeric position on chromosomes. Hereditas 52:201–210
Li MX, Chen RY (1985) A suggestion on the standardization of karyotype analysis in plants. J Wuhan Bot Res 3:297–302
Liu HM, Cheng YJ, Lu GE, Li L, Wu FZ (2010) Karyotype features of 17 species of Spiraea and karyotype parameters analysis. Acta Hortic Sin 27:1456–1462
Meurman O, Therman E (1939) Studies in the chromosome morphology and structural hybridity in genus Clematis. Cytologia 10:1–2
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold, London, pp 87–89
Tamura M (1987) A classification of genus Clematis. Acta Phytotaxon Geobot 38:33–44
Tamura M (1995) Archiclematis and Clematis. In: Hiepko P (ed) DieNatürlichen Pflanzenfamilien. Zwei. Aufl. 17a (4). Duncker and Humblot, Berlin, pp 366–387
Wang WT, Bartholomew B (2001) Clematis. In: Wu Z-Y and Raven P (eds) Flora of China, vol 6. Beijing: Science Press, St. Louis, p 97–165
Wang WT, Li LQ (2005) A new system of classification of the genus Clematis (Ranunculaceae). Acta Phytotaxon Sin 43:431–488
Xie L, Wen J, Li LQ (2011) Phylogenetic analyses of Clematis (Ranunculaceae) based on sequences of nuclear ribosomal ITS and three plastid regions. Systematic Botany 36:907–921 (15)
Yang QE (1998) Does Actaea asiatica have the most symmetric and primitive karyotype in the Ranunculaccae? Acta Phytotax Sin 36:490–495
Zhang YL, He SY (1990) Chromosome studies on 6 species of Clematis in China. J Wuhan Bot Res 8:115–121
Zhang YL, He SY (1991) Chromosome studies on 7 species of Clematis in China. J Wuhan Bot Res 9:107–113
Acknowledgments
We would like to thank Wang LP who provided technical assistance and Wu DD who gave us helpful suggestions. This study was supported by A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sheng, L., Ji, K. & Yu, L. Karyotype analysis on 11 species of the genus Clematis . Braz. J. Bot 37, 601–608 (2014). https://doi.org/10.1007/s40415-014-0099-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-014-0099-5