Abstract
The study examines the karyotype and somatic chromosome numbers of five species of Murdannia Royle (Commelinaceae). The karyotype details of the two species namely M. blumei and M. lanceolata endemic to India are reported for the first time. The karyotype formula of the examined species are: M. blumei - 2n (36) = 1M+12m+5sm; M. lanceolata - 2n (20) = 2m+8sm; M. crocea subsp. ochracea - 2n (36) =7m+8sm+3st; M. spirata - 2n (40) = 9m+8sm+3st and M. triquetra - 2n (40) = 16m+4sm. Further details on karyomorphology including estimates of asymmetry indices, total form percent, ratio of mean length of short arms to long arms, intrachromosomal / interchromosomal asymmetry indices, and centromeric indices are provided. The karyomorphological parameters thus analysed suggest that M. blumei and M. lanceolata fit into the 2A category while M. crocea subsp. ochracea, M. spirata and M. triquetra belonged to the 2B category of Stebbins’ classification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Murdannia Royle is one of the largest genera of family Commelinaceae consisting of about 60 species worldwide [13, 32]. It is one of the only six genera of the family that has native species in both the Old World and the New World [11], with its centre of diversity in India [2, 32] representing 52% of the global diversity. Twenty four species of Murdannia have been studied for chromosome number till date [39].
Previous reports indicated the basic chromosome numbers, x = 6, 9, 10, 11, 12 [6, 18, 23, 41, 35, 24, 33, 34] for Murdannia. Different authors reported various numbers: 2n = 18 [18, 42], 40 [15] and 42 [33] for M. edulis (Stokes) Faden, 2n = 20 for M. assamica Nampy & Ancy [29], M. dimorpha (Dalzell) G.Brückn. [44, 18], M. esculenta (Wall. ex C.B.Clarke) R.S.Rao & Kammathy [18], M. hookeri (C.B.Clarke) G.Brückn. [19], M. nudiflora (L.) Brenan [1, 5, 38, 17, 25] and M. striatipetala Faden [29], 2n = 36 for M. crocea subsp. ochracea (Dalzell) Faden [35, 36]. The smallest chromosome number, n = 6 is reported for M. semiteres (Dalzell) Santapau [19, 44] while n = 12 also reported for the same species as well as M. juncoides (Wight) R.S.Rao & Kammathy [19]. An uncommon number, 2n = 22 was reported for M. gigantea (Vahl) G.Brückn. by Rao et al. [36] and Panigrahi and Kammathy [30]. Rao et al. [36] described 2n = 64 and 42 for M. japonica (Thunb.) Faden. Most species investigated previously have n = 10 or its multiples [36]. According to Lewis [23], the basic number for Murdannia is x = 5, which occurs as a dominant line of x = 10, and basic numbers like x = 9 and 11 may have been formed subsequently from the dominant line, x = 10. M. zeylanica exhibits a bimodal complement with an essentially equal number of chromosomes of two sizes [10].
During our study we found that karyomorphological data of some species of Muradannia are not yet reported. In this study, the karyomorphological analysis of M. blumei (Hassk.) Brenan, M. lanceolata (Wight) Kammathy, M. crocea subsp. ochracea, M. spirata (L.) G.Brückn. and M. triquetra is done, of which, M. blumei and M. lanceolata are studied for the first time.
Materials and methods
The samples were collected from different parts of India and grown in the Calicut University Botanical Garden (details provided in Table 1). The voucher specimens were deposited in the Calicut University Herbarium (CALI).
Karyomorphological studies were done using mitotic squash preparation. Five metaphase cells were counted for finalizing the chromosome number for each species. Root tips of 5–8 mm length were pre-treated in saturated solution of para-dichloro benzene (PDB) mixed with 1% saponin (1:0.01) at 12–16℃ for 3 h, followed by washing in distilled water and fixed in modified Carnoy’s fluid (4 chloroform: 3 ethanol: 1 glacial acetic acid) at room temperature for about 3 h. Root tips were hydrolysed in 1.2 M HCl for 15–20 min at room temperature. They were then stained with saturated solution of aceto-orcein. Micro slides were prepared in 45% glacial acetic acid and photographs were taken with a DMC 4500 camera attached to DM 2000 compound microscope (Leica, Switzerland). Chromosomes were identified and classified based on the length of chromosome, and centromeric indices according to Levan et al. [22]. Number of chromosomes was finalized by counting five somatic cells and karyotype analyses were based on three mitotic metaphase preparations of each species. Karyotypic formula was based on position of centromere and number of chromosomes [22]. Total form percentage (TF%) [16], dispersion index (DI) [21], karyotype asymmetry index (As K%) [3], syi index [14], rec index [46] intrachromosomal asymmetry index (A1), interchromosomal asymmetry index (A2) [48] and asymmetry index (AI) [31] were calculated.
Results
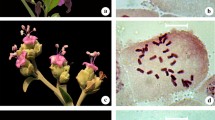
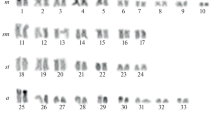
Murdannia blumei is easily recognized by its axillary lilac flowers, hardly exerted from the sheath, and biseriate arrangement of seeds in each capsule locule (Fig. 1a). It has a somatic chromosome number of 2n = 36 (Fig. 1b). The shortest chromosome was 1.03 μm and the longest 1.97 μm. The average chromosome length (ACL) was 1.50 μm and the total haploid chromosome length 27.09 μm. M. blumei had 26 metacentric (M and m) and 10 submetacentric (sm) chromosomes with a karyotype formula of 2M+24m+10sm. The karyogram is shown in Fig. 2a.
Murdannia lanceolata is distinguished by its quite larger flowers and numerous small seeds arranged biseriately in each capsule locule (Fig. 1c). It showed 2n = 20 as somatic chromosome number (Fig. 1d). The range of chromosome length was 1.79–3.15 μm. The average chromosome length (ACL) was 2.3 μm and the total haploid chromosome length of 23 μm. It had 4 metacentric (m) and 16 submetacentric (sm) chromosomes with a karyotype formula of 4m+16sm. The karyogram is given in Fig. 2b.
Murdannia crocea subsp. ochracea is endemic to South India and readily identified by its ochre flowers, obovate–orbicular petals and biseriately arranged seeds in the capsule locule (Fig. 1e). It had 2n = 36, and a haploid number, n = 18 (Fig. 1f). The range of chromosome length lay between 1.17 and 4.56 μm while the average chromosome length was 2.72 μm and the total haploid chromosome length 49.06. Median (m), sub-median (sm) and sub-terminal (st) centromeric chromosomes are with a karyotype formula 14m+16sm+6st. The karyogram is given in Fig. 2c.
Murdannia spirata is a widespread species in India, easily recognized by its axillary or terminal thyrse inflorescence with 1–3 alternate cincinni having lilac to lavender flowers (Fig. 1g) while M. triquetra is distributed from Assam to China and Indo-China. The latter is readily identified by its white–lavender petals, uniseriate arrangement of seeds in the capsule locule and a recurved fruiting pedicel (Fig. 1i). Both the species showed 2n = 40 with a basic chromosome number x = 10 and haploid number n = 20 (Fig. 1 h and 1j). The range of chromosome length of M. spirata was between 1.74 and 3.58 μm and the average chromosome length 2.30 μm while the total haploid chromosome length 46.16. Median (m), sub-median (sm) and sub-terminal (st) centromeric chromosomes are found with a karyotype formula 18m+16sm+6st. M. triquetra has chromosome length ranging from 0.73 to 2.58 μm with an average of 1.63 μm and the total haploid chromosome length 32.64. Median (m) and sub-median (sm) centromeric chromosomes were found with a karyotype formula 32m+8sm. Their karyograms are given in Fig. 2d and e, respectively.
Considering Stebbins [45] definition of symmetry, the karyotypes of M. blumei and M. lanceolata fit into the 2A category while M. crocea subsp. ochracea, M. spirata and M. triquetra belonged to the 2B category as the relation between the longest and shortest arms was lower than 2:1.
Discussion
Chromosome numbers of 24 species of Murdannia have been reported so far [39]. The size of chromosomes in this genus is particularly small and the numbers vary from n = 6 to 30 [10]. Most species studied previously showed x = 10 or its multiple, though other number such as x = 9, 11, 12 had also been recorded [6, 18, 35, 23, 24, 26, 39].
In this work, we reported the chromosome numbers of five species of Murdannia from the root tips: 2n = 36 for M. blumei and M. crocea subsp. ochracea, 2n = 20 for M. lanceolata and 2n = 40 for M. spirata and M. triquetra. Based on our study, in an evolutionary line there are eight species with axillary fascicled inflorescence reported from India [2, 8, 28]. Among them, M. blumei, M. crocea subsp. ochracea, M. versicolor and M. pauciflora have somatic chromosome numbers n = 18, 18, 9 and 9, respectively with basic chromosome number x = 9. Whereas, M. triquetra, M. lanuginosa and M. keisak showed, n = 20, 10 and 10, respectively, with a basic chromosome number x = 10. M. sanjappae is another axillary flowered species whose chromosome number is not available. From an evolutional conception it is surmised that low basic numbers had given rise to higher ones, and the taxa with variable chromosome number are considered young and still in evolutionary flux [40, 47]. In another evolutionary line M. lanceolata and M. spirata with terminal or axillary thyrse having one or two opposite cincinni grouped with M. dimorpha and M. striatipetala [2]. The chromosome number for M. lanceolata is reported for the first time (2n = 20). Different chromosome counts were reported for M. spirata: 2n = 18 [35]; 2n = 20, 40 [37]; 2n = 40 [18] and 2n = 24 [20] while our results corroborate with Kammathy and Rao [18]. For M. dimorpha and M. striatipetala, the chromosome number is 2n = 20 [18, 29, 44]. Thus, basic chromosome number is concluded as x=10 for this group, with two exceptions n = 9 and 12 for M. spirata.
The karyotype analysis revealed that all the species have small sized chromosomes. Among them the largest chromosome size was observed in M. crocea subsp. ochracea (2.72 μm) and the smallest in M. blumei (1.50 μm). Karyotypes of three species possess predominantly median chromosomes and remaining possesses submedian chromosomes. M. crocea subsp. ochracea and M. spirata showed subtelocentric chromosomes. The primitiveness of karyotype is expressed by the presence of symmetrical karyotype, having longer chromosomes, median centromere with chromosome arms of equal size and low basic chromosome number, while more advanced karyotype depicts asymmetrical karyotype having, shorter chromosomes, submedian or other centromere, unequal length of chromosome arms and higher basic chromosome numbers [45, 47].
The karyotype asymmetry was assessed based on fourteen parameters given in Table 2. In which TF%, Syi, A1, A2 and AI have been formulated to evaluate the variation in centromere position in a chromosome complement. The total chromosome length (TCL) varied from 23 to 49.06 μm and the highest and the lowest values are present in M. lanceolata and M. crocea subsp. ochracea respectively. The A1 index was used to estimate karyotype asymmetry for the relationships between the chromosomal arms, with values ranging from zero to one. The A1 index is unaffected by the number of chromosomes or their size. The A1 index obtained for M. blumei is 0.29, for M. lanceolata 0.47, M. crocea subsp. ochracea 0.43, M. spirata 0.42 and M. triquetra 0.26. The A2 index is also represented as 0.17, 0.20, 0.30, 0.22 and 0.30, respectively. The lowest value for A1 and A2 indicates there is a small difference in length of chromosome arms and least variation in chromosome length, whereas highest value of A1 and A2 indicates greatest difference in length of chromosome arm and large variation in chromosome length. Low values of TF%, Syi and Rec indices and increased value of AsK%, A1, A2, AI indicates asymmetry of chromosomes. M. crocea subsp. ochracea has lower TF% and higher AsK% (34.49, 65.50 respectively), indicating an asymmetrical karyotype than the others (Table 2). This information is also supported by high A1, A2 and AI values indicating the existence of asymmetrical karyotypes. M. blumei which showed high value for TF%, Syi index, Rec index and low value for A1, A2, AI and DI indicated the symmetry of chromosomes than the rest. Dispersion index which facilitates quantitative gradation between the closely related karyotypes, that fall under the same class of karyotype asymmetry and its higher value indicate higher level of karyotype specialization [21]. M. crocea subsp. ochracea, M. spirata and M. triquetra falls under 2B category of asymmetry. M. triquetra showed highest DI value than M. crocea subsp. ochracea and M. spirata. Based on the DI value, M. triquetra and M. crocea subsp. ochracea are highly specialized in their karyotype. M. spirata also showed asymmetry in their karyotype formula, due to the presence of subterminal chromosomes and this information is supported by all the morphometric analysis. The information generated from the study of karyotype constitution, chromosome number when combined with morpho-taxomonic features could help differentiate and elucidate species affinities, for which data on more species is desirable to arrive at a meaningful inferences [21, 47].
References
Alam N, Sharma AK. Trends of chromosome evolution in family Commelinaceae. Nucleus. 1984;27:231–41.
Ancy AA, Nampy S. Murdannia Royle (Commelinaceae) of India. Centre for Research in Indigenous Knowledge, Science & Culture (CRIKSC), D-37, Jawahar Nagar, Calicut-673006, Kerala, India; 2015.
Arano H. Cytological studies in subfamily Carduoideae (Compositae) of Japan. IX. The karyotype analysis and phylogenetic considerations on Pertya and Ainsliaea. Bot Mag Tokyo. 1963;76:32–39.
Bai L, Kuriachan PI, Ninan CA. Chromosome Number Reports LXXXII. Taxon. 1984;33:126–34.
Bhattacharya B. Cytological studies on some Indian members of Commelinaceae. Cytologia. 1975;40:285–99.
Briggs BG. Chromosome numbers of some Australian monocotyledons. Contrib New S Wales Nat Herb. 1966;4:24–34.
Chimphamba BB. Chromosome numbers in Malawian Commelinaceae. Bot J Linn Soc. 1973;66:303–6.
Chowdhuri A, Chowdhuri M, Das AP. Murdannia keisak (Hasskarl) Handel-Mazzetti (Commelinaceae): a new record for India. Pleione. 2015;9:531–4.
Darlington CD, Wylie AP. Chromosome atlas of flowering plants. 2nd ed. London: George Allen and Unwin Ltd.; 1956.
Faden RB, Suda Y. Cytotaxonomy of Commelinaceae: chromosome counts of some African and Asiatic species. Bot J Linn Soc. 1980;81:301–25.
Faden RB. Commelinaceae. In: Kubitzki K, editor. The families and genera of vascular plants. 4. Berlin: Springer-Verlag; 1998. pp. 109–28.
Fujishima H. Karyological studies in Commelinaceae II. Karyotypes of Pollia japonica and Aneilema keisak. Bot Mag. 1970;83:21–26.
Govaerts R, Faden RB. World checklist of selected plant families. The Board of Trustees of the Royal Botanical Gardens, Kew. 2016. http://apps.kew.org/wcsp/ Accessed: 27 October 2020.
Greilhuber J, Speta F. C-banded karyotypes in the Scilla hohenackeri group, S. persica and Puschkinia (Liliaceae). Plant Syst Evol. 1976;126:149–188.
Guervin C, Le Coq C. Caryologie des Commelinacees: Application a quelques problems relatifs a leur evolution. Rev Cytol Biol Veg. 1966;29:267–328.
Huziwara Y. The karyotype analysis in some genera of Compositae X: the chromosome of some European species of Aster. Bot Mag. 1962;75:143–50.
Jones K, Jopling C. Chromosomes and classification of the Commelinaceae. Bot J Linn Soc. 1972;65:129–62.
Kammathy RV, Rao RS. Notes on Indian Commelinaceae. II Cytological observations. Bull Bot Surv India. 1961;3:167–9.
Kammathy RV, Rao RS. Notes on Indian Commelinaceae. IV: Cytotaxonomic observations. Bull Bot Surv India. 1964;6:1–6.
Kaur H. Cytological studies on some members of Commelinaceae Mirb. from Kangra valley (Himachal Pradesh) with a short summary of karyological data on the analyzed genera. Acta Biol Cracov Ser Bot. 2018;1:95–103.
Lavania UC, Srivastava S. Quantitative delineation of karyotype variation in Papaver as a measure of phylogenetic differentiation and origin. Curr Sci. 1999;77:429–435.
Levan A, Fredga K, Sandberg AA. Nomenclature for centromeric position of chromosomes. Hereditas. 1964;52:201–20.
Lewis WH. Meiotic chromosomes in African Commelinaceae. SIDA, Contributions to Botany, 1964;1:274–293.
Mehra PN, Sachdeva SK. In: IOPB chromosome reports. XXXIII Taxon. 1971;20:609–14.
Mitra K, Datta N. In IOPB chromosome number reports XIII. Taxon. 1967;16:445–61.
Mohanty BD, Ghosh PD, Maity S. Chromosome analysis in cultured cells of barley (Hordeum vulgare L.): structural alterations in chromosomes. Cytologia. 1991;56:191–7.
Morton JK. A revision of the genus Aneilema R.Br. (Commelinaceae) with a cytotaxonomic account of West African species. J Linn Soc Lond Bot. 1966;59:431–78.
Naik MC, Rao BR. A new species of dew flower Murdannia sanjappae (Commelinaceae) from Andaman Islands, India. J Threat Taxa. 2017;9:10909–13.
Nair STK, Nampy S. New report on karyotypes in three endemic species of Murdannia Royle (Commelinaceae) from India. Cytologia. 2021;86:207–13.
Panigrahi G, Kammathy RV. Studies in the cytomorphology of Aneilema sensu lato in eastern India. Proc Ind Sci Congr Roorkee, 1961;Part 4: 13–14.
Paszko AA. Critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol. 2006;258:39–48.
Pellegrini MOO, Faden RB, Almeida RF. Taxonomic revision of Neotropical Murdannia Royle (Commelinaceae). PhytoKeys. 2016;4:35–78.
Peng CI, Chen YJ, Wang JC. Notes on Commelinaceae of Taiwan: Cyanotis and Belosynapsis, and the rediscovery of Murdannia edulis. Biol Bull Natl Taiwan Norm Univ. 2000;35:77–93.
POWO. Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. 2020. http://www.plantsoftheworldonline.org/ Accessed on 2 November 2020.
Raghavan RS, Rao RS. Cytological observations on the Indian species of Commelinaceae. Curr Sci. 1961;30:310–1.
Rao RS, Kammathy RV, Raghavan RS. Cytotaxonomic studies on Indian Commelinaceae: A review. J Linn Soc Bot. 1968;60:357.
Rao RS, Panigrahi G, Kammathy RV. Cytotoxonomic studies on Indian Commelinaceae. Proc Ind Sci Congr Mumbai. 1960:366.
Renugadevi K, Sampathkumar R. On the karyomorphological delineations in some taxa of Commelinaceae. J Cytol Genet. 1986;21:115–32.
Rice A, Glick L, Abadi S, Einhorn M, Kopelman NM, Salman-Minkov A, Mayzel J, Chay O, Mayrose I. The chromosome count Database (CCDB)- a community resource of plant chromosome number. New Phytol. 2015;206:19–26.
Sharma A. Taxonomy as related to genetic diversity in plants. J Indian Bot Soc. 1990;69:1–3.
Shetty BV, Subramanyam K. Cytological studies in Commelinaceae. Nucleus. 1962;5:39–50.
Shetty BV, Subramanyam K. In IOPB chromosome reports XXXIV. Taxon. 1971;20:785–97.
Sokolovskaya AA. Geograficheskoe rasprostranenie polyploidnykh vidov rasteniy in scledovanie flory primorskogo kraya. Vestrikleninger Univ Ser Biol. 1966;3:96–106.
Sreenath KP, Rama G. Chromosome Number Reports LXXXIII. Taxon. 1984;33:351–4.
Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold (Publishers) Ltd; 1971.
Venora G, Blangiforti S, Castiglione MR, Pignone D, Losavio F, Cremonini R. Chromatin organization and computer aided karyotyping of Triticum durum Desf. Cv Timilia Caryologia. 2002;55:91–8.
Vimala Y, Lavania S, Lavania UC. Chromosome change and karyotype differentiation–implications in speciation and plant systematics. Nucleus. 2021;64:1–22.
Zarco CR. New method for estimating karyotype asymmetry. Taxon. 1986;35:526–30.
Acknowledgements
Authors are grateful to Head, Department of Botany, University of Calicut for facilities and Sreekutty T.K. is grateful to CSIR- HRDG, New Delhi for fellowship.
Funding
Funding was provided by CSIR-HRDG, New Delhi (File No: 09/043(0193)/2019-EMR-1).
Author information
Authors and Affiliations
Contributions
All authors are univocally approved for the work. STK collected the plant specimens, carried out the experiments and prepared manuscript. SN designed the study, supervised experimentation, helped in data interpretation and revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that there is no conflict of interest.
Consent for publication:
Authors approve of publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding Editor: Umesh C Lavania; Reviewers: Anath Bandhu Das, Surochita Basu, M M Lekhak, Geeta Sharma
Rights and permissions
About this article
Cite this article
Nair, S.T.K., Nampy, S. Karyomorphological analysis of five species of Murdannia Royle (Commelinaceae), including two endemics to India. Nucleus 65, 187–192 (2022). https://doi.org/10.1007/s13237-022-00386-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-022-00386-3