Abstract
Background
Up to now, many publications about the Chinese population have evaluated the correlation between interleukin-10 (IL-10) −1082 and −592 polymorphisms and persistent hepatitis B virus (HBV) infection. However, the results remain inconclusive. In order to resolve this conflict, a meta-analysis was performed.
Methods
Seven studies were included and dichotomous data are presented as the odds ratio (OR) with a 95% confidence interval (CI).
Results
The results of our study suggest that carriers of the IL-10 −592A allele were more likely to clear HBV spontaneously in the Chinese pooled population (A vs. C: OR = 0.799, 95% CI = 0.678–0.941, P = 0.007; AC vs. AA: OR = 1.343, 95% CI = 1.017–1.684, P = 0.011; AA vs. AC + CC: OR = 0.736, 95% CI = 0.594–0.912; AA + AC vs. CC: OR = 0.588, 95% CI = 0.408–0.848, P = 0.004) and the IL-10 −1082A allele was associated with significantly reduced persistent HBV infection risk in Chinese (A vs. G: OR = 0.701, 95% CI = 0.494–0.996, P = 0.047; AA vs. GG + GA: OR = 0.684, 95% CI = 0.476–0.982, P = 0.040).
Conclusions
Persistent HBV infection susceptibility is associated with the gene polymorphism IL-10 −1082GA in the Chinese population and the clearance of HBV is associated with the gene polymorphism IL-10 −592CA in the Chinese population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) is the most common cause of acute and chronic liver disease worldwide. Thus, HBV infects more than 350 million people worldwide, especially in several areas of Asia and Africa [1]. HBV is transmitted via contact with infected body fluids, including blood, saliva, and semen. Approximately 5% of HBV-infected adults develop chronic hepatitis B [2], which may result in liver cirrhosis or hepatocellular carcinoma. The risk of HBV persistence is related to two major factors: host immunological factors and genetic factors [3]. Family studies in China provide some evidence that the host genetic factors affect viral persistence, as higher concordance rates were found for HBeAg persistence in identical compared with non-identical twins [4]. Recent studies have shown that cytokine genetic polymorphisms have an association with the development of chronic HBV infection and the progression of the infection [5–8].
Interleukin-10 (IL-10) is an important immunoregulatory cytokine in human beings. It is involved in the regulation of inflammatory responses through direct influence over tumor necrosis factor production [9]. The IL-10 gene region was examined for an association with HBV infection outcome, either chronic or recovered, including three biallelic polymorphisms at positions −1082 and −592 from the transcription start site which have been reported to affect IL-10 expression [10, 11]. Recent studies about the Chinese population have shown that IL-10 −592 and −1082 are associated with increased risk of persistent HBV infection or the clearance of HBV infection [12, 13]. However, other studies have shown conflicting results [14].
Therefore, in this meta-analysis, the purpose was to quantitatively assess the association of IL-10 −592 and IL-10 −1082 genetic polymorphisms with persistent HBV infection cases in the Chinese population. Meta-analysis is a powerful method for quantitatively summarizing the results from different studies. One of the advantages is to increase the sample size, which may reduce the probability that random error will produce false-positive or false-negative association [15].
Materials and methods
Literature search strategy
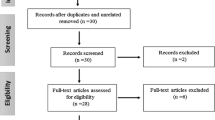
The Medline, PubMed, Embase, Web of Science, and Chinese Biomedical Literature Database on disc (CBM disc) databases were searched (the last search was updated on January, 19, 2010, using the search terms: “hepatitis B” or “HBV”, “polymorphism”, “interleukin-10” or “IL-10”). All searched studies were retrieved and their bibliographies were checked for other relevant publications. Review articles and bibliographies of other relevant identified studies were hand-searched in addition to eligible studies. Only published studies with full-text articles were included. When more than one of the same patient population was included in several publications, only the most recent or complete study was used in this meta-analysis. A flow diagram of the study selection process is shown in Fig. 1.
Inclusion and exclusion criteria
The inclusion and exclusion criteria were drawn up on the basis of discussion. The inclusion criteria were as follows: (1) the diagnosis criteria are as follows: individuals who spontaneously recovered without treatment and had serologic evidence of prior infection (antibodies against hepatitis B core antigen, anti-HBc, and against HBsAg, anti-HBs) with HBsAg undetectable at two time points separated by a minimum of 6 months were self-limiting infection controls, vaccinated patients were excluded; the patients who had been positive for HBV surface antigen (HBsAg) for more than 6 months were included as persistent HBV infection cases; the patients who were without detectable HBV infection were defined as healthy controls; (2) the design type of the study was a case–control study; (3) the study aimed to examine the association between the IL-10 −592 polymorphism and clearance and/or susceptibility of persistent HBV infection, or the study aimed to examine the association between the IL-10 −1082 polymorphism and clearance and/or susceptibility of persistent HBV infection; (4) the study provided the number of persistent HBV infection cases or controls and the frequency of IL-10 −592 or IL-10 −1082 genotypes; (5) the genotype was tested in controls and cases to ensure their fitting with the Hardy–Weinberg equilibrium (HWE). The exclusion criteria were as follows: (1) the study did not fit the diagnosis criteria; (2) the study was conducted on animals; (3) the study was not a case–control study; (4) the study reported useless data; (5) studies in which the control and case groups deviated from the HWE; (6) the study population was not Chinese.
Data extraction
All of the data were extracted independently by two reviewers (T.-C. Zhang and L.-Z. Zhang) according to the prespecified selection criteria. Disagreement was resolved by discussion. The following data were extracted: study design and period, statistical methods, population, number of persistent HBV infection cases (including asymptomatic carriers and chronic liver diseases), self-limiting infection controls and health controls studied, and the study results.
Statistical analysis
Allele frequencies at the IL-10 −1082GA and IL-10 −592CA single nucleotide polymorphisms (SNPs) from the respective studies were determined by the allele counting method. A χ2 test was used to determine if observed frequencies of genotypes conformed to the HWE. The statistical analysis was conducted using Stata 9.0 (StataCorp, College Station, TX) and a P-value ≤0.05 was considered to be statistically significant. Dichotomous data are presented as the odds ratio (OR) with a 95% confidence interval (CI). Statistical heterogeneity was measured using the Q-statistic (P ≤ 0.10 was considered to be representative of statistically significant heterogeneity). We also quantified the effect of heterogeneity using the I 2 statistic, which measures the degree of inconsistency in the studies by calculating what percentage of the total variation across studies is due to heterogeneity rather than by chance. A fixed effects model was used when there was no heterogeneity of the results of the trials; otherwise, the random effects model was used. For dichotomous outcomes, patients with incomplete or missing data and small sample studies were included in the sensitivity analyses by counting them as treatment failures. To establish the effect of clinical heterogeneity between studies on the meta-analyses’ conclusions, subgroup analysis was conducted on the basis of race. Several methods were used to assess the potential for publication bias. Visual inspection of asymmetry in funnel plots was conducted. Begg’s rank correlation method and Egger’s weighted regression method were also used to statistically assess the publication bias (P ≤ 0.05 was considered to be representative of statistically significant publication bias).
Main results
Characteristics of studies
There were seven relevant studies evaluating IL-10 −1082GA or IL-10 −592CA SNPs found in our search, and the characteristics of each study are presented in Tables 1 and 2.
Quantitative data synthesis
The aim of this paper was to use the meta-analysis method to quantitatively summarize the results from the selected different studies, so we compared the persistent HBV infection cases with healthy controls, persistent HBV infection cases with self-limiting infection controls. We also compared persistent HBV infection cases with healthy controls to discover the relation of IL-10 −1082GA and IL-10 −592CA polymorphisms to persistent HBV infection susceptibility, and compared persistent HBV infection cases with self-limiting infection controls to discover the relationship between IL-10 −1082GA and IL-10 −592CA polymorphisms and HBV infection clearance. As the comparison is between two or three genotypes, such as IL-10 −1082AA versus IL-10 −1082GG, we adjusted the test level to 0.0167 (α = 0.05/3).
As Table 3 shows, the IL-10 −1082A allele was associated with significantly reduced persistent HBV infection risk in Chinese (A vs. G: OR = 0.701, 95% CI = 0.494–0.996, P = 0.047; AA vs. GG + GA: OR = 0.684, 95% CI = 0.476–0.982, P = 0.040). As Table 4 shows, the IL-10 −1082GA polymorphisms had no association with the clearance for persistent hepatitis B infection in Chinese.
We found no evidence of an association between IL-10 −592CA SNPs and the susceptibility of persistent hepatitis B infection in the Chinese pooled population (Table 5). As Table 6 shows, the results of our meta-analysis suggest that carriers of the IL-10 −592A allele were more likely to clear HBV spontaneously compared with those carrying the IL-10 −592C allele genotypes in the Chinese pooled population (A vs. C: OR = 0.799, 95% CI = 0.678–0.941, P = 0.007; AC vs. AA: OR = 1.343, 95% CI = 1.017–1.684, P = 0.011; AA vs. AC + CC: OR = 0.736, 95% CI = 0.594–0.912, P = 0.005; AA + AC vs. CC: OR = 0.588, 95% CI = 0.408–0.848, P = 0.004).
Heterogeneity
The heterogeneity was calculated among all studies using the Q-statistic (Q > 0.10) and the I 2 statistic (I = 0.0%); heterogeneity in the overall studies group was not found.
Sensitivity analysis
A single study involved in the meta-analysis was deleted each time to investigate the influence of the individual dataset on the pooled ORs. The corresponding pooled ORs were not materially altered (data not shown), indicating that our results were statistically robust.
Publication bias
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of the literature and we found no asymmetry of the funnel plot, suggesting that there was no publication bias in our meta-analysis.
Discussion
In recent investigations, a number of studies have identified polymorphisms that affect susceptibility to infectious diseases [20, 21]. It is believed that the host genetic factors involving genetic polymorphisms are responsible for the susceptibility and clinical outcomes of infectious diseases [22], because the differences in the susceptibility to infection or severity of disease cannot be solely attributed to the virulence of an organism. The natural outcome of HBV infection varies among individuals. Patients who are positive for HBsAg or HBV DNA for more than 6 months become HBV carriers and it may induce intrahepatic inflammation and, subsequently, the development of persistent (chronic) infection, such as CH, progressive fibrosis, and even liver cancer. However, some cases will never progress to any significant liver disease [23, 24]. Cytokines play an essential role on the pathogenesis of virus-associated hepatitis. Clearance of hepatitis viruses following acute infection is associated with a vigorous cytotoxic T-cell response, partially mediated and augmented through the inhibition of viral replication and gene expression by the proinflammatory and Th1 cytokines [25]. IL-10 is an important immune regulatory cytokine produced mainly in Th2 cells and macrophage cells. IL-10 negatively regulates the expression of cytokines and suppresses proinflammatory cytokine (TNF-, IFN-, IL-6, IL-8, IL-12) production and the antigen-presenting capacity of monocytes/macrophages [24]. Several studies have suggested the association of IL-10 gene polymorphisms with infectious diseases and autoimmune diseases [26, 27].
Many publications about the Chinese population have evaluated the correlation between IL-10 −1082 and −592 polymorphisms and persistent HBV infection, but the results are not uniform. Such inconsistencies are probably induced by inadequate sample content. Some studies suggested that the −1082GG genotype could be associated with susceptibility to chronic HBV infection, while Zhang et al. have shown that no statistically significant differences were observed between patients and controls [19]. Interestingly, Gao et al. suggested that the −1082AA genotype could increase the risk of HBV infection [14], but we found that their study was not fitting with the HWE. Chen et al. advised that the IL-10 −592AA genotype was easy to clear HBV [12], but Zhang et al. showed no association. Therefore, we adopt a meta-analysis method, expanding the samples content to explore the relation of polymorphism at sites −1082 and −592 of the IL-10 gene promoter with susceptibility and clearance to persistent HBV infection only in the Chinese population.
In our study of the relationship between IL-10 −1082GA polymorphisms and persistent HBV infection, our meta-analysis results indicated that there was significantly reduced risk of persistent HBV infection with IL-10 −1082AA genotypes in the Chinese pooled population (A vs. G: OR = 0.701, 95% CI = 0.523–1.024, P = 0.047; AA vs. GG + GA: OR = 0.684, 95% CI = 0.476–0.982, P = 0.040). We believe that the reason for this is that the individuals with IL-10 −1082A (low expression allele) have lower IL-10 secretion than the other genotypes [28], leaving increased local inflammatory cell infiltration in the liver and enhancing the activity of inflammatory cytokines [19], but we did not find IL-10 −1082 to be associated with the clearance of HBV.
The results of the present study suggest that carriers of the IL-10 −592A allele were more likely to clear HBV spontaneously. IL-10 is a regulatory cytokine that potentially inhibits T-cell activation and affects the outcome of HBV infection [29]. The A allele at position 592 in the IL-10 gene promoter is associated with lower IL-10 levels [28] and is more inclined to remove the virus. Chen et al.’s study showed that 592AA were much higher in Chinese than in Caucasians [12]. It seems that the protective effects of these genotypes are overtaken by other risk factors in Chinese.
We analyze the reasons for inconsistent results as the following two points: firstly, the sample size is not large enough, since the proportion of GG genotype in the Chinese population was very small, which may lead to false-positive results; secondly, in some experiments, the sample does not fit the HWE, making the sample representative poorly, which may cause results to bias.
It should be noted that there were some limitations in this study. Firstly, because only published studies were included in the meta-analysis, publication bias may have occurred, even though it was not found by making use of statistical tests. Secondly, as the relevant research articles are scarce in number, our sample size in this study was not large. Thirdly, this meta-analysis was based on unadjusted estimates, while a more precise analysis could be performed if individual data were available. Fourthly, meta-analysis essentially remains with observational study that was subject to the methodological deficiencies of the included studies. Fifthly, for the reason of inclusion and exclusion criteria, there are not adequate studies for us to analyze the population of European, USA, and other countries of Asia. Finally, the major genotypes of HBV in Chinese are B and C, but most of the studies did not analyze them separately, and for the methodological limit of the meta-analysis, we could not solve this shortcoming.
In conclusion, in spite of several limitations mentioned above, this meta-analysis also suggests that the IL-10 −592 polymorphism is associated with the clearance for persistent hepatitis B infection in the Chinese population, and the IL-10 −1082 polymorphism is associated with persistent hepatitis B infection susceptibility in the Chinese population.
References
Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–45.
Vildózola Gonzales H, Salinas JL. Natural history of chronic hepatitis B virus infection. Rev Gastroenterol Peru. 2009;29:147–57.
Wang FS. Current status and prospects of studies on human genetic alleles associated with hepatitis B virus infection. World J Gastroenterol. 2003;9:641–4.
Lin TM, Chen CJ, Wu MM, Yang CS, Chen JS, Lin CC, Kwang TY, Hsu ST, Lin SY, Hsu LC. Hepatitis B virus markers in Chinese twins. Anticancer Res. 1989;9:737–41.
Ben-Ari Z, Mor E, Papo O, Kfir B, Sulkes J, Tambur AR, Tur-Kaspa R, Klein T. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98:144–50.
Miyazoe S, Hamasaki K, Nakata K, Kajiya Y, Kitajima K, Nakao K, Daikoku M, Yatsuhashi H, Koga M, Yano M, Eguchi K. Influence of interleukin-10 gene promoter polymorphisms on disease progression in patients chronically infected with hepatitis B virus. Am J Gastroenterol. 2002;97:2086–92.
Basturk B, Karasu Z, Kilic M, Ulukaya S, Boyacioglu S, Oral B. Association of TNF-alpha −308 polymorphism with the outcome of hepatitis B virus infection in Turkey. Infect Genet Evol. 2008;8:20–5.
Kummee P, Tangkijvanich P, Poovorawan Y, Hirankarn N. Association of HLA-DRB1*13 and TNF-alpha gene polymorphisms with clearance of chronic hepatitis B infection and risk of hepatocellular carcinoma in Thai population. J Viral Hepat. 2007;14:841–8.
Eskdale J, Keijsers V, Huizinga T, Gallagher G. Microsatellite alleles and single nucleotide polymorphisms (SNP) combine to form four major haplotype families at the human interleukin-10 (IL-10) locus. Genes Immun. 1999;1:151–5.
Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, Woo P. Polymorphic haplotypes of the interleukin-10 5′ flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum. 1999;42:1101–8.
Mäurer M, Kruse N, Giess R, Toyka KV, Rieckmann P. Genetic variation at position −1082 of the interleukin 10 (IL10) promoter and the outcome of multiple sclerosis. J Neuroimmunol. 2000;104:98–100.
Chen DQ, Zeng Y, Zhou J, Yang L, Jiang S, Huang JD, Lu L, Zheng BJ. Association of candidate susceptible loci with chronic infection with hepatitis B virus in a Chinese population. J Med Virol. 2010;82:371–8.
Li C, Zhi-Xin C, Li-Juan Z, Chen P, Xiao-Zhong W. The association between cytokine gene polymorphisms and the outcomes of chronic HBV infection. Hepatol Res. 2006;36:158–66.
Gao QJ, Liu DW, Zhang SY, Jia M, Wang LM, Wu LH, Wang SY, Tong LX. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World J Gastroenterol. 2009;15:5610–9.
Wang J, Pan HF, Hu YT, Zhu Y, He Q. Polymorphism of IL-8 in 251 allele and gastric cancer susceptibility: a meta-analysis. Dig Dis Sci. 2010;55:1818–23.
Tseng LH, Lin MT, Shau WY, Lin WC, Chang FY, Chien KL, Hansen JA, Chen DS, Chen PJ. Correlation of interleukin-10 gene haplotype with hepatocellular carcinoma in Taiwan. Tissue Antigens. 2006;67:127–33.
Peng XM, Huang YS, Ma HH, Gu L, Xie QF, Gao ZL. Interleukin-10 promoter polymorphisms are associated with the mode and sequel of HBeAg seroconversion in patients with chronic hepatitis B virus infection. Liver Int. 2006;26:326–33.
Yan Z, Tan W, Zhao W, Dan Y, Wang X, Mao Q, Wang Y, Deng G. Regulatory polymorphisms in the IL-10 gene promoter and HBV-related acute liver failure in the Chinese population. J Viral Hepat. 2009;16:775–83.
Zhang PA, Li Y, Yang XS. Associated study on interleukin 10 gene promoter polymorphisms related to hepatitis B virus infection in Chinese Han population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2006;23:410–4.
Helminen M, Lahdenpohja N, Hurme M. Polymorphism of the interleukin-10 gene is associated with susceptibility to Epstein–Barr virus infection. J Infect Dis. 1999;180:496–9.
Mozzato-Chamay N, Mahdi OS, Jallow O, Mabey DC, Bailey RL, Conway DJ. Polymorphisms in candidate genes and risk of scarring trachoma in a Chlamydia trachomatis-endemic population. J Infect Dis. 2000;182:1545–8.
Powell EE, Edwards-Smith CJ, Hay JL, Clouston AD, Crawford DH, Shorthouse C, Purdie DM, Jonsson JR. Host genetic factors influence disease progression in chronic hepatitis C. Hepatology. 2000;31:828–33.
Kim YJ, Viana AC, Curtis KM, Orrico SR, Cirelli JA, Mendes-Junior CT, Scarel-Caminaga RM. Association of haplotypes in the IL8 gene with susceptibility to chronic periodontitis in a Brazilian population. Clin Chim Acta. 2010;411:1264–8.
Guidotti LG, Guilhot S, Chisari FV. Interleukin-2 and alpha/beta interferon down-regulate hepatitis B virus gene expression in vivo by tumor necrosis factor-dependent and -independent pathways. J Virol. 1994;68:1265–70.
Guidotti LG, Ando K, Hobbs MV, Ishikawa T, Runkel L, Schreiber RD, Chisari FV. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc Natl Acad Sci USA. 1994;91:3764–8.
Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25–36.
Romero R, Lavine JE. Cytokine inhibition of the hepatitis B virus core promoter. Hepatology. 1996;23:17–23.
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8.
Brooks DG, Trifilo MJ, Edelmann KH, Teyton L, McGavern DB, Oldstone MB. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–9.
Acknowledgments
This work was supported by grants from the Ministry of Science and the 973 pre-basic research project “Study on genetic epidemiology of chronic hepatitis B” (2009CB526411) and the National and Provincal Natural Science Foundation of China (30771849, 30972530, 090413133).
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Additional information
T.-C. Zhang and L.-Z. Zhang contributed equally to this work and should be considered as co-first authors.
Rights and permissions
About this article
Cite this article
Zhang, TC., Pan, FM., Zhang, LZ. et al. A meta-analysis of the relation of polymorphism at sites −1082 and −592 of the IL-10 gene promoter with susceptibility and clearance to persistent hepatitis B virus infection in the Chinese population. Infection 39, 21–27 (2011). https://doi.org/10.1007/s15010-010-0075-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s15010-010-0075-3