Abstract
This study aimed to explore the role of miR-222-3p in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC). MiR-222-3p expression in tumor tissues of HBV (+) or HBV (−) HCC patients and corresponding cell lines was detected by quantitative reverse transcription PCR (qRT-PCR). Cell proliferation was assessed by cell counting kit-8 (CCK-8) and colony formation assays. Cell apoptosis was evaluated by flow cytometry. The potential targets of miR-222-3p were predicted by Targetscan, and the binding relationship between miR-222-3p and thrombospondin-1 (THBS1) was determined by luciferase reporter assay and RNA immunoprecipitation (RIP) assay. MiR-222-3p was significantly upregulated in HCC tissues and cell lines and further elevated by HBV infection. MiR-222-3p downregulation effectively inhibited the proliferation and induced the apoptosis of HBV (−) HepG2 cells, HBV (+) HepG2.2.15 cells, Huh7-V cells, and Huh7-HBV cells. In addition, miR-222-3p overexpression enhanced the proliferation of these cell lines but exhibited no obvious effect on their apoptosis. Mechanistically, miR-222-3p was directly bound to the 3’-UTR of THBS1 and acted as its competing endogenous RNA (ceRNA). Interestingly, THBS1 silencing attenuated the inhibitory effect of miR-222-3p downregulation on the proliferation of these cell lines in vitro. Our results revealed that HBV infection further increased miR-222-3p expression and promoted HCC progression via miR-222-3p-mediated THBS1 downregulation. Our findings suggest that miR-222-3p might be a potential diagnostic and therapeutic target for HCC and HBV-related HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide [1, 2]. One reason for HCC is hepatitis B virus (HBV) infection, which is also the most severe and prevalent chronic viral infection [3]. It is estimated that HBV has infected a total of two billion people in the world, and chronic carriers of HBV are approximately more than 10–20% of total HBV cases [4, 5]. Increasing evidences demonstrate that people infected with HBV are more susceptible to HCC [6]. Therefore, it is more urgent to understand the pathogenesis of HBV-caused HCC, which may contribute to identifying effective targets for HCC diagnosis and treatment.
MicroRNAs (miRNAs) are a type of conserved non-coding RNAs with approximately 20–22 nucleotides in length and can bind to the 3’-untranslated region (UTR) of target mRNAs to inhibit the transcription of downstream genes [7]. Increasing studies have revealed that miRNAs are closely associated with pathological processes, including cell proliferation, apoptosis, and metastasis [8]. The dysregulation of miRNAs plays essential roles as tumor suppressors and oncogenes in the development of human malignancies, including breast cancer [9], lung cancer [10], osteosarcoma [11], and leukemia [12]. In HBV-related HCC, previous studies identified many dysregulated miRNAs, such as miR-325-3p [13], miR-2 [14], and miR-122 [15], to name a few, and revealed their crucial roles. MiR-222-3p was upregulated and identified as an important oncogene in various types of human cancers, including osteosarcoma [16], diffuse large B-cell lymphoma [17], endometrial carcinoma [18], and renal cell carcinoma [19]. A recent study found that miR-222-3p was upregulated in HCC tissues [20] and further upregulated in HBV-infected HCC tissues compared with that in HBV negative HCC tissues [21]. However, the role of miR-222-3p in HCC and HBV-related HCC remains unclear.
The trombospondin family is composed of five secreted proteins that have diverse roles in regulating cellular function [22]. Thrombospondin-1 (THBS1), a member of the thrombospondin family, is the first recognized endogenous antiangiogenic factor [23]. More and more studies demonstrate that THBS1 could inhibit tumor growth by blocking tumor angiogenesis, a key element during cancer development [24]. Previous studies reported that miRNAs acted their regulatory functions in cancer progression through binding to the 3’-UTR of THBS1, such as miR-19a in colorectal cancer [25] and miR-194 in colon cancer [26]. In addition, THBS1 methylation promoted tumor angiogenesis in HCC and mediated the inhibitory effects of silencing long non-coding RNA BZRAP1-AS1 on tumor angiogenesis in HCC [23]. These reports suggested that THBS1 might be direct targets of miRNAs to participate in the progression of human cancer, including HCC.
In this study, we demonstrated that miR-222-3p was upregulated in HCC. Moreover, HBV infection further enhanced miR-222-3p expression, indicating the oncogenic role of miR-222-3p in HCC and HBV-related HCC. Further, function assays revealed that miR-222-3p downregulation effectively inhibited proliferation and induced apoptosis of four HCC cell lines (HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells) and miR-222-3p overexpression significantly enhanced their proliferation. Taken together, we demonstrated that miR-222-3p downregulation inhibited the progression of HCC and HBV-related HCC in vitro by directly targeting THBS1, suggesting that miR-222-3p might be an effective therapeutic target for HCC, especially HBV-related HCC.
Materials and methods
Tissue specimens
A total of 30 pairs of HCC tissues and adjacent para-cancerous tissues (without cirrhosis) were obtained from 30 HCC patients at Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University. Meanwhile, these para-carcinoma tissues are from the area without cirrhosis as authenticated by two professional pathologists. Informed consent was obtained from all participants. This study was approved by the Human Ethics Committee of Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University (Approval number: KY2016-02,633). The relationships between miR-222-3p expression level and clinicopathological characteristics of HCC patients were shown in Table 1.
Cell culture
HBV-related cell lines HepG2 (HBV−), HepG2.2.15 (HBV+), and Huh7, normal hepatocyte cell line (LO2), and HEK-293 T cells were purchased from American Type Culture Collection (ATCC, Manassas, USA). All cells were cultured in DMEM (Gibco, USA) with 10% FBS (Gibco, USA) and 1% penicillin–streptomycin (Invitrogen, Shanghai, China) at 37 °C with an additional 5% CO2.
HBV plasmid transfection
An HBV replicon plasmid (HBV 1.3- mer WT replicon, Wuhan SeqHealth Tech Co., Ltd. China) was used as a standard to quantify absolute HBV copy numbers in the culture supernatant. Huh7 cells were transfected with HBV plasmid or empty vector to establish Huh7-HBV and Huh7-V cells (control), respectively, for subsequent experiments.
Cell transfection
The miR-222-3p mimic, miR-222-3p inhibitor, and their corresponding negative control (miRNA and inhibitor NC), as well as two small interfering RNA targeting THBS1 (si-THBS1-1 and -2) and si-NC were purchased from RiboBio (Guangzhou, China). These mimics/inhibitor and si-RNAs were transfected into HepG2 and HepG2.2.15, Huh7-V, and Huh7-HBV cells at 50 nM by Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The sequences used in this study were listed in Table 2. After transfection for 48 h, cells were collected and used for the subsequent experiments.
RNA isolation and qRT-PCR
Total RNAs of HCC tissues and cultured cells were extracted by using TRIzol reagent (Invitrogen, USA). Complementary DNA was obtained by a PrimeScript RT reagent kit (Takara, Tokyo, Japan). Then all PCR reactions were performed on a 7300 real-time PCR system (Applied Biosystems) with SYBR Green Master (Roche, USA). Relative expression changes of miR-222-3p and THBS1 were calculated by using the 2−ΔΔCt method with U6 and GAPDH as the internal references, respectively. The primers used in this study were miR-222-3p forward 5’- CTCAGTAGCCAGTGTAG -3’ and reverse GAACATGTCTGCGTATCTC-3’, U6 forward 5’-ACAGATCTGTCGGTGTGGCAC-3’ and reverse 5’-GGCCCCGGATTATCCGACATTC-3’, THBS1 forward 5’-GCTGGAAA TGTGGTGCTTGTCC-3’ and reverse 5’-CTCCATTGTGGTTGAAGCAGGC-3’, GAPDH forward 5’-ATCCACGGGAGAGCGACAT-3’ and reverse 5’-CAGCTGCTTGT AAAGTGGAC-3’.
Western blot
Total protein of cultured cells was extracted by using RIPA lysis buffer (Beyotime, Shanghai, China). Approximately equal amounts of protein samples were separated by 10% SDS-PAGE and then transferred onto PVDF membranes. The membranes were incubated with specific primary antibodies against THBS1 (1:1000, LS-C88176, LSBio) and GAPDH (1:5000, ab181602, Abcam) overnight 4 °C. On the following day, the membranes were incubated with HRP-conjugated secondary antibody (1: 10,000, ZSGB-BIO, China) for 2 h at room temperature. Protein bands were visualized by using enhanced chemiluminescence (ECL) kit (Amersham Pharmacia Biotech, Amersham, UK). Then, the X-ray film (Sangon, Shanghai, China) was exposed for 10–60 s, and the contents of protein bands were analyzed by using Image Lab 5.1 (Bio-Rad, USA).
CCK-8 assay
Cell viability was evaluated by using the Cell Counting Kit-8 (CCK-8) (Dojindo, Japan). In brief, 2 × 104 transfected or un-transfected HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were seeded into a 96-well plate and cultured overnight. After transfection for 48, 72, and 96 h, 10 μL CCK-8 solution was added into each well and incubated for another 2 h. Finally, cell viability was shown by the OD450 values under a microplate reader (Bio-Rad, USA).
Colony formation assay
Colony formation assay was evaluated as previously described [27]. Briefly, approximately 1000 transfected HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were seeded into a six well plate and cultured for two weeks. Cells were fixed in 4% paraformaldehyde for 20 min and stained with 0.5% crystal violet for 20 min. Then the stained cells were observed and counted by using an inverted microscope.
RNA immunoprecipitation assay
RNA immunoprecipitation (RIP) assay was carried out using the Magna RIP RNA‐binding protein immunoprecipitation kit (Millipore, Billerica, Massachusetts) according to the manufacturer’s protocol. Antibodies against Ago2 and IgG were purchased from Millipore. The co-precipitated RNAs (total RNA) were extracted for the detection of THBS1 expressions by real-time PCR.
Luciferase reporter assay
The putative binding site between miR-222-3p and THBS1 was predicted by using Targetscan. Wild type (WT) and mutant type (MUT) fragments of THBS1 containing the miR-222-3p putative binding site were amplified and cloned into the downstream renilla luciferase gene of psiCHECK-2 luciferase reporter vector (Promega, USA). The recombinant luciferase reporter vectors were co-transfected with miR-222-3p mimics or miR-NC into 293 T cells using Lipofectamine 3000. At 48 h after transfection, cells were collected, and the relative luciferase activity of THBS1-WT or THBS-MUT was detected by using the dual luciferase reporter system (Promega).
Statistical analysis
Statistical analyses were performed using SPSS 20.0 (SPSS Inc., Armonk, NY, USA), and data were presented as means ± standard deviation (SD). All experiments were repeated five independent times. Comparisons between two or multiple groups were performed by using Student’s t test or one-way ANOVA, respectively. p < 0.05 was considered as the significant threshold.
Results
HBV infection significantly enhanced the expression of miR-222-3p in HCC
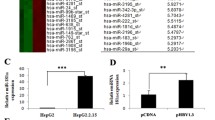
It has been reported that HBV infection upregulated miR-222-3p expression in HCC [21]. To explore the role of miR-222-3p in HBV-related HCC, we firstly detected miR-222-3p expression in HBV-related HCC cell lines and tumor tissues. The results showed that miR-222-3p mRNA level was higher in HCC tissues than that in para-carcinoma tissues (p < 0.001, Fig. 1A). Meanwhile, high miR-222-3p expression was observed in most tissues of HBV (+) HCC patients, and low miR-222-3p expression was observed in most tissues of HBV (−) HCC patients (Fig. 2B). The copies of HBV DNA were higher in HBV (+) HepG2.2.15 cells than in HBV (−) HepG2 cells (p < 0.001) and LO2 cells (p < 0.001, Fig. 1C) and higher in Huh7-HBV cells than in Huh7 cells (p < 0.001) and Huh7-V cells (p < 0.001, Fig. 1D). Moreover, miR-222-3p expression was higher in HBV (−) HepG2 cells and HBV (+) HepG2.2.15 cells than in LO2 cells (p < 0.01 vs HBV (−) HepG2 and p < 0.001 vs HBV (+) HepG2.2.15), while miR-222-3p the expression was further increased in HBV (+) HepG2.2.15 cells than in HBV (−) HepG2 cells (p < 0.001, Fig. 1E). Similarly, miR-222-3p expression was higher in Huh7-HBV cells than in Huh7 cells (p < 0.001) and Huh7-V cells (p < 0.001, Fig. 1F). These results confirmed that HBV infection significantly enhanced miR-222-3p expression in HCC.
HBV infection significantly enhanced miR-222-3p expression in HCC. A MiR-222-3p expression in HCC tissues and adjacent para-cancerous tissues was detected by qRT-PCR (n = 30). B The percentage of HBV (\(-\)) and HBV (+) HCC tissues was evaluated according to the median miR-222-3p expression (n = 30). C The copies of HBV DNA in HepG2, HepG2.2.15, and LO2 cells. D HBV DNA copies in Huh7-V, Huh7-HBV, and Huh7 cells. E MiR-222-3p expression in HepG2, HepG2.2.15, and LO2 cells was assessed by qRT-PCR. F MiR-222-3p expression in Huh7-V, Huh7-HBV, and Huh7 cells was assessed by qRT-PCR. Data were presented as mean ± SD. **p < 0.01, ***p < 0.001
MiR-222-3p downregulation inhibited proliferation and induced apoptosis of HBV(−) and HBV(+) related HCC cells. A and B HepG2 and HepG2.2.15 cells were transfected with miR-222-3p inhibitor and inhibitor NC. A The transfection efficiency was confirmed by qRT-PCR. B Cell viability was evaluated by CCK-8 assay. C and D Huh7-V and Huh7-HBV cells were transfected with miR-222-3p inhibitor and inhibitor NC. C The transfection efficiency was confirmed by qRT-PCR. D Cell viability was evaluated by CCK-8 assay. E and F HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were transfected with miR-222-3p inhibitor and inhibitor NC. E Cell proliferation was evaluated by colony formation assay. F Cell apoptosis was evaluated by flow cytometry. Data were presented as mean ± SD. *p < 0.05, **p < 0.01
MiR-222-3p downregulation effectively inhibited proliferation and induced apoptosis of HBV-related HCC cells
To investigate the role of miR-222-3p in HCC, HepG2 and HepG2.2.15 cells were transfected with miR-222-3p inhibitor and inhibitor NC, respectively. The transfection efficiency was determined by qRT-PCR (p < 0.01, Fig. 2A). By performing CCK-8 assay, we found that miR-222-3p downregulation significantly reduced cell viability compared with inhibitor NC in these two cell lines (p < 0.01, Fig. 2B). Then miR-222-3p inhibitor and inhibitor NC were transfected into Huh7-V and Huh7-HBV cells, and the transfection efficiency was determined by qRT-PCR (p < 0.01, Fig. 2C). Similarly, CCK-8 assay showed that miR-222-3p downregulation significantly reduced cell viability compared with inhibitor NC in these two cell lines (p < 0.001, Fig. 2D). The colony formation assay resultsalso revealed that miR-222-3p downregulation significantly inhibited cell proliferation of HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells compared with inhibitor NC (p < 0.01 for HepG2, HepG2.2.15, and Huh7-V, and p < 0.001 for Huh7-HBV, Fig. 2E). Subsequently, flow cytometry results revealed that miR-222-3p downregulation obviously exacerbated apoptosis of HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells compared with inhibitor NC (p < 0.01 for HepG2, HepG2.2.15, and p < 0.001 for Huh7-V and Huh7-HBV, Fig. 2F). These results demonstrated that miR-222-3p inhibition effectively inhibited the progression of HBV (−) and HBV (+) HCC in vitro.
MiR-222-3p overexpression promoted proliferation of HBV-related HCC cells
To further determine the function of miR-222-3p in HCC, HepG2 and HepG2.2.15 cells were transfected with miR-222-3p mimics and miR-NC. The transfection efficiency was confirmed by qRT-PCR (p < 0.01, Fig. 3A). CCK-8 assay showed that miR-222-3p overexpression obviously enhanced the viability of HepG2 and HepG2.2.15 cells compared with negative control miR-NC (p < 0.01 for HepG2, p < 0.05 for HepG2.2.15, Fig. 3B). Then miR-222-3p mimics and miR-NC were transfected into Huh7-V and Huh7-HBV cells, and the transfection efficiency was determined by qRT-PCR (p < 0.01, Fig. 3C). The results of CCK-8 assay revealed that miR-222-3p overexpression obviously enhanced the viability of Huh7-V and Huh7-HBV cells compared with miR-NC (p < 0.01 for Huh7-V, p < 0.05 for Huh7-HBV, Fig. 3D). By performing colony formation assay (Fig. 3E), we found that miR-222-3p overexpression significantly increased the number of colonies in HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells compared with miR-NC group (p < 0.01 for HepG2, p < 0.01 for HepG2.2.15, p < 0.001 for Huh7-V and Huh7-HBV; Fig. 3E). However, there was no obvious effect of miR-222-3p mimics and miR-NC on apoptosis of these four cell lines (Fig. 3D). These results indicated that miR-222-3p overexpression effectively promoted the proliferation of HBV (−) and HBV (+) HCC cells in vitro.
MiR-222-3p overexpression promoted the proliferation of HBV(−) and HBV(+) related HCC cells. A and B HepG2 and HepG2.2.15 cells were transfected with miR-222-3p mimics and miR-NC. A The transfection efficiency was confirmed by qRT-PCR. B Cell viability was evaluated by CCK-8 assay. C and D Huh7-V and Huh7-HBV cells were transfected with miR-222-3p mimics and miR-NC. C The transfection efficiency was confirmed by qRT-PCR. D Cell viability was evaluated by CCK-8 assay. E and F HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were transfected with miR-222-3p mimics and miR-NC. E Cell proliferation was evaluated by colony formation assay. F Cell apoptosis was evaluated by flow cytometry. Data were presented as mean ± SD. *p < 0.05, **p < 0.01
THBS1 was a target of miR-222-3p
Correlation analysis revealed an obvious negative correlation between the level of miR-222-3p and THBS1 in HCC patients (p < 0.05, Fig. 4A). To further explore the mechanism of miR-222-3p in HBV-related HCC, we predicted the interaction of miR-222-3p and THBS1 using Targetscan and found that THBS1 might be a target of miR-222-3p (Fig. 4B). Then we performed luciferase reporter assays, and the results showed that miR-222-3p overexpression significantly reduced the relative luciferase activity of THBS1-WT compared with miR-NC (p < 0.05) but had no effect on the relative luciferase activity of THBS1-MUT (Fig. 4C). Further, RIP assay was performed in HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells. More THBS1 was pulled down in the anti-AGO2 group than the IgG group and input group (all p < 0.001), further indicating that THBS1 was a target of miR-222-3p. Next, HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were transfected with miR-222-3p mimics, miR-NC, miR-222-3p inhibitor and inhibitor NC, and THBS1 expression was detected. The results showed that miR-222-3p mimics significantly reduced THBS1 expression in these four cells compared with miR-NC (p < 0.01), while miR-222-3p inhibitor significantly increased THBS1 expression in these four cell lines compared with inhibitor NC (p < 0.01, Fig. 4E-G). These results demonstrated that THBS1 was a target of miR-222-3p.
THBS1 was a target of miR-222-3p. A The correlation analysis of miR-222-3p and THBS1 levels in HCC patients. B The putative binding site between miR-222-3p and THBS1 was predicted by Targetscan. C Relative luciferase activity of THBS1-WT and THBS1-MUT in 293 T cells was detected by dual luciferase reporter system. D THBS1 enrichment in HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells by using anti-Ago2 and negative control anti-IgG antibody. E–G HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were transfected with miR-222-3p mimics, miR-NC, miR-222-3p inhibitor and inhibitor NC. THBS1 expression in HepG2 (E), HepG2.2.15 cells (E), Huh7-V (F), and Huh7-HBV cells (F) was evaluated by qRT-PCR. G THBS1 expression in the four cell lines was evaluated by Western blot. Data were presented as mean ± SD. *p < 0.05, **p < 0.01
MiR-222-3p exerted its function in HBV-related HCC through targeting THBS1
To determine whether the effects of miR-222-3p on HBV-related HCC were mediated by THBS1, HepG2, and HepG2.2.15 cells were transfected with inhibitor NC, miR-222-3p inhibitor, or co-transfected with miR-222-3p inhibitor and si-NC, or co-transfected with miR-222-3p inhibitor and si-THBS1-1 or THBS1-2. The transfection efficiency of si-THBS1-1 and 2 was determined by Western blot, and the results revealed that si-THBS1 significantly reduced THBS1 at the protein level compared with si-NC in both HepG2 and HepG2.2.15 (Fig. 5A). By performing CCK-8 assay (Fig. 5B), we found that miR-222-3p inhibition significantly inhibited the viability of HepG2 and HepG2.2.15 cells (p < 0.05), while co-transfection of miR-222-3p inhibitor and si-THBS1-1 or si-THBS1-2 obviously attenuated the inhibitory effect of miR-222-3p inhibitor on the viability of these two cell lines (p < 0.05 for HepG2, p < 0.01 for HepG2.2.15, Fig. 5B). The same treatments were performed in Huh7-V and Huh7-HBV cells, and the transfection efficiency was confirmed by Western blot (Fig. 5C). The results of CCK-8 assay showed that miR-222-3p inhibition significantly inhibited the viability of Huh7-V and Huh7-HBV cells (p < 0.05), while co-transfection of miR-222-3p inhibitor and si-THBS1-1 or si-THBS1-2 obviously attenuated the inhibitory effect of miR-222-3p inhibitor on the viability of these two cell lines (p < 0.01 for Huh7-V, p < 0.001 for Huh7-HBV, Fig. 5D). The results of colony formation assay revealed that miR-222-3p inhibition significantly inhibited the proliferation of HepG2, HepG2.2.15, Huh7-V, and Huh-HBV cells, while co-transfection of miR-222-3p inhibitor and si-THBS1-1 or si-THBS1-2 obviously attenuated the inhibitory effect of miR-222-3p inhibitor on the proliferation of these four cell lines (p < 0.01 for all, Fig. 5E and F). These results demonstrated that miR-222-3p inhibition suppressed the progression of HCC and HBV-related HCC through targeting THBS1 in vitro.
MiR-222-3p exerts its function in HBV-related HCC through targeting THBS1. A and B HepG2 and HepG2.2.15 cells were transfected with two si-THBS1 (−1 and −2) and si-NC. The expression of THBS1 was detected by Western blot. B HepG2 and HepG2.2.15 cells were transfected with inhibitor NC, miR-222-3p inhibitor, or co-transfected with miR-222-3p inhibitor and si-NC, or co-transfected with miR-222-3p inhibitor and si-THBS1-1. Cell proliferation was evaluated by CCK-8 assay. C Huh7-V and Huh7-HBV cells were transfected with two si-THBS1 (−1 and −2) and si-NC. The expression of THBS1 was detected by Western blot. D Huh7-V and Huh7-HBV cells were transfected with inhibitor NC, miR-222-3p inhibitor, or co-transfected with miR-222-3p inhibitor and si-NC, or co-transfected with miR-222-3p inhibitor and si-THBS1-1. Cell proliferation was evaluated by CCK-8 assay. E and F HepG2, HepG2.2.15, Huh7-V, and Huh7-HBV cells were transfected with inhibitor NC, miR-222-3p inhibitor, or co-transfected with miR-222-3p inhibitor and si-NC, or co-transfected with miR-222-3p inhibitor and si-THBS1-1. Cell proliferation of HepG2 E, HepG2.2.15 cells E and Huh7-V F, and Huh7-HBV cells F was evaluated by colony formation assay. Data were presented as mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
In the past decades, the lack of effective early‐stage diagnostic modalities has led to a poor prognosis for HCC patients [28]. Due to the huge threat of HBV infection to HCC patients, it is necessary to identify more specific targets for the early diagnosis of HBV-related HCC and develop new therapeutic drugs to prevent its outcomes [29, 30]. Interestingly, several previous reviews demonstrated that miRNAs exerted great potentials in the diagnosis and prognosis of HCC, including HBV-related HCC, and even in HCC therapy both as miRNA mimic and antimir [31, 32]. Hence, more and more attentions have focused on the functions and molecular mechanisms of miRNAs in HCC. In the present study, we aimed to investigate the role of miR-222-3p in HBV-related HCC and revealed that miR-222-3p served as an oncogene in HBV-related HCC by targeting THBS1.
We firstly revealed that miR-222-3p was upregulated in HCC cell lines. Moreover, HBV (+) cell lines exhibited a higher miR-222-3p level than HBV (−) cell lines. In addition, high miR-222-3p expression was observed in HCC tissues of HBV (+) patients than in HCC tissues of HBV (−) patients. These results indicated that HBV infection enhanced miR-222-3p expression, consistent with a previous study showing that miR-222-3p level was elevated in HCC tissues of HBV (+) and HBV (−) patients [20, 21]. In previous studies, miR-222-3p was identified to play an oncogenic role in different types of cancers, including osteosarcoma [16], ovarian cancer [33], renal clear cell carcinoma [34], and non-small cell lung cancer [35]. Here, we investigated the function of miR-222-3p and found that miR-222-3p downregulation significantly inhibited the progression of HCC and HBV-related HCC. Meanwhile, miR-222-3p overexpression enhanced the proliferation of HCC and HBV-related HCC cells. In comparison with the roles of miR-222-3p in other cancers, our results revealed a novel role of miR-222-3p in HCC, including HBV-related HCC.
Increasing evidences reported that miRNAs could act as competitive endogenous RNAs (ceRNAs) by directly binding to the 3'-UTR of target mRNAs and then suppress the transcription of their downstream mRNAs [36]. Therefore, we explored the underlying molecular mechanisms of miR-222-3p and indicated that THBS1 might be a target of miR-222-3p. THBS1 exerts fundamental functions in inhibiting tumor angiogenesis to prevent cancer development [37]. In addition, THBS1 was identified to act as direct targets of many miRNAs in diverse human diseases. For example, miR-194 reduces inflammatory response and permeability of human dermal microvascular endothelial cells by targeting THBS1 in chronic idiopathic urticaria [38]. MiR-19a enhances cell proliferation, migration, and invasion by promoting lymphangiogenesis through targeting THBS1 in colorectal cancer [25]. In this study, luciferase reporter assay and RIP assay revealed that miR-222-3p could negatively regulate THBS1 by directly binding to the 3’-UTR of THBS1. Moreover, miR-222-3p downregulation increased THBS1 expression, and miR-222-3p overexpression reduced THBS1 expression. Rescue experiments further demonstrated that the role of miR-222-3p in HCC and HBV-related HCC was partially mediated by THBS1.
Except for THBS1, miR-222-3p may also target other genes in various diseases. For example, miR-222-3p/CDKN1B axis is involved in the progression of intervertebral disc degeneration [39]. MiR-222-3p overexpression promotes the development of diffuse large B-cell lymphoma cells by directly downregulating PPP2R2A [17]. MiR-222-3p participates in the development of oral squamous cell carcinoma through targeting CDKN1B [40]. In addition, FOXP2 [41], HIPK2 [42], HMGA2 and CDKN1B [43] also potential targets of miR-222-3p. These reports suggested that these targets of miR-222-3p might also participate in the progression of HBV-related HCC, which needed to be investigated in the subsequent experiments.
In summary, we revealed that miR-222-3p level was elevated in HCC, and HBV infection further enhanced miR-222-3p expression. Moreover, miR-222-3p downregulation inhibited the progression of both HBV (−) and HBV (+) HCC by upregulating THBS1 in vitro. Our study provided a potential therapeutic target for HCC and HBV-related HCC.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
References
Craig AJ, von Felden J. Tumour evolution in hepatocellular carcinoma. Nat Rev Gastro Hepat. 2020;17:139–52.
Schwabe RF, Greten TF. Gut microbiome in HCC—mechanisms, diagnosis and therapy. J Hepatol. 2020;72:230–8.
Trépo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet. 2014;384:2053–63.
Yuen MF, Chen DS, Dusheiko GM, et al. Hepatitis B virus infection. Nat Rev Dis Primers. 2018;4:18035.
Yuen Man-Fung, Chen Ding-Shinn, Dusheiko Geoffrey M, Janssen Harry L A, Lau Daryl T Y, Locarnini Stephen A, Peters Marion G, Lai Ching-Lung. Hepatitis B virus infection. Nat Rev Dis Primers. 2018. https://doi.org/10.1038/nrdp.2018.35.
Marrone A, Ciotti M, Rinaldi L. Hepatitis B and C virus infection and risk of haematological malignancies. J Viral Hepat. 2020;27:4–12.
Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451–65.
Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World J Biol Chem. 2017;8:45–56.
Fridrichova I, Zmetakova I. MicroRNAs Contribute to Breast Cancer Invasiveness. Cells. 2019. https://doi.org/10.3390/cells8111361.
Du X, Zhang J, Wang J, Lin X, Ding F. Role of miRNA in lung cancer-potential biomarkers and therapies. Curr Pharm Des. 2018;23:5997–6010.
Wang J, Liu S, Shi J, et al. The role of miRNA in the diagnosis, prognosis, and treatment of osteosarcoma. Cancer Biother Radiopharm. 2019;34:605–13.
Mardani R, Jafari Najaf Abadi MH, Motieian M, et al. MicroRNA in leukemia: tumor suppressors and oncogenes with prognostic potential. J Cell Physiol. 2019;234:8465–86.
Li R, Xu T, Wang H, et al. Dysregulation of the miR-325-3p/DPAGT1 axis supports HBV-positive HCC chemoresistance. Biochem Biophys Res Commun. 2019;519:358–65.
Yao L, Zhou Y, Sui Z, et al. HBV-encoded miR-2 functions as an oncogene by downregulating TRIM35 but upregulating RAN in liver cancer cells. EBioMedicine. 2019;48:117–29.
Quoc NB, Phuong NDN, Ngan TK, Linh NTM, Cuong PH, Chau NNB. Expression of plasma hsa-miR122 in HBV-related hepatocellular carcinoma (HCC) in Vietnamese patients. Microrna. 2018;7:92–9.
Guo J, Liu Q, Li Z, Guo H, Bai C, Wang F. miR-222-3p promotes osteosarcoma cell migration and invasion through targeting TIMP3. Onco Targets Ther. 2018;11:8643–53.
Sun S, Wang H, Ji M. Overexpression of miR-222-3p promotes the proliferation and inhibits the apoptosis of diffuse large B-Cell lymphoma cells via suppressing PPP2R2A. Technol Cancer Res Treat. 2019;18:1533033819892256.
Liu B, Che Q, Qiu H, et al. Elevated MiR-222–3p promotes proliferation and invasion of endometrial carcinoma via targeting ERα. PLoS One. 2014. https://doi.org/10.1371/journal.pone.0087563.
Zhao L, Quan J, Li Z, et al. MicroRNA-222-3p promotes tumor cell migration and invasion and inhibits apoptosis, and is correlated with an unfavorable prognosis of patients with renal cell carcinoma. Int J Mol Med. 2019;43:525–34.
Wang X, Liao X, Huang K, et al. Clustered microRNAs hsa-miR-221-3p/hsa-miR-222-3p and their targeted genes might be prognostic predictors for hepatocellular carcinoma. J Cancer. 2019;10:2520–33.
Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in Hepatitis B virus-related hepatocellular carcinoma: potential as biomarkers and therapeutic targets. Front Oncol. 2020;10:1271.
Sipes JM, Murphy-Ullrich JE, Roberts DD. Thrombospondins: purification of human platelet thrombospondin-1. Methods Cell Biol. 2018;143:347–69.
Wang W, Chen G, Wang B, et al. Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation. J Transl Med. 2019;17:421.
Zaslavsky A, Baek KH, Lynch RC, et al. Platelet-derived thrombospondin-1 is a critical negative regulator and potential biomarker of angiogenesis. Blood. 2010;115:4605–13.
Yin Q, Wang PP, Peng R, Zhou H. MiR-19a enhances cell proliferation, migration, and invasiveness through enhancing lymphangiogenesis by targeting thrombospondin-1 in colorectal cancer. Biochem Cell Biol. 2019;97:731–9.
Sundaram P, Hultine S, Smith LM, et al. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71:7490–501.
Wang Y, Yang L, Chen T, et al. A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194–5p/FOXA1 axis. Mol Cancer. 2019. https://doi.org/10.1186/s12943-019-0957-7.
Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–53.
Yan SY, Fan JG, Qio L. Hepatitis B Virus (HBV) infection and hepatocellular carcinoma—new insights for an old topic. Curr Cancer Drug Targets. 2017;17:505–11.
Li Y, Chen X, Huang H, et al. Identification of novel lncrnas for detection of HBV-associated hepatocellular carcinoma. Onco Targets Ther. 2019;12:10199–211.
Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr Gene Ther. 2015;15:142–50.
Xie KL, Zhang YG, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176–92.
Fan L, Lei H, Zhang S, et al. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics. 2020;10:5895–913.
Lyu J, Zhu Y, Zhang Q. An increased level of MiR-222-3p is associated with TMP2 suppression, ERK activation and is associated with metastasis and a poor prognosis in renal clear cell carcinoma. Cancer Biomark. 2020;28:141–9.
Chen W, Li X. MiR-222–3p Promotes cell proliferation and inhibits apoptosis by targeting PUMA (BBC3) in non-small cell lung cancer. Technol Cancer Res Treat. 2020. https://doi.org/10.1177/1533033820922558.
Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83.
Lawler PR, Lawler J. Molecular basis for the regulation of angiogenesis by thrombospondin-1 and -2. Cold Spring Harb Perspect Med. 2012;2(5):a006627. https://doi.org/10.1101/cshperspect.a006627.
Qu S, Yang L, Liu Z. MicroRNA-194 reduces inflammatory response and human dermal microvascular endothelial cells permeability through suppression of TGF-β/SMAD pathway by inhibiting THBS1 in chronic idiopathic urticaria. J Cell Biochem. 2020;121:111–24.
Liu J, Yu J, Jiang W, He M, Zhao J. Targeting of CDKN1B by miR-222-3p may contribute to the development of intervertebral disc degeneration. FEBS Open Bio. 2019;9:728–35.
Yang K, Chen Y, Cui Z, et al. MicroRNA-222-3p participates in the development of oral squamous cell carcinoma by targeting CDKN1B. J Oral Pathol Med. 2020;49:621–9.
Wang H, Deng Z, Chen X, et al. Downregulation of miR-222-3p reverses doxorubicin-resistance in LoVo cells through upregulating forkhead box protein P2 (FOXP2) Protein. Med Sci Monit. 2019;25:2169–78.
Tan X, Tang H, Bi J, Li N, Jia Y. MicroRNA-222–3p associated with Helicobacter pylori targets HIPK2 to promote cell proliferation, invasion, and inhibits apoptosis in gastric cancer. J Cell Biochem. 2018;119:5153–62.
Di Fazio P, Maass M, Roth S, et al. Expression of hsa-let-7b-5p, hsa-let-7f-5p, and hsa-miR-222-3p and their putative targets HMGA2 and CDKN1B in typical and atypical carcinoid tumors of the lung. Tumour Biol. 2017;39:1010428317728417.
Acknowledgments
The authors would like to express our gratitude to those who have critically reviewed this manuscript and those who give us help during this experiment.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
HC: guarantor of integrity of the entire study, study concepts. YS, PS, QW: study design, study perform, original manuscript writing. BL, ZY, HJ: definition of intellectual content, literature research, experimental study, data acquisition.
Corresponding author
Ethics declarations
Conflicts of interests
The authors declare they have no conflict of interest.
Ethical approval
This study was approved by the Ethics Committee of the Shandong Provincial Hospital, Cheeloo College of Medicine, Shandong University. All procedures were followed the guideline of this hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sun, Y., Shi, P., Wu, Q. et al. MiR-222-3p induced by hepatitis B virus promotes the proliferation and inhibits apoptosis in hepatocellular carcinoma by upregulating THBS1. Human Cell 34, 1788–1799 (2021). https://doi.org/10.1007/s13577-021-00577-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00577-1