Abstract
Neuropeptides are structurally highly diverse messenger molecules that act as regulators of many physiological processes such as development, metabolism, reproduction or behavior in general. Differentiation of neuropeptidergic cells often corresponds with the presence of the transcription factor DIMMED. In the central nervous system of the fruit fly Drosophila melanogaster, DIMMED commonly occurs in neuroendocrine neurons that release peptides as neurohormones but also in interneurons with complex branching patterns. Fly strains with green fluorescence protein (GFP)-expressing dimmed cells make it possible to systematically analyze the processed neuropeptides in these cells. In this study, we mapped individual GFP-expressing neurons of adult D. melanogaster from the dimmed (c929)>GFP line. Using single cell mass spectrometry, we analyzed 10 types of dimmed neurons from the brain/gnathal ganglion. These cells included neuroendocrine cells with projection into the retrocerebral complex but also a number of large interneurons. Resulting mass spectra not only provided comprehensive data regarding mature products from 13 neuropeptide precursors but also evidence for the cellular co-localization of neuropeptides from different neuropeptide genes. The results can be implemented in a neuroanatomical map of the D. melanogaster brain.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Neuropeptides are structurally highly diverse messenger molecules which act, mostly via G-protein coupled receptors, as regulators of many physiological processes such as development, metabolism, reproduction or behavior in general. For the fruitfly Drosophila melanogaster, which is an important model organism in neuropeptide research, more than 50 neuropeptide and protein hormone genes and the corresponding receptor genes have been described (e.g., [1,2,3,4]). Although we have only limited knowledge about the coordinated action of neuropeptides within the central nervous system (CNS), specific functions of many neuropeptides are known and these functions are often related to hormone activity (see [5, 6]). In the central nervous system (CNS), a majority of neurohormones is produced in LEAP cells (large, episodically-releasing, amidating peptide producing cells that contain the transcription factor DIMMED [7]); a fundamental controller and promotor of the regulated secretory pathway of many neuropeptides [8,9,10]. It was also shown that DIMMED plays multiple transcriptional roles such as affecting cell growth and inhibiting apoptosis at different developmental stages in a cell type-specific manner [11]
Dimmed cells are characterized typically by their relatively large soma size with an enhanced ability to accumulate and/or release large amounts of secretory granules containing mature amidated peptides [12, 13]. In neuroendocrine cells, for example, this accumulation of neuropeptides is necessary to raise hormone levels in the hemolymph to a physiologically effective dosage. Peptidergic interneurons with complex branching patterns also express dimmed [9].
The c929>GAL4 Drosophila strain with green fluorescence protein (GFP)-expressing dimmed cells makes it possible to systematically analyze processed neuropeptides in neurosecretory neurons of the CNS, either by methods such as immunocytochemistry or by means of single cell mass spectrometry (SCMS) [7, 14]. The mapping of DIMMED in the CNS of 3rd instar larvae in combination with immunocytochemistry revealed that most dimmed cells were peptidergic, but not all peptidergic cells synthesize DIMMED [7]. Using affinity cell-capture mass spectrometry, pooled GFP-expressing dimmed cells from adult Drosophila brains yielded not less than 42 peptides from 16 neuropeptide precursors [14]. However, a distinct correlation to specific dimmed neurons is missing so far.
In this study, we mapped individual gfp-expressing neurons from the c929>GAL4 driver line by direct MALDI-TOF mass spectrometry. Presence of GFP expression was necessary to identify somata of neurosecretory neurons or large peptidergic interneurons throughout dissections. Whereas such neurons can frequently be visualized in larger insects by retrograde dye filling via nerves (e.g., [15]), retrograde filling is not a suitable technique to detect specific neurons in insects as small as D. melanogaster. Instead, in D. melanogaster, numerous Gal4 lines are available, which allow the identification of different neuron populations by synthesis of marker proteins such as GFP under UAS control. The c929>GFP driver line is one of these Gal4 lines and resulting mass spectra provided data on (1) possible co-localization of neuropeptides from different neuropeptide genes and (2) major processing products of prepropeptides translated from these genes. As it turned out, SCMS was a reliable method to reveal the neuropeptidome of dimmed cells in the brain of adult flies. In our study, we targeted cells with a soma size ≥7 μm; the results can be implemented in a neuroanatomical map of the D. melanogaster brain.
Experimental
Fly Strain
Adults (2–10 d old) of both sexes of D. melanogaster were used for experiments. Flies were raised on standard agar, cornmeal, and yeast food at 25 °C and 60% relative humidity on a 12 h:12 h light–dark cycle. For cell identification, w;c929-Gal4;+ (provided by Christian Wegener, University Würzburg, Germany) [13] was crossed to y* w* P{UAS-mCD8::GFP.L}LL4; P{UAS-mCD8::GFP.L}LL5; P{UAS-mCD8::GFP.L}LL6 (provided by Manuela Ruppert, University of Cologne) [16].
Chemicals
Chemicals were purchased from SIGMA-Aldrich (Steinheim, Germany).
Single Cell Dissection for Mass Spectrometry
Adult flies of both sexes were immobilized on ice and brains were removed in ice-cold saline buffer (125.94 mM NaCl, 5.37 mM KCl, 0.17 mM NaH2PO4, 0.22 mM KH2PO4, pH 7.2) with forceps under a stereofluorescence microscope (SteREO Lumar V12, Carl Zeiss AG, Goettingen, Germany) equipped with a GFP filter set (488 nm). The dissected brains were transferred to a fresh drop of either saline buffer as described above or saline buffer containing 33% glycerol. Both approaches showed similar results. Without any enzyme treatment, only one individual cell soma per brain sample was manually (without any manipulators) removed using a glass capillary that was fitted to a tube connected to a mouthpiece [see 15]. Before, the ganglionic sheath of the brain was carefully opened close to the position of the cell of interest using a fine scissor and without obvious changes in the position of surrounding cells. Then, the tip of the glass capillary was positioned at the cell body. Subsequently, the soma was removed from the surrounding tissue and completely absorbed into the tip of the capillary by applying gentle negative pressure. Each isolated soma was transferred onto a stainless steel target plate for MALDI-TOF mass spectrometry analysis. Using the same glass capillary, the buffer solution around the cell was removed and the respective cells dried at room temperature. The area around the cell was then carefully rinsed with ice-cold 50% methanol/water using another glass capillary and dried again. Only cell bodies without visible contamination from attached tissue or other cells were analyzed. Finally, cells and surrounding area were covered with matrix; resulting in a matrix spot of about 500 μm. Many somata of dimmed cells are located near the surface of the brain but the decision to analyze a specific cell type was based solely on cell size and frequent traceability.
Matrix Application
Solutions of 10 mg/ml 2,5-dihydroxybenzoic acid (DHB) dissolved in 20% acetonitrile/1% formic acid, 79% HPLC grade water or, alternatively, 10 mg/mL α–cyano-4-hydroxycinnamic acid (CHCA) dissolved in 60% ethanol, 36% acetonitrile, 4% HPLC grade water were used as matrices. Prior to analysis, CHCA stock solution was diluted 1:3 with 50% methanol/water. Depending on cell size, 10–20 nL matrix solution was pipetted onto a cell sample using a nanoliter injector (World Precision Instruments, Berlin, Germany). For an even distribution of DHB matrix crystals samples were dried using a regular hairdryer.
MALDI-TOF Mass Spectrometry
Mass spectra were acquired in reflector positive ion mode under manual control on an UltrafleXtreme TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) in a detection range of m/z 600–10,000. The instrument settings were optimized for the mass ranges of m/z 600–4000 and 3000–10,000, and calibrated using a suitable synthetic peptide mixture, respectively. Laser fluency was adjusted to provide the optimal signal-to-noise ratio. Data were collected using a laser beam diameter of 50 μm and random walk laser setting over the entire matrix spot. The data obtained in these experiments were processed with the FlexAnalysis 3.4 software package. MS/MS was performed with LIFT technology. LIFT acceleration was set at 1 kV. The number of laser shots used to obtain a spectrum varied from 5000 to 10,000, depending on signal quality. Peptide identities were verified using MS/MS fragmentation of the molecules, determination of the molecular mass of the fragments, and comparison of predicted (http://prospector.ucsf.edu) and experimentally obtained fragmentation patterns.
Drosophila Precursor Sequences
Neuropeptide precursor sequences were obtained from blast searches at NCBI database (https://www.ncbi.nlm.nih.gov/). Signal peptides were predicted using the SignalP 4.1 server (www.cbs.dtu.dk/services/SignalP/). Potential cleavage sites were manually assigned according to Veenstra 2000 [17].
Documentation and Single Cell Verification
For documentation, microphotographs were taken before and after cell dissection procedure using a stereofluorescence microscope (SteREO Lumar V12) equipped with a digital camera (AxioCam MRc, Zeiss, Germany). For a more detailed mapping, samples were processed according to [18]. For that, brain samples were fixed in 4% paraformaldehyde dissolved in phosphate buffer solution (PBS) (1.86 mM NaH2PO4, 8.41 mM Na2HPO4, 175 mM NaCl) for 120 min at room temperature under vacuum. Then, samples were washed three times for 10 min in PBS containing 0.1% Triton-X 100 and two times for 10 min in PBS, respectively. After mounting in glycerol containing 20% PBS and 50 mg/ml 1,4-diazabicyclo(2.2.2)octane, samples were analyzed using a Zeiss LSM 510 Meta confocal microscope equipped with a Plan-Apochromat 20x/0.75 objective. GFP was excited with an Argon laser at 488 nm and emission collected via a BP 505–550 filter. Serial optical sections were analyzed with a thickness of 0.3 to 0.8 μm. Contrast adjusted image stacks were used to reconstruct the brain surface whereas GFP expression was visualized by voltex function using Amira 5.4.2 software package (FEI, Hillsboro, OR). The final figures were exported and processed to adjust brightness and contrast with Adobe Photoshop CS6 software (Adobe Systems, San Jose, CA).
Results and Discussion
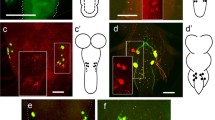
A reconstruction of dimmed cells visualized in an adult brain of D. melanogaster is shown in Figure 1a. The number of dimmed cells detectable with the confocal microscope varied considerably between different brain preparations (Figure 1b–d). Only dimmed cells with sufficient GFP expression in the majority of brain samples and an average soma size of ≥7 μm were selected for SCMS experiments (Figure 1c, d blue marker). These cells could be identified with certainty also during dissections, which simplified the repeatability of mass spectra from the same type of neurons. Differences between images recorded with the fluorescence stereomicroscope used for single cell dissection and a common confocal laser scanning microscope are illustrated in Figure 1e.
Drosophila c929>GFP driver line. (a) 3D-reconstruction of GFP-positive signals (dark red) in an adult brain with gnathal ganglion. (b) Maximum projection view of dimmed cells recorded with a confocal microscope. (c), (d) Schematic reconstruction of dimmed cells in two different brains illustrating the variation in cell numbers between different brain preparations. Spots marked in blue are reproducibly identified as dimmed cells and were used for single cell mass spectrometry. Remaining dimmed cells, which were not detected in the majority of brain preparations, are marked in yellow. (e) Brain as it was seen under the fluorescence stereomicroscope used for manual cell picking. Scale bar: 50 μm. PC: protocerebrum, DC: deutocerebrum, GG: gnathal ganglion, OL: optic lobe.
In the following, we describe 10 different cell types from the c929>GFP strain using direct SCMS. In contrast to FACS-sorting and similar approaches, we show in this study the only way to identify and map single neurons on peptidomics differences within a living brain and by thus exploit the advantage of invertebrate brains. A summary of all peptides observed in these cells is listed in Table 1.
Cell Type #1: Allatostatin C (Ast-C) neuron (n = 6); FlyBase ID: FBgn0032336
A single large bilateral cell (#1; soma size: 18–20 μm) in the posterior medio-lateral protocerebrum is classified as Ast-C neuron (Figure 2a-I). The Ast-C precursor contains a single putative Ast-C sequence. The processing of this peptide was confirmed in mass spectra (Figure 2a-II), which suggested almost equimolar amounts of Ast-C with N-terminal Gln and N-terminal pyroglutamate (pGln). Both forms were identified earlier by mass spectrometry using an immunoaffinity cell enrichment of brain cells from the same D. melanogaster c929>GFP line with GFP expression under UAS control [14]. MS/MS fragmentation of the ion signal at m/z 1921.8 yielded fragments typical of Ast-C with N-terminal Gln (Figure 3a). The type #1 cell is probably identical to the PMP2 neuron described from adult D. melanogaster protocerebrum [19]. An assignment of our analyzed neuron to the mapped Ast-C immunopositive dimmed neurons of larval brain [7] is not possible; this cell type was obviously not part of the GAL4 pattern described from larvae. The type #1 cell could belong to a cell lineage described as neuroendocrine CA-LP 1 cell with projection into the ring gland of larva [20]. Co-localization with other neuropeptides was not found in cell type #1 neurons. The only detected neuropeptide, Ast-C, is a non-amidated peptide (Figure 2a-III). Therefore, these dimmed cells do not likely process amidated neuropeptides.
Mapping of cell type #1-3. Arrows mark the soma location in one brain hemisphere whereas the circle depicts the position of the dissected soma in the contralateral brain hemisphere. (I) Reconstruction of dimmed cells after dissection of an individual dimmed neuron. (II) Mass spectra obtained from dissected dimmed cells. (III) Precursor sequences with signal peptides (highlighted in grey), identified peptides (underlined), confirmed cleavage sites (bold), and amidation signals (italic). (a) Cell type #1: Allatostatin-C (AST-C). The mass spectrum shows the Gln form (Q) and the pyroglutamate form (pQ) of Ast-C. (b) Cell type #2: myosuppressin (MS). The mass spectrum shows the pyroglutamate form of MS and the C-terminal precursor peptide (PP). (c) Cell type #3: allatostatin-A (AST-A)/Myoinhibiting peptide (MIP)/natalisin (Nat). The mass spectrum verified the co-localization of products from three neuropeptide genes. Sodium adduct ions are marked with an asterisk
Cell Type #2: Myosuppressin (MS) neuron (n = 20); FlyBase ID: FBgn0011581
A single large bilateral cell (#2; soma size: 24 μm) in the medio-lateral protocerebrum anterior of the type #1 cell is classified as MS neuron (Figure 2b-I). The MS precursor contains a single putative MS sequence. The processing of this peptide and a precursor peptide that covers the remaining propeptide sequence was confirmed by mass spectra (Figure 2b-II, b-III). This precursor peptide contains an internal disulfide bridge and its presence confirmed indirectly the length of the suggested signal peptide. The type #2 cell is likely identical to the medial protocerebral cells (MP2) described from D. melanogaster. In the CNS, MP2 cells are the first MS-immunopositive neurons during the development [21]. These MP2 neurons do not project into the retrocerebral complex but develop extensive arborizations along the ventral nerve cord. One of the two bilateral MP2 neurons of larva disappears during metamorphosis [21]. Therefore, the type #2 cell analyzed here likely resembles a cell type represented by a single cell only, just as it was described above for type #1 cell. MP2 dimmed neurons with anti-MS immunoreactivity, likely from the same cell lineage, were described from larval D. melanogaster brain [7]. A suggested co-localization with short neuropeptide F [7] or any other neuropeptide was not found in cell type #2 mass spectra of adults.
Cell Type #3: Allatostatin-A (Ast-A) + myoinhibiting peptide (MIP) + Natalisin (Nat) neuron (n = 7); FlyBase IDs: FBgn0015591, FBgn0036713, FBgn0085417
A single bilateral cell (#3; soma size: 14 μm) anteriorly in the ventro-lateral brain is classified as AST-A/MIP/natalisin neuron (Figure 2c-I). This is the only dimmed neuron analyzed with co-expression of three neuropeptide genes (Figure 2c-II, c-III). The AST-A precursor contains four AST-A paracopies. Ion signals of all paracopies were observed in mass spectra of cell type #3 preparations. In addition, an ion signal mass-identical with the suggested C-terminal precursor peptide was found in mass spectra (Figure 2c-II). The MIP precursor contains five MIP paracopies; all of these predicted neuropeptides were detected in mass spectra from type #3 cells. For the first time, MIP-1 could be biochemically confirmed (Figure 3b). This peptide is mass-identical with MIP-2 and, like the other MIPs that do not contain Arg (MIP-4, 5), MIP-1 and 2 were primarily detected as sodium and potassium adducts. The natalisin precursor is the third multiple copy precursor processed in type #3 cells and contains five putative natalisin paracopies. Ion signals of four of these peptides (see Table 1) were observed in mass spectra of cell type #3 preparations. The predicted natalisin-1 was only indicated by a N-terminal fragment (natalisin-11-14), which suggested use of an internal monobasic Lys cleavage site. The analyzed type #3 cell is likely identical with the inferior contralateral interneuron (ICLI), which is one of the few natalisin-immunopositive neurons in the brain [22]. Apparently identical cells were later described as MIP-expressing cells of adult Drosophila brain; also named ICLI neurons [23] or inferior posterior slope neurons with extensive projection processes to different brain neuropiles (IPS; [24]). Analysis of anti-Ast-A immunostaining in the brain of adult D. melanogaster [25] suggests that a cell described as ALT2 (anterolateral tritocerebrum) is identical with the cell type #3 neuron discussed here. These bilateral ALT2 neurons first appeared during metamorphosis, which explains the absence of this cell type in the thorough mapping of larval Ast-A and MIP dimmed cells [7].
Cell Type #4: CAPA Neurons (n = 6); FlyBase ID: FBgn0039722
Capa and pyrokinin (hugin; see cell type #5, Figure 4a-I) genes share a common evolutionary origin and likely result from a gene duplication at the origin of hexapods [26]. The ancestor in hexapods was a single gene encoding three types of receptor ligands [26]. These ligands are designated in insects as pyrokinins (PKs), periviscerokinins (PVKs, CAP2b), and tryptoPKs (new name; [27]), and each activate different receptors in D. melanogaster [28, 29]. The derived capa gene of D. melanogaster encodes two PVKs and a single tryptoPK (CAPA-PK, CAPA-3). Earlier studies suggested that the CAPA-precursor is differentially processed in neuroendocrine cells of abdominal ganglia and gnathal ganglion. Whereas PVKs and tryptoPK were observed in abdominal hormone release sites (perisympathetic organs), only truncated tryptoPK was found in the retrocerebral complex [30, 31]. Two CAPA cells (16 μm) in the gnathal ganglion (see [7, 32]), identical to earlier described CC-MS 2 neurons [20], project into the retrocerebral complex (see Figure 4a-I). These DIMMED-positive neurons were analyzed here and resulting mass spectra confirmed (1) the absence of PVKs, and (2) the presence of N-terminally truncated tryptoPK (Figure 4a-II). In addition, the precursor peptide N-terminal of tryptoPK in the precursor sequence was detected (Table 1). Since transcriptome data did not support different CAPA transcripts in abdominal ganglia and gnathal ganglion (Neupert, Ragionieri, Predel; unpublished), different enzyme complements in the respective CAPA cells are the most likely scenario to yield the different products. This is the only well-supported case for a differential neuropeptide precursor processing in D. melanogaster. The finding of PVKs and full length tryptoPK in brain extracts of adult D. melanogaster [14] can be attributed to two CAPA-immunopositive cerebral cells which were in fact not considered as real CAPA cells at that time [32]. However, these cells, which were not detected in the c929>GFP line, express capa and in this case the processing of the CAPA precursor yielded PVKs and tryptoPK (Neupert, Predel, Wegener, unpublished).
Mapping of cell type #4-6. Arrows mark the soma location in one brain hemisphere while the circle depicts the position of the dissected soma in the contralateral brain hemisphere. (I) Reconstruction of dimmed cells after dissection of an individual dimmed neuron. (II) Mass spectra obtained from dissected dimmed cells. (III) Precursor sequences with signal peptides (highlighted in grey), identified peptides (underlined), confirmed cleavage sites (bold), and amidation signals (italic). (a) Cell type #4: CAPA. The mass spectrum shows signals of truncated CAPA-tryptopyrokinin (tPK) and a CAPA-precursor peptide (PP) and confirms the differential processing of the CAPA precursor in neuroendocrine cells of the gnathal ganglion (shown here) and abdominal ganglia. (b) Cell type #5 hugin-pyrokinin (PK). The mass spectrum shows a prominent ion signal of PK. The weak ion signal at m/z 3307.8 is mass-identical with a PK precursor peptide (PP). (c) Cell type #6: sulfakinin (SK). The mass spectrum shows prominent signals of SK-1 and SK-2. Sulfation of Tyr cannot be detected with MALDI-TOF MS in positive ion mode. Sodium adduct ions are marked with an asterisk
Cell Type #5: Hugin Pyrokinin (huginPK) Neurons (n = 10); FlyBase ID: FBgn0028374
HuginPK expression in D. melanogaster is restricted to several cell populations in the gnathal ganglion. These hugin neurons have projections to pharyngeal muscles, the protocerebrum and also to the retrocerebral complex (CC-MS 1 cells; [20, 33,34,35]), and receive input from external gustatory receptor-expressing neurons as well as pharyngeal chemosensory organs [36]. The huginPK precursor of D. melanogaster contains only two potential PKs. We have analyzed the different huginPK neurons in earlier experiments already [37] and found only PK (SVPFKPRLamide) in single cell mass spectra. Here, we repeated these experiments and collected randomly isolated dimmed neurons (cell size 8–10 μm) from different subpopulations in the gnathal ganglion (see Figure 4b-I). Subsequent mass spectrometric analyses confirmed the presence of Drm-PK (Figure 4b-II). In addition, an ion signal mass-identical with the N-terminal precursor peptide was found, which supported the length of the predicted signal peptide (Figure 4b-III). The second PK, which was originally predicted from the huginPK precursor and named hug γ [38], was not detected in mass spectra.
Cell Type #6: Sulfakinin (SK) Neuron (n = 3); FlyBase ID: FBgn0000500
Two bilateral dimmed neurons (#6, soma size: 10–14 μm) in the medium protocerebrum are classified as SK neurons (Figure 4c-I). The SK precursor contains two SK paracopies (SK-1 and 2) and a further amidated peptide without Arg ("SK-0") (Figure 4c-III). The presence of SK-1 and 2 was confirmed in mass spectra (Figure 4c-II) albeit Tyr sulfation cannot be detected with MALDI-TOF mass spectrometry in positive ion mode. The sulfation of these peptides, essential for proper activation of their receptors [39], was confirmed in earlier studies [14]. Ion signals of SK-0 or other precursor peptides were not observed. The analyzed type #6 cell is likely identical with one of the two medial protocerebrum cells (MP1) with positive SK-immunostaining [40]. These cells are also known as protocerebral descending neurons, which are highly conserved in insects and show extensive arborizations along the entire ventral nerve cord [41]. Putative homologous MP1 dimmed neurons with anti-SK immunoreactivity were described from larval D. melanogaster brain [7]. The suggested co-localization with Ast-C [7] was not found in cell type #6 mass spectra of adults.
Cell Type #7: Corazonin/Short Neuropeptide F (sNPF) Neurons (n = 7); FlyBase ID: FBgn0013767
Three to four dorso-lateral dimmed neurons in the protocerebrum that could not be differentiated from each other during single cell dissections (#7, soma size: 7–9 μm) are classified here together as corazonin/sNPF neurons (Figure 5a-I). The corazonin precursor contains a single corazonin sequence next to the signal peptide (Figure 5a-III). The presence of the pyroglutamyl form of corazonin was confirmed in mass spectra (Figure 5a-II). No ion signal of corazonin with N-terminal Gln could be detected. The sNPF precursor contains four sNPF paracopies, which were confirmed to be processed (Figure 5a-II). The sequence of sNPF-2 is identical with truncated sNPF-1 if the optional internal monobasic cleavage site (Arg) of sNPF-1 is used. Relative signal intensities of sNPFs in type #7 cells (Figure 5a-I) suggest that this cleavage site is indeed efficiently working although minor signals of complete sNPF-1 are present as well. The detection of the precursor peptide that follows the signal peptide in the precursor sequence, with and without the N-terminal amino acid, indicates that the signal peptide might function with different lengths. In all mass spectra of type #7 cells, the corazonin signal was much more prominent than signals of the different sNPFs. Type #7 neurons in the lateral protocerebrum are described as dorso-latero-posterior (DLP) neurons with positive corazonin immunostaining and mRNA expression in adult D. melanogaster [42]. For a number of these lateral neurosecretory cells, a co-localization of corazonin and sNPFs was suggested (adult brains: [43]; larval brains: [7]). Our mass spectra corroborated the expression of the two respective genes in type #7 neurons; other known neuropeptides were not found in these cells.
Mapping of cell type #7-9. Arrows mark the soma location in one brain hemisphere while the circle depicts the position of the dissected soma in the contralateral brain hemisphere. (I) Reconstruction of dimmed cells after dissection of an individual dimmed neuron. (II) Mass spectra obtained from dissected dimmed cells. (III) Precursor sequences with signal peptides (highlighted in grey), identified peptides (underlined), confirmed cleavage sites (bold), and amidation signals (italic). (a) Cell type #7: corazonin (Cor.)/short neuropeptide F (sNPF). The mass spectrum shows the prominent ion signal of the pyroglutamyl form of corazonin, all predicted sNPFs sequences, and an additional sNPF precursor peptide (PP). (b) Cell type #8: pigment dispersing factor (PDF). The mass spectrum shows ion signals of PDF ([M+H]+, [M+Na]+). (c) Cell type #9: neuropeptide-like precursor 1 (NPLP1). The mass spectrum shows the presence of many NPLP1 peptides. Several of these peptides are represented by truncated forms as well
Cell Type #8: Pigment Dispersing Factor (PDF) Neurons (n = 5); FlyBase ID: FBgn0023178
Four bilateral neurons (#8, soma size: 10 μm) in the lateral protocerebrum are classified as PDF neurons (Fig. 5b-I). These dimmed neurons represented the large ventral lateral clock neurons (l-LNv-cells; [44]) that were already analyzed at the single cell level in earlier experiments using a pdf-GAL4 line [37]. The mass spectra of type #10 dimmed cells, which confirmed the presence of the mature PDF (Figure 5b-II, b-III), are nearly identical to the mass spectra obtained from the pdf-GAL4 line. Ion signals of other neuropeptides were not detected in mass spectra of type #8 cells.
Cell Type #9: Neuropeptide-Like Precursor 1 (NPLP1) Neuron (n = 2); FlyBase ID: FBgn0035092
Two bilateral dimmed neurons (#9, soma size: 10 μm) in the dorso-lateral protocerebrum are classified as NPLP1 neurons (Figure 5d-I). The NPLP1 precursor of D. melanogaster contains a number of neuropeptide sequences with potential amidation motifs but the sequences show little sequence similarity among themselves (Figure 5d-III). Mass spectra of type #9 cells (Figure 5d-II) confirmed the presence of all NPLP1 peptides identified before in comprehensive peptidomic analyses of the Drosophila CNS [14, 30, 45, 46]. In addition, an ion signal mass-identical to a further peptide (YPPFHTWGTPPRITALH-OH) was observed. Ion signal intensity was not sufficient to confirm the sequence of this peptide. Different from most other neuropeptides in single cell mass spectra, distinct signals of truncated forms of several NPLP1 peptides were observed in spectra of #9 cells. Interestingly, a single NPLP1 peptide (NIATMARLQSAPSTHRDPK-OH) contains the Lys from the common Lys-Arg cleavage site; this peptide was already reported earlier [45]. The predicted peptide without Lys was not detectable. Similar data exist for a possibly homologous NPLP1 peptide (NPLP1-6) from Aedes aegypti [47]. The analyzed type #9 cell likely corresponds with the large dorso-lateral IPN immunopositive interneuron described without projection from larval and adult D. melanogaster [48]. Mapping of larval D. melanogaster dimmed cells did not reveal NPLP1 neurons in the brain [7].
Cell Type #10: Neuropeptide F (NPF) Neuron (n = 3); FlyBase ID: FBgn0027109
A single bilateral dimmed neuron (#10, soma size: 14 μm) in the dorsal medium protocerebrum, slightly postero-lateral of the dimmed neurons of the pars intercerebralis, is classified as NPF neuron (Figure 6-I). The NPF precursor of D. melanogaster contains a single NPF sequence [49], which has never been identified by peptidomics. As shown in Figure 6-II, the processed D. melanogaster NPF (Table 1) is shorter than predicted and was confirmed by MSMS (Figure 6-IV). The N-terminus of mature NPF indicates a complete proteolytic cleavage at a monobasic Arg cleavage site. Two further precursor peptides of the NPF precursor were detected in mass spectra as well (Figure 6-II). The analyzed type #10 cell corresponds with the large medial anti-NPF immunoreactive interneuron in the dorsal protocerebrum of adult D. melanogaster, which innervates the dorsal and lateral protocerebrum (e.g., [49,50,51]). The type #10 neuron is likely also identical with the DIMMED-positive NPF cell #8 described in larval brains [7]. A suggested co-localization of NPF and DH-31 in these NPF cells #8 [7] could not be convincingly verified with our mass spectra although a minor ion signal, mass-identical with DH-31, was found in a single spectrum (Figure 6-II).
Mapping of cell type #10: neuropeptide F (NPF). The arrow marks the soma location in one brain hemisphere while the circle depicts the position of the dissected soma in the contralateral brain hemisphere. (I) Reconstruction of the dimmed cell after dissection of an individual dimmed neuron. (II) Mass spectra in lower and higher mass range show ion signals of NPF and a precursor peptide (PP). A weak ion signal mass-identical with diuretic hormone 31 (DH-31) is marked as well. This signal was not seen in other preparations of cell type #10 and contamination cannot be ruled out. (III) Precursor sequence with signal peptide (highlighted in grey), identified peptides (underlined), confirmed cleavage sites (bold), and amidation signals (italic). (IV) MS/MS spectrum of NPF from the same sample. The fragments confirmed NPF with a sequence shorter than originally predicted
Conclusions
Using SCMS of identified neurons, we analyzed 10 types of dimmed neurons from the brain/gnathal ganglion of adult D. melanogaster (Figure 7). These cells included neuroendocrine cells with projection into the retrocerebral complex but also a number of large interneurons. Resulting mass spectra provided comprehensive data regarding mature products from 13 neuropeptide precursors. Interestingly, data from these single cell analyses consistently revealed all mature neuropeptides, which have been described for these precursors in earlier publications. Some neuropeptides such as Drosophila natalisins could be biochemically confirmed for the first time. The mass spectra presented clear evidence for a co-expression of up to three neuropeptide precursors in some of the dimmed cells. The majority of dimmed neurons, however, each yielded only products from a single neuropeptide gene. Although ion suppression is a general issue in MALDI-TOF mass spectrometry, single cell mass spectra with low peptide complexity are probably less affected than tissue samples by this problem. In addition, our experimental approach did exclude cell lysis during cell dissection and therefore avoided possible changes of the peptidome in the course of sample preparation. It has to be noted in this context that our approach did not cover larger protein hormones such as insulin-like peptides. Moreover, closely packed dimmed cells of the pars lateralis and pars intercerebralis in the protocerebrum and many neurons, which were detectable in only few brain preparations, have not been analyzed yet. The already available data may be implemented in neuroanatomical maps of the D. melanogaster brain; e.g., virtual fly brain (http://www.virtualflybrain.org/).
References
Hewes, R.S., Taghert, P.H.: Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 11, 1126–1142 (2001)
Vanden Broeck, J.: Neuropeptides and their precursors in the fruitfly. Drosophila melanogaster. Peptides. 22, 241–254 (2001)
Hauser, F., Cazzamali, G., Williamson, M., Blenau, W., Grimmelikhuijzen, C.J.: A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog. Neurobiol. 80, 1–19 (2006)
Pauls, D., Chen, J., Reiher, W., Vanselow, J.T., Schlosser, A., Kahnt, J., Wegener, C.: Peptidomics and processing of regulatory peptides in the fruit fly Drosophila. EuPA Open Proteomics. 3, 114–127 (2014)
Nässel, D.R., Winther, A.M.: Drosophila neuropeptides in regulation of physiology and behavior. Prog. Neurobiol. 92, 42–104 (2010)
Taghert, P.H., Nitabach, M.N.: Peptide neuromodulation in invertebrate model systems. Neuron. 76, 82–97 (2012)
Park, D., Veenstra, J.A., Park, J.H., Taghert, P.H.: Mapping peptidergic cells in Drosophila: where DIMM fits in. PLoS ONE. 3, e1896 (2008)
Park, D., Taghert, P.H.: Peptidergic neurosecretory cells in insects: organization and control by the bHLH protein DIMMED. Gen. Comp. Endocrinol. 162, 2–7 (2009)
Hamanaka, Y., Park, D., Yin, P., Annangudi, S.P., Edwards, T.N., Sweedler, J., Meinertzhagen, I.A., Taghert, P.H.: Transcriptional orchestration of the regulated secretory pathway in neurons by the bHLH protein DIMM. Curr. Biol. 20, 9–18 (2010)
Hadžić, T., Park, D., Abruzzi, K.C., Yang, L., Trigg, J.S., Rohs, R., Rosbash, M, Taghert, P.H.: Genome-wide features of neuroendocrine regulation in Drosophila by the basic helix-loop-helix transcription factor DIMMED Nucl. Acids Res: gku 1377 (2015)
Liu, Y., Luo, J., Nässel, D.R.: The Drosophila transcription factor Dimmed affects neuronal growth and differentiation in multiple ways depending on neuron type and developmental stage. Front Mol Neurosci. 13, 97 (2016)
Gauthier, S.A., Hewes, R.S.: Transcriptional regulation of neuropeptide and peptide hormone expression by the Drosophila dimmed and cryptocephal genes. J. Exp. Biol. 209, 1803–1815 (2006)
Hewes, R.S., Park, D., Gauthier, S.A., Schaefer, A.M., Taghert, P.H.: The bHLH protein Dimmed controls neuroendocrine cell differentiation in Drosophila. Development. 130, 1771–1781 (2003)
Yew, J.Y., Wang, Y., Barteneva, N., Dikler, S., Kutz-Naber, K.K., Li, L., Kravitz, E.A.: Analysis of neuropeptide expression and localization in adult Drosophila melanogaster central nervous system by affinity cell-capture mass spectrometry. J. Proteome Res. 8, 1271–1284 (2009) Erratum in: J. Proteome Res. 8, 3786 (2009)
Neupert, S., Predel, R.: Mass spectrometric analysis of single identified neurons of an insect. Biochem. Biophys. Res. Commun. 327, 640–645 (2005)
Lee, T., Luo, L.: Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 22, 451–461 (1999)
Veenstra, J.A.: Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect Biochem. Physiol. 2, 49–63 (2000)
Pitman, J.L., Huetteroth, W., Burke, C.J., Krashes, M.J., Lai, S.L., Lee, T., Waddell, S.: A pair of inhibitory neurons are required to sustain labile memory in the Drosophila mushroom body. Curr. Biol. 21, 855–861 (2011)
Zitnan, D., Sehnal, F., Bryant, P.J.: Neurons producing specific neuropeptides in the central nervous system of normal and pupariation-delayed Drosophila. Dev. Biol. 156, 117–135 (1993)
Siegmund ,T., Korge, G.: Innervation of the ring gland of Drosophila melanogaster. J. Comp. Neurol. 431, 481–491 (2001)
McCormick, J., Nichols, R.: Spatial and temporal expression identify dromyosuppressin as a brain-gut peptide in Drosophila melanogaster. J. Comp. Neurol. 338, 278–288 (1993)
Jiang, H., Lkhagva, A., Daubnerová, I., Chae, H.S., Šimo, L., Jung, S.H., Yoon, Y.K., Lee, N.R., Seong, J.Y., Žitňan, D., Park, Y., Kim, Y.J.: Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc. Natl. Acad. Sci. USA. 110, E3526–E3534 (2013)
Min, S., Chae, H.S., Jang, Y.H., Choi, S., Lee, S., Jeong, Y.T., Jones, W.D., Moon, S.J., Kim, Y.J., Chung, J.: Identification of a peptidergic pathway critical to satiety responses in Drosophila. Curr. Biol. 26, 814–820 (2016)
Huang, J., Liu, W., Qi, Y.X., Luo, L., Montell, C.: Neuromodulation of courtship drive through tyramine-responsive neurons in the Drosophila brain. Current Biology. 26, 2246–2256 (2016)
Yoon, J.G., Stay, B.: Immunocytochemical localization of Diploptera punctata allatostatin-like peptide in Drosophila melanogaster. J. Comp. Neurol. 363, 475–488 (1995)
Derst, C., Dircksen, H., Meusemann, K., Zhou, X., Liu, S., Predel, R.: Evolution of neuropeptides in non-pterygote hexapods. BMC Evol. Biol. 16, 51 (2016)
Veenstra, J.A.: The contribution of the genomes of a termite and a locust to our understanding of insect neuropeptides and neurohormones. Front. Physiol. 19, 454 (2014)
Predel, R., Wegener, C.: Biology of the CAPA peptides in insects. Cell Mol. Life Sci. 63, 2477–2490 (2006)
Rosenkilde, C., Cazzamali, G., Williamson, M., Hauser, F., Søndergaard, L., DeLotto, R., Grimmelikhuijzen, C.J.: Molecular cloning, functional expression, and gene silencing of two Drosophila receptors for the Drosophila neuropeptide pyrokinin-2. Biochem. Biophys. Res. Commun. 309, 485–494 (2003)
Predel, R., Wegener, C., Russell, W.K., Tichy, S.E., Russell, D.H., Nachman, R.J.: Peptidomics of CNS-associated neurohemal systems of adult Drosophila melanogaster: a mass spectrometric survey of peptides from individual flies. J. Comp. Neurol. 474, 379–392 (2004)
Wegener, C., Reinl, T., Jänsch, L., Predel, R.: Direct mass spectrometric peptide profiling and fragmentation of larval peptide hormone release sites in Drosophila melanogaster reveals tagma-specific peptide expression and differential processing. J. Neurochem. 96, 1362–1374 (2006)
Kean, L., Cazenave, W., Costes, L., Broderick ,K.E., Graham, S., Pollock, V.P., Davies, S.A., Veenstra, J.A., Dow, J.A.: Two nitridergic peptides are encoded by the gene capability in Drosophila melanogaster. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282, 1297–1307 (2002)
Bader, R., Wegener, C., Pankratz, M.J.: Comparative neuroanatomy and genomics of hugin and pheromone biosynthesis activating neuropeptide (PBAN). Fly (Austin). 4, 228–231 (2007)
Schlegel, P., Texada, M.J., Miroschnikow, A., Schoofs, A., Hückesfeld, S., Peters, M., Schneider-Mizell, C.M., Lacin, H., Li, F., Fetter, R.D., Truman, J.W., Cardona, A., Pankratz, M.J.: Synaptic transmission parallels neuromodulation in a central food-intake circuit. Elife. 5, e16799 (2016)
Hückesfeld, S., Peters, M., Pankratz, M.J.: Central relay of bitter taste to the protocerebrum by peptidergic interneurons in the Drosophila brain. Nat. Commun. 7, 12796 (2016)
Melcher, C., Pankratz, M.J.: Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3, e305 (2005)
Neupert, S., Johard, H.A., Nässel, D.R., Predel, R.: Single-cell peptidomics of Drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal. Chem. 79, 3690–3694 (2007). Erratum in: Anal. Chem. 84, 5164 (2012)
Meng, X., Wahlström, G., Immonen, T., Kolmer, M., Tirronen, M., Predel, R., Kalkkinen, N., Heino, T.I., Sariola, H., Roos, C.: The Drosophila hugin gene codes for myostimulatory and ecdysis-modifying neuropeptides. Mech. Dev. 117, 5–13 (2002)
Kubiak, T.M., Larsen, M.J., Burton, K.J., Bannow, C.A., Martin, R.A., Zantello, M.R., Lowery, D.E.: Cloning and functional expression of the first Drosophila melanogaster sulfakinin receptor DSK-R1. Biochem. Biophys. Res. Commun. 291, 313–320 (2002)
Nichols, R., Lim, I.A.: Spatial and temporal immunocytochemical analysis of drosulfakinin (Dsk) gene products in the Drosophila melanogaster central nervous system. Cell Tissue Res. 283, 107–116 (1996)
Agricola, H.J., Bräunig, P.: Comparative aspects of peptidergic signaling pathways in the nervous systems of arthropods. EXS. 72, 303–327 (1995)
Choi, Y.J., Lee, G., Hall, J.C., Park, J.H.: Comparative analysis of corazonin-encoding genes (Crz's) in Drosophila species and functional insights into Crz-expressing neurons. J. Comp. Neurol. 482, 372–385 (2005)
Nässel, D.R., Enell, L.E., Santos, J.G., Wegener, C., Johard, H.A.: A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 19, 9–90 (2008)
Helfrich-Förster, C.: The neuroarchitecture of the circadian clock in the brain of Drosophila melanogaster. Microsc. Res. Technol. 62, 94–102 (2003)
Schoofs, L., Baggerman, G.: Peptidomics in Drosophila melanogaster. Brief Funct. Genomic Proteom. 2, 114–120 (2003)
Salisbury, J.P., Boggio, K.J., Hsu, Y.W., Quijada, J., Sivachenko, A., Gloeckner, G., Kowalski, P.J., Easterling, M.L., Rosbash, M., Agar, J.N.: A rapid MALDI-TOF mass spectrometry workflow for Drosophila melanogaster differential neuropeptidomics. Mol. Brain. 6, 60 (2013)
Predel, R., Neupert, S., Garczynski, S.F., Crim, J.W., Brown, M.R., Russell, W.K., Kahnt, J., Russell, D.H., Nachman, R.J.: Neuropeptidomics of the mosquito Aedes aegypti. J. Proteome Res. 9, 2006–2015 (2010)
Verleyen, P., Baggerman, G., Wiehar,t U., Schoeters, E., Van Lommel, A., De Loof, A., Schoofs, L.: Expression of a novel neuropeptide, NVGTLARDFQLPIPNamide, in the larval and adult brain of Drosophila melanogaster. J. Neurochem. 88, 311–319 (2004)
Brown, M.R., Crim, J.W., Arata, R.C., Cai, H.N., Chun, C., Shen, P.: Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides. 20, 1035–1042 (1999)
Wen, T., Parrish, C.A., Xu, D., Wu, Q., Shen, P.: Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc. Natl. Acad. Sci. USA. 102, 2141–2146 (2005)
Krashes, M.J., DasGupta, S., Vreede, A., White, B., Armstrong, J.D., Waddell, S.: A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 139, 416–427 (2009)
Acknowledgment
The authors thank the German Research Foundation (NE911/3-1; PR766/11-1) for supporting this study. The authors also thank Christian Wegener (University Würzburg) and Manuela Ruppert (University Cologne) for providing Drosophila strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Diesner, M., Predel, R. & Neupert, S. Neuropeptide Mapping of Dimmed Cells of Adult Drosophila Brain. J. Am. Soc. Mass Spectrom. 29, 890–902 (2018). https://doi.org/10.1007/s13361-017-1870-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1870-1