Abstract
Being encoded by hepatitis B, hepatitis B X (HBx) protein plays crucial roles in hepatitis B-related hepatocellular carcinoma (HCC) occurrence, development, and metastasis. miRNAs also function in the progression of hepatitis B-related HCC. Hence, the objective of this study was to explore the impacts of miR-3677-3p on tumor progression and sorafenib resistance in hepatitis B-related HCC and the related underlying mechanisms. Our research revealed that miR-3677-3p and FOXM1 was up-regulated and FBXO31 was down-regulated in HBV+ HCC cells and tumor tissues from nude mice. After miR-3677-3p overexpression, cell proliferative, invasive, and migrating potentials and stemness-related protein (CD133, EpCAM, and OCT4) levels were enhanced, and cell apoptosis was reduced in Huh7 + HBx/SR cells and HepG2.2.15/SR cells. Besides, miR-3677-3p promoted the drug resistance of Huh7 + HBx/SR cells and HepG2.2.15/SR cells to sorafenib and lifted IC50. In vivo experiments, miR-3677-3p down-regulation suppressed the tumor growth in the hepatitis B HCC nude mouse models. Mechanistically, miR-3677-3p targeted and negatively-regulated FBXO31 and FBXO31 could enrich FOXM1 protein. miR-3677-3p down-regulation or FBXO31 overexpression promoted the ubiquitylation of FOXM1. In a word, miR-3677-3p bound to FBXO31 and inhibited FBXO31 expression, thus curtailing the ubiquitylation degradation of FOXM1, contributing to HCC development and sorafenib resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Primary liver cancer nowadays ranks 6th and 3rd on the list of the most frequent tumors and that of the major cancer-associated death causes respectively around the world in 2020, causing estimated 830 thousand deaths and 906,000 new cases per year [1]. Hepatocellular carcinoma (HCC) is the major pathological pattern of this cancer with a approximate ratio of 80% [2, 3]. The dominant risk factors for HCC include chronic infection with hepatitis C virus (HCV) or hepatitis B virus (HBV), heavy alcohol use, diabetes, and smoking [4, 5]. Accumulated evidence has reported that HBV X (HBx) protein accelerates the occurrence and progressive course of HCC [6, 7]. The therapeutic management of HCC is various and the decision is made dependent on the tumor progression, with the options of surgical resection, liver transplantation, transarterial embolization and radiotherapy as well as systemic pharmacological treatment [8]. Sorafenib is marketed as a small-molecule multiple-target tyrosine kinase inhibitor that possesses the function of anti-angiogenesis and anti-proliferation as the first-line systemic treatment for HCC [9, 10]. However, only about 30% of HCC patients may obtain benefits from sorafenib, and usually drug resistance occurs within 6 months [10]. Previous evidence has shown that HBx protein may initiate epigenetic modifications, such as abnormal microRNA (miRNA) expression, which in turn regulates the downstream epigenetic changes in the pathogenesis of HBV-HCC [11]. Therefore, we try to figure out the mechanism of HBx-induced dysregulated miRNA in sorafenib resistance of HCC.
Deregulated miRNAs participate in the progression and metastasis of tumors and drug resistance phenotype [12]. In a previous study, 5 miRNAs closely related to overall survival of HCC patients were screened through the TCGA database, with miR-3677-3p, miR-744*, miR-210, and miR-3613-5p negatively correlated with the overall survival, while miR-148a positively linked with the overall survival [13]. It was also reported that miR-3677-3p was implicated in the progression of HCC [14]. F‐box protein 31 (FBXO31), initially reported in 2005, belongs to the F‐box family and participates in the ubiquitin‐proteasome system [15, 16]. In addition, FBXO31 exerts a tumor suppressor function in HCC [17]. Through our analysis in Starbase database and Jefferson database, miR-3677-3p was found to have a negative relation with FBXO31 in HCC, likely having a direct binding site for FBXO31. Forkhead Box M1 (FOXM1) serves as a typical proliferation-related transcription factor with the function of stimulating cell proliferation [18]. It has been evinced that FBXO31 is a component of the SCF ubiquitination complex and is able to promote FOXM1 ubiquitination and degradation [19]. Based on the previous evidence, the current paper sought to investigate the influences of miR-3677-3p on development and sorafenib resistance of hepatitis B-related HCC and to explore its molecular mechanism related with FBXO31/FOXM1.
Materials and methods
Cell culture
HBV+ hepatoma HepG2.2.15 cells were obtained from the laboratory of the Brain Hospital of Hunan Province, with the parental HepG2 cells (HBV−) and normal human hepatocyte THLE-2 cells purchased from ATCC (Manassas, VA, USA). Huh7 cells and HEK293T cells were purchased from Procell Life Science & Technology (Wuhan, China). THLE-2 cells were cultured in BEGM medium (Lonza/Clonetics Corporation, Walkersville, USA), HepG2 and HepG2.2.15 in MEM medium (GIBCO BRL, Grand Island, NY, USA), and Huh7 and HEK293T cells in DMEM medium (GIBCO BRL, Grand Island, NY, USA), and all cells were cultured at 37 °C in an incubator (5% CO2). To establish resistant tumor cell lines, HepG2.2.15 cells and Huh7 + HBx cells were exposed to escalated concentrations (low to high) of sorafenib (SR) solution until HepG2.2.15/SR cells and Huh7 + HBx/SR cells grew stably in medium with the selected concentration of SR.
Cell transfection
Cells were transfected or co-transfected with miR-3677-3p mimic, miR-3677-3p inhibitor (50 nM, GenePharma, Shanghai, China), pcDNA3.1-HBx, pcDNA3.1-FBXO31, sh-FBXO31, sh-FOXM1 and their corresponding negative controls (NC) (2 μg, GenePharma, Shanghai, China). Transfection was conducted using LipoFiter™ transfection reagent (HanBio, Shanghai, China) as per the instructions.
Tumorigenesis in nude mice
Thirty-six SPF BALB/c nude mice (4–6 weeks old, weighing 16 ± 20 g) were purchased from Shanghai Laboratory Animal Center, Chinese Academy of Sciences. All animals were housed in a SPF-grade sterile laminar flow room under constant temperature (22–26 ℃) and humidity (55 ± 5%). All animal experiments were performed with the consent from the Ethics Committee of our hospital and abide by applicable rules and regulations and operating procedures as well as ethical requirements related to experimental animals.
Nude mice were randomly assigned into 6 groups (Huh7 + pcDNA3.1 group, Huh7 + HBx group, Huh7 + HBx + inhibitor NC group, Huh7 + HBx + miR-3677-3p inhibitor group, HepG2.2.15 + inhibitor NC group, and HepG2.2.15 + miR-3677-3p inhibitor group) and intraperitoneally injected with pentobarbital sodium (60 mg/kg) for anesthetization. Subsequently, conventional iodophor was utilized to disinfect the right back skin of the nude mice. The Huh7 + HBx and the Huh7 + pcDNA3.1 group of mice received injections of 0.2 mL suspension of Huh7 cells (1 × 106) that stably overexpressed HBx or NC, respectively. The mice in the Huh7 + HBx + miR-3677-3p inhibitor group and the Huh7 + HBx + inhibitor NC group were injected with the same amount of suspension of HBx-overexpressing Huh7 cells (1 × 106) with stably down-regulated miR-3677-3p or its NC, respectively. For mice in the HepG2.2.15 + inhibitor NC and HepG2.2.15 + miR-3677-3p inhibitor groups, an equal amount of suspension of HBx-overexpressing HepG2.2.15 cells (1 × 106) with stably down-regulated miR-3677-3p or its NC was injected. After injection, the syringe was pulled out slowly, and the injection site was pressed gently with an alcohol cotton ball for about 10 s to prevent cell suspension from flowing out. Three weeks later, the mice were sacrificed by cervical dislocation and the subcutaneous tumors were isolated and weighed, with the tumor volume calculated (tumor volume = 1/2 × long diameter × short diameter2).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol (15,596,026, Invitrogen, Carlsbad, USA), and RNA was reverse transcribed into cDNA with reverse transcription kit (RR047A, Takara, Japan). MiRNA was reverse transcribed into cDNA with TaqMan™ MicroRNA Reverse Transcription Kit (4,366,596, Thermo Fisher Scientific, Waltham, MA, USA). qRT-PCR reactions were conducted in a real-time PCR instrument (ABI7500, ABI, Foster City, CA, USA) with reaction system: SYBR Mix 9 μL, positive primer 0.5 μL, negative primer 0.5 μL, cDNA 2 μL, RNase Free dH2O 8 μL, and reaction conditions: 95 ℃ for 10 min and 40 consecutive cycles of 95 ℃ for 15 s and 60 ℃ for 1 min. Three replicates were prepared for each sample and the primers were synthesized by Sangon Biotech (Shanghai, China) (primer sequence shown in Table 1). The Ct value of each well was recorded, and the relative expression of the product was calculated using the 2−ΔΔCt method with U6 or GAPDH as the internal reference. The formula was as follows: ΔΔCt = (mean Ct of target gene in experimental group—mean Ct of housekeeping gene in experimental group)—(mean Ct of target gene in control group—mean Ct of housekeeping gene in control group).
Western blot
Cells or tissues or from each group underwent washing three times with pre-cooled PBS buffer and lysed by adding protein extraction lysis buffer (100 μL/50 mL culture flask). After BCA kit (Beyotime) measurement of protein concentration, appropriate volume of protein was mixed with loading buffer (Beyotime) and denatured for 3 min in a boiling water bath. After 30-min 80 V electrophoresis, the voltage was switched to 120 V for another 1–2-h electrophoresis when bromophenol blue entered the separation gel. Subsequent to the transferring of the protein to a PVDF membrane in 60 min ice bath (300 mA), the membrane was washed and placed in blocking solution for 60 min at ambient temperature overnight at 4 °C. followed by 1-h culture with diluted primary antibodies against FBXO31 (ab86137, 1:1000), FOXM1 (ab207298, 1:1000), HBx (ab39716, 1:1000), E-cadherin (ab40772, 1:1000), N-cadherin (ab76011, 1:1000), Vimentin (ab92547, 1:1000), CD133 (ab284389, 1:1000), EpCAM (ab223582, 1:1000), OCT4 (ab200834, 1:10,000) or GAPDH (ab9485, 1:2500) (Abcam, Cambridge, UK) at room temperature on a shaker and 3 times washing (10 min/time). Then the PVDF membrane was transferred into secondary antibody solution (rabbit IgG, 1:10,000, Abcam, Cambridge, UK) and washed three times (10 min/time) after incubation for 1 h at room temperature. The image of blots treated with color developer was taken using a chemiluminescence imaging system (Bio-Rad).
Luciferase reporter assay
By using Jefferson database (https://cm.jefferson.edu/rna22/), a binding site for miR-3677-3p and FBXO31 was predicted. Based on the predicted results, wild and mutated sequences (wt-FBXO31 and mut-FBXO31) of the binding site were designed and synthesized. The wild sequence or mutated sequence of the binding site was inserted into a luciferase reporter vector (pGL3-Promoter, Promega, MADISON, WI, USA) and then co-transfected with miR-3677-3p mimic or mimic NC into cells. After transfection, Firefly luciferase activity and Renilla luciferase activity were measured, with Renilla luciferase activity as an internal reference, and the ratio of Firefly luciferase activity to Renilla luciferase activity was calculated as the relative activity of luciferase.
CCK-8 assay
HepG2.2.15 and Huh7 + HBx cells (100 μL) transfected for 24 h were seeded into 96-well plates, and three replicates were set for each sample. CCK-8 reagent (10 μL, Dojindo, Tokyo, Japan) was supplemented to each well 0 h, 24 h, 48 h, 72 h and 96 h after cell incubation in the incubator, followed by another 2-h incubation and the measurement of absorbance (450 nm).
HepG2.2.15/SR and Huh7 + HBx/SR cells (100 μL each) transfected for 24 h were inoculated into 96-well plates, and three replicates were set for each sample. HepG2.2.15/SR and Huh7 + HBx/SR cells were treated with various concentrations (0, 2.5, 5, 7.5, 10, 12.5, 15, 17.5, and 20 μM) of sorafenib solution for 48 h [20]. CCK-8 reagent (10 μL, Dojindo) was then added to each well, after which the plates were incubated for 2 h, with the absorbance measured at a wavelength of 450 nm.
Colony formation assay
After transfection, cells underwent 2–3 week-incubation in 6-well plates (37 °C, 5% CO2), which was ceased as cell clones were visible to the naked eye. Cells were washed with PBS after the culture medium was discharged, after which 4% paraformaldehyde was supplemented for 15-min cell fixing in each well. The fixative was removed, and Giemsa staining solution (1 mL) was slowly added along the well wall for 20-min staining away from light. Then the Giemsa staining solution was washed away with running water, and the 6-well plate was inverted on clean absorbent paper. The number of colonies was counted.
Transwell
The Transwell insert (Corning, New York, USA) with a pore size of 8 μm plated with Matrigel was pre-incubated with 0.5 mL of serum-free culture medium at 37 ℃ and 5% CO2 for 2 h. Cells in the logarithmic growth phase were obtained and prepared into single cell suspension to be evenly seeded in a 6-well plate, with three replicate wells set up in each group, after which the plate was placed in an incubator at 37 ℃ in 5% CO2. When the cell confluence reached 70–90%, the cells in each group were treated according to the experimental grouping, and the incubation continued for 24 h at 37 °C in 5% CO2. The cells were digested by trypsin, and washed twice with PBS after collection. Then the cells were resuspended with serum-free DMEM medium, with the cell concentration adjusted. Culture medium (600 μL) containing 10% fetal bovine serum was supplemented to the lower Transwell chamber, and 100 μL of prepared cells to the upper chamber and cultured for 24 h in an incubator at 37 °C in 5% CO2. Upon removing the insert, the supernatant was discarded and the residual cells on the inner side of the insert membrane were wiped off with a cotton swab. Invasive cells on the outer side of the insert membrane were fixed by 4% paraformaldehyde for 20 min. The number of invasive cells in 5 random fields was counted at high magnification after the cells were stained with Wright-Giemsa staining solution.
Scratch assay
Cells in the logarithmic growth phase were obtained and prepared into single cell suspension (5 × 105 cells/mL) and evenly seeded on a 6-well plate. After 24 h, the cells in each group were treated correspondingly and cultured in an incubator with 5% CO2 at 37 °C for 24 h. A sterile tip (100 μL) was used to scratch in the 6-well plate. When scratching, the tip was vertical and it should be ensured that the scratch width of each group was basically the same. Next, the culture medium in the 6-well plate was discarded, and the cells were washed twice with PBS and continued to be cultured with serum-free culture medium, followed by observation and photographing to record the scratch area of cells at 0 h as the control. The 6-well plate was placed in an incubator with 5% CO2 at 37 ℃ for 24 h, after which migrating cells were observed and photographed, with the cell migration rate calculated as (scratch width at 0 h—scratch width at 24 h)/scratch width at 0 h × 100%.
Flow cytometry
The cell density of each group was adjusted to 105 cells/mL and 3 mL of cell suspension for each group was collected in a centrifuge tube (10 mL), followed by twice centrifugation at 500 rpm for 5 min and removal of the culture medium. Cells were resuspended with 100 μL of binding buffer, after which 5 μL of Annexin V-FITC and 5 μL of PI were added and mixed gently, and the mixture was placed at room temperature away from light for 15 min. FITC fluorescence and PI fluorescence were detected by flow cytometry to analyze the apoptosis rate.
Co-immunoprecipitation (Co-IP)
Cells were collected and lysed with 0.6 mL of lysis buffer plus protease inhibitors (Roche Applied Science) for 40 min on ice. After centrifugation at 12,000×g for 15 min, the lysate was immunoprecipitated with 1 μg of specific antibody overnight at 4℃, followed by the addition of washed Protein A Agarose beads (Invitrogen) or Protein G Agarose beads (Santa Cruz). The loading samples were boiled for 5 min and centrifuged at 14,000×g, with the remaining Agarose beads collected. Subsequently, the supernatant was boiled again for 5 min for denaturation and electrophoresed, followed by western blot detection of FBXO31 and FOXM1 protein expression.
Ubiquitination assay
HEK293T cells were treated with transfection of miR-3677-3p inhibitor/inhibitor NC or pcDNA3.1-FBXO31/empty plasmid (GenePharma, Shanghai, China) together with Myc-FOXM1 and HA-ubiquitin (Genomeditech, Shanghai, China) using LipoFiter™ transfection reagent (HanBio, Shanghai, China) according to the transfection instructions. The cells were then treated with 20 μM of the proteasome inhibitor MG132 for 6 h, washed twice with cooled PBS, and lysed with RIPA lysis buffer. Cytosolic protein was obtained by centrifugation of the lysate and incubated overnight with anti-FOXM1 antibody (ab207298, Abcam, Cambridge, UK), followed by the addition of 100 μL of Protein A/G Agarose beads to capture antigen–antibody complexes for 4 h at 4 ℃ on a shaker. After being washed 3 times with lysis buffer, the complexes were boiled in 2 × SDS loading buffer to release the protein, and immunoblotting was performed by using antibody against ubiquitin (ab134953, 1:2000, Abcam, Cambridge, UK).
Statistical analysis
All collected raw data were statistically analyzed with GraphPad prism 7.0 software, and exhibited as mean ± standard deviation. For data conforming to normal distribution, T test was used for comparison between two groups, one-way analysis of variance for comparison between multiple groups, and Tukey’s multiple comparisons test for post-hoc multiple comparisons. Where data did not conform to normal distribution, for two and multiple independent samples, Mann–Whitney U and Kruskal–Wallis tests were respectively performed. The correlation of genes was evaluated through Pearson’s correlation analysis. P < 0.05 was deemed significant in statistical difference.
Results
MiR-3677-3p was highly expressed in HBV+ HCC cells and tumor tissues from nude mice
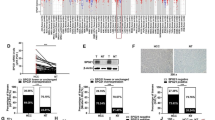
We first constructed Huh7 cells with high HBx expression and verified the transfection efficiency by Western blot (Fig. 1A). qRT-PCR for detecting the expression of five miRNAs (miR-3677-3p, miR-744*, miR-210, miR-3613-5p, and miR-148a) demonstrated to have associations with the overall survival of HCC patients [13] showed that miR-3677-3p, miR-744*, miR-210, and miR-3613-5p expression levels were elevated and miR-148a declined in HepG2, HepG2.2.15, Huh7 + pcDNA3.1, and Huh7 + HBx cells relative to the expression in THLE-2 cells, and the miRNA expression was more significantly different in HepG2.2.15 and Huh7 + HBx cells compared with HepG2 or Huh7 + pcDNA3.1 cells; in particular, the fold up-regulation of miR-3677-3p was more prominent than that of the other miRNAs (Fig. 1B). Based on those results, we finally selected miR-3677-3p for subsequent experiments. The HCC nude mouse model was constructed by subcutaneous injection of Huh7 cells stably transfected with HBx, and it was revealed miR-3677-3p had higher expression in the Huh7 + HBx group than that in the Huh7 + pcDNA3.1 group (Fig. 1C). miR-3677-3p up-regulation in HCC was found by searching the Starbase database (Fig. 1D), which also indicated poor prognosis (Fig. 1E). Kaplan–Meier Plotter database analysis also unveiled miR-3677-3p overexpression was linked with poor prognosis in HCC (Fig. 1F), which further clarified the research value of miR-3677-3p.
MiR-3677-3p was highly expressed in HBV+ HCC cells and tumor tissues. Huh7 cells were transfected with pcDNA3.1-HBx. A Expression of HBx in Huh7 cells measured by Western blot, *P < 0.05 versus the pcDNA3.1 group; B Expression of miR-3677-3p, miR-744*, miR-210, miR-3613-5p, and miR-148a in different cell lines detected by qRT-PCR; C Expression of miR-3677-3p in tumor tissues from nude mice detected by qRT-PCR, *P < 0.05 versus the Huh7 + pcDNA3.1 group; D Expression of miR-3677-3p in HCC in Starbase database. LIHC group: 370, normal group: 50, *P < 0.05 versus the normal group; E Starbase database analysis of the relation between miR-3677-3p expression and overall survival of HCC patients. High Num: 184, Low Num: 185; F Kaplan–Meier Plotter database analysis of the relation between miR-3677-3p expression and overall survival of HCC patients. High Num: 155; Low Num: 216. *P < 0.05. n = 6 in panel C. Three replicates were set up for the rest of the experiments
MiR-3677-3p promoted tumorigenesis and sorafenib resistance of HBV+ HCC cells
To further explore the participation of miR-3677-3p in hepatitis B-related HCC, we constructed Huh7 + HBx and HepG2.2.15 cell models with miR-3677-3p expression alterations, and the transfection efficiency was tested by qRT-PCR (Fig. 2A). Cell proliferation was tested by CCK-8 and colony formation assay, cell migration and invasion by scratch assay and Transwell, and apoptosis by flow cytometry, and the results revealed that cell migration, invasion and proliferation were enhanced (Fig. 2B-E) and apoptosis was lowered (Fig. 2F) in the miR-3677-3p mimic group relative to the mimic NC group. Western blot revealed an evident reduction of E-cadherin protein level and enhancement in N-cadherin and Vimentin protein levels in the miR-3677-3p mimic group relative to the mimic NC group (Fig. 2G). Like other cancer stem cells, liver cancer stem cells (LCSCs) have unlimited self-renewal capacity, strong tumorigenicity, and drug resistance and can be labeled by stemness-related proteins CD133, EpCAM, and OCT4 [21]. miR-3677-3p up-regulation markedly increased stemness-related protein (CD133, EpCAM, and OCT4) levels (Fig. 2H), while also promoted sorafenib resistance and increased IC50 in Huh7 + HBx/SR cells and HepG2.2.15/SR cells (Fig. 2I). Cell migration, invasion and proliferation were reduced (Fig. 2B-E) and apoptosis was enhanced (Fig. 2F) in the miR-3677-3p inhibitor group relative to the inhibitor NC group. Western blot revealed that E-cadherin protein expression was obviously elevated and N-cadherin and Vimentin protein levels were obviously reduced (Fig. 2G); the expression of stemness-related proteins was suppressed (Fig. 2H) in the miR-3677-3p inhibitor group. After inhibition of miR-3677-3p, sorafenib resistance was also inhibited, with IC50 reduced in Huh7 + HBx/SR cells and HepG2.2.15/SR cells (Fig. 2I). The above findings evinced that miR-3677-3p promoted development and sorafenib resistance of HBV+ HCC cells.
MiR-3677-3p promoted development and sorafenib resistance of HBV+ HCC cells. Huh7 + HBx and HepG2.2.15 cell models with overexpression or knockdown of miR-3677-3p were constructed. A The expression of miR-3677-3p in Huh7 + HBx cells and HepG2.2.15 cells detected by qRT-PCR; B, C Cell proliferation detected by CCK-8 and colony formation assay; D, E Cell migration and invasion detected by scratch assay and Transwell; F Apoptosis detected by flow cytometry; G The expression of E-cadherin, N-cadherin and Vimentin in Huh7 + HBx and HepG2.2.15 cells measured by Western blot; H The expression of CD133, EpCAM and OCT4 measured by Western blot; I The activity of Huh7 + HBx/SR cells and HepG2.2.15/SR cells treated with different concentrations of sorafenib and the IC50 tested by CCK-8. *P < 0.05 versus the mimic NC or inhibitor NC group, three replicates were set up for all experiments
MiR-3677-3p targeted and negatively regulated FBXO31
Starbase database exposed the negative correlation between miR-3677-3p and FBXO31 in HCC (Fig. 3A); the Jefferson database showed that miR-3677-3p had a direct binding site for FBXO31. qRT-PCR and Western blot revealed that FBXO31 was lowly expressed in the HepG2.2.15 and Huh7 + HBx cells and tumor tissues from nude mice subcutaneously injected with Huh7 cells overexpressing HBx (Fig. 3B–E). Wild and mutant sequences (wt-FBXO31 and mut-FBXO31) of the binding site were designed and synthesized based on the predicted results (Fig. 3F). Dual-luciferase reporter assay revealed that the luciferase activity of the miR-3677-3p mimic group inserted with the mut-FBXO31 sequence was not different from that of the mimic NC group, while the luciferase activity of the miR-3677-3p mimic group inserted with the wt-FBXO31 sequence was reduced relative to that of the mimic NC group (Fig. 3G). In Huh7 + HBx and HepG2.2.15 cells transfected with miR-3677-3p mimic or miR-3677-3p inhibitor, results evinced that miR-3677-3p could negatively regulate the expression of FBXO31 (Fig. 3H–I). The above findings revealed that miR-3677-3p targeted and negatively regulated FBXO31 in hepatitis B-related HCC.
MiR-3677-3p targeted and negatively regulated FBXO31. A Correlation between miR-3677-3p and FBXO31 in HCC in Starbase database; B-E The expression of FBXO31 in cultured cells and tumor tissues from nude mice detected by qRT-PCR (B, D) and Western blot (C, E), for the D and E, P < 0.05 versus the Huh7 + pcDNA3.1 group; F The binding site between miR-3677-3p and FBXO31 and wild sequence and mutant sequence of FBXO31; G The binding of miR-3677-3p to FBXO31 detected by dual-luciferase reporter assay; H, I The expression of FBXO31 in Huh7 + HBx and HepG2.2.15 cells transfected with miR-3677-3p mimic or miR-3677-3p inhibitor detected by qRT-PCR and Western blot. *P < 0.05. n = 6 in panels D and E. Three replicates were set up for the rest of the experiments
FBXO31 inhibited development and sorafenib resistance in HBV+ HCC cells
To investigate the influence of miR-3677-3p/FBXO31 on tumor development and sorafenib resistance in HBV+ HCC, sh-FBXO31 and its sh-NC were first transfected into HepG2.2.15 cells and Huh7 + HBx cells, and the transfection efficiency was verified by qRT-PCR and Western blot (Fig. 4A, B). For following experiments, sh2-FBXO31 presenting the best efficiency was chosen. Subsequently, miR-3677-3p inhibitor and sh2-FBXO31/sh2-NC were co-transfected into HepG2.2.15 cells and Huh7 + HBx cells. It was also presented that cell migration, invasion and proliferation were markedly enhanced (Fig. 4C–F) and apoptosis was significantly reduced (Fig. 4G) in the inhibitor + sh2-FBXO31 group relative to the inhibitor + sh2-NC group. Western blot revealed downgraded E-cadherin expression and upgraded N-cadherin and Vimentin expression (Fig. 4H). Cancer stem cell-related factor expression was markedly up-regulated in the inhibitor + sh2-FBXO31 group (Fig. 4I), and additionally, knockdown of FBXO31 promoted sorafenib resistance and increased IC50 in Huh7 + HBx/SR cells and HepG2.2.15/SR cells in the presence of miR-3677-3p inhibitor (Fig. 4J). The above findings illustrated that FBXO31 could reverse the cancer-promoting effect of miR-3677-3p on hepatitis B-related HCC cells.
FBXO31 inhibited development and sorafenib resistance in HBV+ HCC cells. A, B The expression of FBXO31 in HepG2.2.15 and Huh7 + HBx cells transfected with sh-FBXO31 or sh-NC was detected by qRT-PCR and Western blot, *P < 0.05 versus the sh-NC group. Next, miR-3677-3p inhibitor and sh-FBXO31/sh-NC were co-transfected into HepG2.2.15 and Huh7 + HBx cells. C, D Cell proliferation detected by CCK-8 and colony formation assay; E, F Cell migration and invasion detected by scratch assay and Transwell; G Apoptosis detected by flow cytometry; H The expression of E-cadherin, N-cadherin and Vimentin in Huh7 + HBx and HepG2.2.15 cells measured by Western blot; I The expression of CD133, EpCAM and OCT4 measured by Western blot; J The activity of Huh7 + HBx/SR cells and HepG2.2.15/SR cells treated with different concentrations of sorafenib and the IC50 tested by CCK-8. *P < 0.05 versus the inhibitor + sh2-NC group, three replicates were set up for all experiments
MiR-3677-3p inhibited FOXM1 ubiquitination and degradation by binding FBXO31
It was known from Starbase database that FOXM1 expression was high in HCC (Fig. 5A), and was linked with poor prognosis (Fig. 5B). Negative correlation between the expression of FBXO31 and FOXM1 was discovered in patients with HCC through analysis in the Starbase database (Fig. 5C). Based on the evidence, we speculated that FBXO31, as a component of the SCF ubiquitination complex, might inhibit HBV+ HCC chemoresistance through promoting FOXM1 ubiquitination and degradation. Western blot revealed that FOXM1 was highly expressed in the HepG2.2.15 and Huh7 + HBx cells, and tumor tissues from nude mice injected with Huh7 cells overexpressing HBx (Fig. 5D–E). The expression of FOXM1 in HepG2.2.15 and Huh7 + HBx cells detected by Western blot was definitely reduced after inhibition of miR-3677-3p, on the basis of which knockdown of FBXO31 could reverse the result (Fig. 5F), indicating that miR-3677-3p could regulate the expression of FOXM1 post-translationally. Co-IP assay found that FBXO31 could enrich FOXM1 (Fig. 5G). Further validation of the ubiquitination modification of FOXM1 by Co-IP assay showed that miR-3677-3p knockdown or FBXO31 up-regulation markedly enhanced the ubiquitination of FOXM1 (Fig. 5H, I). The above findings illustrated that miR-3677-3p targeted and downregulated FBXO31 expression, which in turn inhibited the ubiquitination and degradation of FOXM1.
MiR-3677-3p inhibited FOXM1 ubiquitination and degradation by binding FBXO31. A FOXM1 expression in HCC in Starbase database, *P < 0.05 versus the normal group; B Analysis of FOXM1 expression and patient survival; C Correlation of FBXO31 and FOXM1 expression in patients with HCC; D, E The expression of FOXM1 in cells and tumor tissues from nude mice measured by Western blot, *P < 0.05 versus the Huh7 + pcDNA3.1 group; F The expression of FOXM1 in transfected HepG2.2.15 and Huh7 + HB cells measured by Western blot; G FBXO31 and FOXM1 protein binding detected by Co-IP; H, I Ubiquitination modification of FOXM1 detected by Co-IP. *P < 0.05. n = 6 in panel E. Three replicates were set up for the rest of the experiments
FBXO31 promoted FOXM1 ubiquitination and degradation to inhibit HBV+ HCC cell development and sorafenib resistance
To deeply study the effect of FBXO31 on HBV-related HCC development and sorafenib resistance by regulating FOXM1 ubiquitination, we first transfected sh-FOXM1 and its negative control into HepG2.2.15 and Huh7 + HBx cells and the transfection efficiency was verified (Fig. 6A, B), and selected the most efficient sh1-FOXM1 for subsequent experiments. Sh-FBXO31, sh-FOXM1, and their negative controls were transfected or co-transfected into HepG2.2.15 and Huh7 + HBx cells. The expression of FOXM1 was detected by Western blot, and the results evinced that the expression of FOXM1 in the sh2-FBXO31 group was elevated relative to that in the sh2-NC group and the expression of FOXM1 in the sh2-FBXO31 + sh1-FOXM1 group was down-regulated versus the sh2-FBXO31 + sh1-NC group (Fig. 6C). Cell migration, invasion, and proliferation were markedly increased (Fig. 6D–G) and apoptosis was markedly reduced (Fig. 6H) in the sh2-FBXO31 group relative to the sh2-NC group. The reduced E-cadherin protein level and augmented N-cadherin and Vimentin protein levels were observed in the sh2-FBXO31 group (Fig. 6I), and cancer stem cell characteristics were markedly enhanced in the sh2-FBXO31 group (Fig. 6J). When making comparison with the sh2-FBXO31 + sh1-NC group, cell proliferative, migratory, and invasive capacities were markedly reduced (Fig. 6D–G), apoptosis was markedly increased (Fig. 6H), and mesenchymal properties and cancer stem cell characteristics were significantly inhibited (Fig. 6I, J) in the sh2-FBXO31 + sh1-FOXM1 group. Knockdown of FBXO31 promoted cell resistance to sorafenib and increased IC50, while down-regulation of FOXM1 reversed those results (Fig. 6K). The above results illustrated that FBXO31 inhibited the expression of FOXM1 by regulating FOXM1 ubiquitination, which further inhibited HBV+ HCC cell development and sorafenib resistance.
FBXO31 promoted FOXM1 ubiquitination and degradation to inhibit HBV+ HCC cell development and sorafenib resistance. A, B The expression of FOXM1 in HepG2.2.15 and Huh7 + HBx cells transfected with sh-FOXM1 or its negative control was detected by qRT-PCR and Western blot. *P < 0.05 versus the sh-NC group. Next, sh-FBXO31, sh-FOXM1, and their negative controls were transfected or co-transfected into HepG2.2.15 and Huh7 + HBx cells. C The expression of FOXM1 measured by Western blot; D, E Cell proliferation detected by CCK-8 and colony formation assay, *P < 0.05 versus the sh2-NC or sh2-FBXO31 + sh1-NC group; F, G Cell migration and invasion detected by scratch assay and Transwell; H Apoptosis detected by flow cytometry; I The expression of E-cadherin, N-cadherin and Vimentin in Huh7 + HBx and HepG2.2.15 cells measured by Western blot; J The expression of CD133, EpCAM and OCT4 measured by Western blot; K The activity of Huh7 + HBx/SR cells and HepG2.2.15/SR cells treated with different concentrations of sorafenib and the IC50 tested by CCK-8, *P < 0.05 versus the sh2-NC or sh2-FBXO31 + sh1-NC group. * P < 0.05, three replicates were set up for all experiments
Down-regulation of miR-3677-3p inhibited growth of HBV+ HCC in nude mice
To investigate the effect of miR-3677-3p on HBV+ HCC growth in vivo, we constructed HBV+ HCC nude mouse models by injecting Huh7 cells stably overexpressing HBx and under-expressing miR-3677-3p or by injecting HepG2.2.15 cells stably under-expressing miR-3677-3p. After the euthanasia of nude mice 3 weeks later, tumor tissues were obtained and weighed. The tumor weight and volume were significantly reduced after the miR-3677-3p inhibition (Fig. 7A–C). Levels of miR-3677-3p were determined by qRT-PCR, and the results evinced that the expression of miR-3677-3p was successfully down-regulated in tumor tissues (Fig. 7D). It was exhibited that FBXO31 expression was distinctly enhanced and FOXM1 expression was reduced after miR-3677-3p down-regulation (Fig. 7E, F). It was explained that miR-3677-3p knockdown in vivo promoted expression of FBXO31, thereby suppressing FOXM1 expression and ultimately suppressing growth of HBV+ HCC.
Down-regulation of miR-3677-3p inhibited growth of HBV+ HCC in nude mice. A Tumor of nude mice; B Tumor weight; C Tumor volume; D:The expression of miR-3677-3p in tumor tissues detected by qRT-PCR; E, F: The expression of FBXO31 and FOXM1 in tumor tissues measured by Western blot. *P < 0.05 versus the inhibitor NC group. n = 6
Discussion
HCC, as a common fatal malignancy around the world, brings heavy burden and therapeutic challenge to the public [4]. MiR‐3677‐3p is a reported prognosis‐related miRNA in HCC patients. However, the underlying mechanisms of miR‐3677‐3p influencing the cancer development and sorafenib resistance in hepatitis B-related HCC is poorly understood.
Here we observed high expression of miR-3677-3p in HBV+ HCC cells and tumor tissues from nude mice. MiR-3677-3p could promote tumor development in hepatitis B-related HCC, serving as an oncomiR. Consistently, it was recorded miR-3677-3p enhanced HCC cell malignant behaviors through inhibiting SIRT5 [22]. Li et al. also demonstrated high level of miR-3677-3p in primary tumor tissues from HCC subjects and suppression of miR-3677-3p sharply inhibited the cell proliferation and tumor development through GSK3β inhibition [14]. Meanwhile, accumulating studies demonstrated that miRNAs served as regulators of sorafenib resistance in cancers, such as miR-30e-3p, miR-124 and miR-30a-5p [12, 23, 24] but few evidence reported the impacts of miR-3677-3p on sorafenib resistance. Our results that miR-3677-3p facilitated the sorafenib resistance in hepatitis B-related HCC partially filled in this knowledge gap.
FBXO31 is one of the members of the human F-box family, which mediates protein abundance and the function of oncogenes and tumor suppressors through the ubiquitin-dependent proteasome degradation pathway [16]. Huang et al. demonstrated that as a tumor suppressor, FBXO31 was down-regulated in HCC and the ectopic expression of FBXO31 triggered the inhibition of colony growth and lowered the proliferation of HEPG2 cells [17]. Targeting the FBXO31–MAPK pathway might be beneficial to the control of esophageal squamous cell carcinoma development and increase cisplatin sensitivity [25]. However, there is no report on the regulation of drug resistance by FBXO31 in HCC. In this study, we observed that FBXO31 inhibited tumor development and sorafenib resistance in HBV+ HCC. miR-3677-3p targeted and negatively regulated FBXO31 in hepatitis B-related HCC. Specifically, FBXO31 offset the cancer-promoting effect of miR-3677-3p on hepatitis B-related HCC.
JM Jeffery et al. concluded that FBXO31 was a component of the SCF ubiquitination complex and had specific interaction with FOXM1 during the G2/M transition, causing FOXM1 ubiquitination and degradation [19]. Specifically in this study, miR-3677-3p targeted and downregulated the expression of FBXO31, which in turn inhibited the ubiquitination and degradation of FOXM1. A previous study revealed that FOXM1 was a proliferation-associated transcription factor and could promote the progression of HCC via mediating KIF4A expression [26]. Chou et al. found that inhibition on the expression of the transcription factor FOXM1 led to the suppression of liver tumor cell malignant behaviors [27]. Another study concluded that FOXM1 up-regulation could promote the epithelial mesenchymal transition and HCC metastasis [28]. In term of drug resistance, high expression of FOXM1 was observed in sorafenib-resistant HCC cells and addition of FOXM1 inhibitor might promote the cytotoxic effects of sorafenib in HCC [29]. Yan et al. also provided the evidence that knockdown of FOXM1 significantly reduced tumor growth in liver cancer and increased the sorafenib sensitivity in drug‑tolerant cells [30]. Previous reports demonstrated that ubiquitination was a critical mode of regulating the expression of FOXM1 and the reduction of FOXM1 ubiquitination resulted in the increased of FOXM1 transcription expression [31,32,33]. Moreover, the FOXM1 ubiquitination inhibition-caused increase in FOXM1 expression was involved in the promotion of drug resistance in non-small-cell lung cancer cells [34]. Ubiquitination or deubiquitination of proteins had important involvement in the development and progressive course of HCC [35,36,37]. But the impacts of FOXM1 ubiquitination on the drug resistance of HCC cells were barely discussed before. Our study firstly revealed that FBXO31 inhibited the expression of FOXM1 by regulating FOXM1 ubiquitination, which further inhibited HBV+ HCC progression and sorafenib resistance. Nevertheless, the underlying mechanisms of FBXO31-promoted FOXM1 ubiquitination affecting HBV + HCC progression and sorafenib resistance warrant further investigations. Finally, we further validated the in vivo functions of miR-3677-3p on HBV+ HCC growth.
Taken together, our study concluded that miR-3677-3p bound to FBXO31 and inhibited its expression, which in turn suppressed the ubiquitination and degradation of FOXM1 and ultimately promoted the development of HCC and sorafenib resistance. Nevertheless, there are limitations in the current study which warrant solving in following research. Firstly, in this study, we only explored the relationship between miR-3677-3p and FBXO31/FOXM1 ubiquitination, and further studies are warranted to fully elucidate the up/downstream pathways and related mechanism. Secondly, additional tyrosine kinase inhibitors, such as lenvatinib and regorafenib, have been demonstrated to improve the survival of HCC. In terms of drug resistance, more typical drugs may be compared. Nonetheless, for the clinical importance, our findings provide a logical basis of targeting miR-3677-3p for HCC treatment.
Data availability
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49.
Gao YX, Yang TW, Yin JM, et al. Progress and prospects of biomarkers in primary liver cancer (Review). Int J Oncol. 2020;57:54–66.
Wege H, Schulze K, von Felden J, Calderaro J, Reig M. Rare liver tumors working group of the European Reference Network on Hepatological D. Rare variants of primary liver cancer: fibrolamellar, combined, and sarcomatoid hepatocellular carcinomas. Eur J Med Genet. 2021;64:104313.
Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–9.
Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(477–91): e1.
Wang Z, Wu Z, Huang P. The function of miRNAs in hepatocarcinogenesis induced by hepatitis B virus X protein (review). Oncol Rep. 2017;38:652–64.
Zhang XD, Wang Y, Ye LH. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med. 2014;11:182–90.
Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604.
Li ZJ, Dai HQ, Huang XW, et al. Artesunate synergizes with sorafenib to induce ferroptosis in hepatocellular carcinoma. Acta Pharmacol Sin. 2021;42:301–10.
Tang W, Chen Z, Zhang W, et al. The mechanisms of sorafenib resistance in hepatocellular carcinoma: theoretical basis and therapeutic aspects. Signal Transduct Target Ther. 2020;5:87.
Lin X, Zuo S, Luo R, et al. HBX-induced miR-5188 impairs FOXO1 to stimulate beta-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics. 2019;9:7583–98.
Gramantieri L, Pollutri D, Gagliardi M, et al. MiR-30e-3p influences tumor phenotype through MDM2/TP53 axis and predicts sorafenib resistance in hepatocellular carcinoma. Cancer Res. 2020;80:1720–34.
Qin L, Huang J, Wang G, et al. Integrated analysis of clinical significance and functional involvement of microRNAs in hepatocellular carcinoma. J Cell Physiol. 2019;234:23581–95.
Li Y, Zhou Y, Ma L, Liu D, Dai Z, Shen J. miR-3677-3p promotes hepatocellular carcinoma progression via inhibiting GSK3beta. Acta Biochim Biophys Sin. 2020;52:1404–12.
He H, Dai J, Feng J, et al. FBXO31 modulates activation of hepatic stellate cells and liver fibrogenesis by promoting ubiquitination of Smad7. J Cell Biochem. 2020;121(8–9):3711–9.
Tan Y, Liu D, Gong J, Liu J, Huo J. The role of F-box only protein 31 in cancer. Oncol Lett. 2018;15:4047–52.
Huang HL, Zheng WL, Zhao R, Zhang B, Ma WL. FBXO31 is down-regulated and may function as a tumor suppressor in hepatocellular carcinoma. Oncol Rep. 2010;24:715–20.
Wierstra I. The transcription factor FOXM1 (Forkhead box M1): proliferation-specific expression, transcription factor function, target genes, mouse models, and normal biological roles. Adv Cancer Res. 2013;118:97–398.
Jeffery JM, Kalimutho M, Johansson P, Cardenas DG, Kumar R, Khanna KK. FBXO31 protects against genomic instability by capping FOXM1 levels at the G2/M transition. Oncogene. 2017;36:1012–22.
Li W, Dong X, He C, et al. LncRNA SNHG1 contributes to sorafenib resistance by activating the Akt pathway and is positively regulated by miR-21 in hepatocellular carcinoma cells. J Exp Clin Cancer Res. 2019;38:183.
Nio K, Yamashita T, Kaneko S. The evolving concept of liver cancer stem cells. Mol Cancer. 2017;16:4.
Yao B, Li Y, Niu Y, et al. Hypoxia-induced miR-3677-3p promotes the proliferation, migration and invasion of hepatocellular carcinoma cells by suppressing SIRT5. J Cell Mol Med. 2020;24:8718–31.
Feng Y, Jiang W, Zhao W, Lu Z, Gu Y, Dong Y. miR-124 regulates liver cancer stem cells expansion and sorafenib resistance. Exp Cell Res. 2020;394: 112162.
Zhang Z, Tan X, Luo J, Yao H, Si Z, Tong JS. The miR-30a-5p/CLCF1 axis regulates sorafenib resistance and aerobic glycolysis in hepatocellular carcinoma. Cell Death Dis. 2020;11:902.
Liu J, Lv L, Gong J, et al. Overexpression of F-box only protein 31 predicts poor prognosis and deregulates p38alpha- and JNK-mediated apoptosis in esophageal squamous cell carcinoma. Int J Cancer. 2018;142:145–55.
Hu G, Yan Z, Zhang C, et al. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J Exp Clin Cancer Res. 2019;38:188.
Chou LF, Chen CY, Yang WH, et al. Suppression of hepatocellular carcinoma progression through FOXM1 and EMT inhibition via hydroxygenkwanin-induced miR-320a expression. Biomolecules. 2019;10:20.
Meng FD, Wei JC, Qu K, et al. FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma. World J Gastroenterol. 2015;21:196–213.
Su WL, Chuang SC, Wang YC, et al. Expression of FOXM1 and Aurora-A predicts prognosis and sorafenib efficacy in patients with hepatocellular carcinoma. Cancer Biomark. 2020;28:341–50.
Yan D, Yan X, Dai X, et al. Activation of AKT/AP1/FoxM1 signaling confers sorafenib resistance to liver cancer cells. Oncol Rep. 2019;42:785–96.
Karunarathna U, Kongsema M, Zona S, et al. OTUB1 inhibits the ubiquitination and degradation of FOXM1 in breast cancer and epirubicin resistance. Oncogene. 2016;35:1433–44.
Kongsema M, Zona S, Karunarathna U, et al. RNF168 cooperates with RNF8 to mediate FOXM1 ubiquitination and degradation in breast cancer epirubicin treatment. Oncogenesis. 2016;5: e252.
Zhang Z, Liu W, Bao X, et al. USP39 facilitates breast cancer cell proliferation through stabilization of FOXM1. Am J Cancer Res. 2022;12:3644–61.
Wang WD, Shang Y, Wang C, et al. c-FLIP promotes drug resistance in non-small-cell lung cancer cells via upregulating FoxM1 expression. Acta Pharmacol Sin. 2022;43:2956–66.
Shi W, Feng L, Dong S, et al. FBXL6 governs c-MYC to promote hepatocellular carcinoma through ubiquitination and stabilization of HSP90AA1. Cell Commun Signal. 2020;18:100.
Zhang L, Chen J, Ning D, et al. FBXO22 promotes the development of hepatocellular carcinoma by regulating the ubiquitination and degradation of p21. J Exp Clin Cancer Res. 2019;38:101.
Zhu H, Yan F, Yuan T, et al. USP10 promotes proliferation of hepatocellular carcinoma by deubiquitinating and stabilizing YAP/TAZ. Cancer Res. 2020;80:2204–16.
Acknowledgements
We appreciate Hunan Provincial Brain Hospital, Changsha Science and Technology Bureau, Hunan University of Traditional Chinese Medicine for supporting this study.
Funding
This study was funded by the Natural Science Foundation of Changsha (No. kq2014189).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethical approval
All the animal experiments in the current research were conducted in accordance with Guidelines for the care and use of laboratory animals (formulated by the Chinese Commission for Institutional Animal Protection and Use) and approved by the Animal Care Ethics Committee of the Brain Hospital of Hunan Province (approval number: 2019K033).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, H., Zhou, J., Cheng, F. et al. MiR-3677-3p promotes development and sorafenib resistance of hepatitis B-related hepatocellular carcinoma by inhibiting FOXM1 ubiquitination. Human Cell 36, 1773–1789 (2023). https://doi.org/10.1007/s13577-023-00945-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-023-00945-z