Abstract
Background and Objectives
Ibrutinib is an antineoplastic agent that reduces B-cell proliferation by inhibiting Bruton's tyrosine kinase. We describes population pharmacokinetics of ibrutinib in healthy adults, and explores potential patient characteristics associated with ibrutinib pharmacokinetics.
Methods
A population pharmacokinetic modeling approach was applied to 39 healthy subjects. Modeling was performed using Monolix (v.2019R2). Serial blood samples to measure the plasma ibrutinib concentration were collected following the oral administration of 140 mg ibrutinib on two different occasions under fasting conditions. Demographic and clinical information were evaluated as possible predictors of ibrutinib pharmacokinetics during model development. Simulations (using mlxR: R package v.4.0.2) following the administration of therapeutic doses were performed to explore the clinical implications of identified covariates on ibrutinib steady-state concentrations.
Results
A two-compartment model with zero order absorption best fit the data. Inter-individual and inter-occasion variability were quantified by the proposed model. We identified smoking status as a significant covariate associated with ibrutinib clearance. Smoking was found to increase ibrutinib clearance by approximately 60%, which resulted in a reduction in simulated steady-state concentrations by around 40%.
Conclusion
The model can be used to simulate clinical trials or various dosing scenarios. The proposed model can be used to optimize ibrutinib dosing based on the smoking status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Ibrutinib has been demonstrated to have high inter-individual variability in its pharmacokinetic profiles. |
Smoking was identified as a covariate that affects the observed ibrutinib concentrations. |
Smokers are expected to have around a 40% reduction in steady-state ibrutinib concentrations compared to non-smokers. |
1 Introduction
Ibrutinib is considered as a potent, small-molecule irreversible inhibitor of Bruton's tyrosine kinase (BTK). Inhibition of BTK reduces malignant B-cell proliferation. It is also indicated for several types of malignancies, including mantle cell lymphoma (in patients who have received at least one prior therapy), chronic lymphocytic leukemia, small lymphocytic lymphoma, and Waldenström's macroglobulinemia [1]. Ibrutinib acts by forming a covalent bond with a cysteine residue (Cys-481) in the BTK active site, which leads to sustained inhibition of BTK enzymatic activity [2, 3]. As a member of the Tec kinase family, BTK is an important signaling molecule of the B-cell antigen receptor (BCR) and cytokine receptor pathways. The BCR pathway is involved in the pathogenesis of several B-cell malignancies, including mantle cell lymphoma, diffuse large B-cell lymphoma), follicular lymphoma, and chronic lymphocytic leukemia. BTK's pivotal role in signaling through the B-cell surface receptors results in activation of pathways necessary for B-cell trafficking, chemotaxis, and adhesion. Preclinical studies have shown that ibrutinib effectively inhibits malignant B-cell proliferation and survival in vivo, as well as cell migration and substrate adhesion in vitro [4].
After oral administration, ibrutinib is rapidly absorbed with a median time to reach peak concentration of 1–2 h [4, 5]. Absolute bioavailability (F) under fasting condition was only 3.9% while it was doubled when combined with a meal [6]. Pharmacokinetics of ibrutinib do not significantly differ in patients with different B-cell malignancies. Ibrutinib exposure increases with doses up to 840 mg. The steady-state area under the curve (AUC) observed in patients at 560 mg is (mean ± standard deviation) 953 ± 705 ng h/mL [4].
Ibrutinib binds reversibly to human plasma protein in vitro (97.3%). There is no concentration dependence in the range of 50–1000 ng/mL. The apparent oral volume of distribution at steady-state was approximately 10,000 L. Disposition of ibrutinib appears to follow a biexponential pattern. Ibrutinib is metabolized by cytochrome P450 (CYP) 3A4 to produce a less active metabolite (dihydrodiol metabolite) that has an inhibitory activity approximately 1/15th that of ibrutinib. Apparent clearance (CL/F) of ibrutinib is approximately 1000 L/h and it has a half-life of around 4–13 h [4, 5].
Ibrutinib pharmacokinetics in healthy adults have been explored by several research groups [5,6,7,8]. De Jong et al. investigated the effect of omeprazole on ibrutinib pharmacokinetics [5]. Omeprazole decreased ibrutinib maximum concentration with no significant effect on AUC. The effect of food on ibrutinib pharmacokinetics was also explored in healthy adults and patients with chronic lymphocytic leukemia [8]. Food increased ibrutinib concentration by approximately 67% [8]. Similarly, ibrutinib bioavailability in patients with B--ell malignancies under fasting conditions was around two thirds the bioavailability value under fed conditions following a high fat meal [9]. Other research groups utilized labeled ibrutinib to obtain further details about ibrutinib pharmacokinetics in healthy adults [6, 7]. The ibrutinib elimination pathway was explored by Scheers et al. using 14C-labeled ibrutinib [7]. The majority of ibrutinib (as metabolites) was eliminated in feces (81% of radioactivity) with a minor contribution of renal excretion (8% of radioactivity) [7]. Absolute bioavailability of ibrutinib was estimated using a microdose of 13C6- ibrutinib in healthy patients under different meal conditions. Administration of ibrutinib 30 min before breakfast was found to increase absolute bioavailability compared to fasting state (F = 8.4% vs. 3.9%, respectively). The addition of grape fruit juice to breakfast resulted in a further increase in absolute bioavailability (F = 15.9%) [6].
In spite of population pharmacokinetic studies conducted in patients [9, 10], none of the studies conducted in healthy adults implemented a nonlinear mixed effects modeling approach to explore ibrutinib pharmacokinetics from a population pharmacokinetics perspective [5,6,7,8]. This analysis aimed to develop a nonlinear mixed effects pharmacokinetic model to describe ibrutinib plasma concentration–time data obtained in healthy human subjects after oral administration of ibrutinib and to explore relevant relationships of pharmacokinetic parameters with demographic, clinical, and behavioral covariates of the subjects.
2 Subjects and Setting
2.1 Study Design
The study protocol was approved by the institutional review board (IRB) of the International Pharmaceutical Research Center and the clinical trials Committee at the Jordan Food and Drug Administration. A total of 39 healthy human subjects were included in the present analysis. A total of 38 healthy human subjects were dosed on two different occasions and 1 subject was dosed on one occasion with a 140-mg oral dose of ibrutinib (Imbruvika, Batch number HIS4S00, Exp. Date: 08/20; Janssen-Cilag International, Belgium). The subjects were admitted to the clinical site the night before drug administration, supervised for at least 10 h of overnight fasting, confined until the 24-h sample was taken, and then returned to give the rest of the samples. Each subject received two doses of Imbruvika on two different occasions separated by at least 2 weeks, with 240 ml of water. Twenty blood samples were drawn at 0.00 (pre-dose sample), 0.25, 0.50, 0.75, 1.00, 1.33, 1.67, 2.00, 2.50, 3.00, 4.00, 6.00, 8.00, 10.00, 12.00, 24.00, 36.00, 48.00 and 60.00 h (post-dose). A blood volume of 7 ml was drawn for each sample. The blood samples for ibrutinib were collected in lithium-heparinized (LI-Heparin) tubes and centrifuged, and the resulting plasma samples were immediately stored in plain plastic tubes in a freezer at a temperature of – 20 °C.
2.2 Bioanalysis
A selective, sensitive, and rapid liquid chromatography–tandem mass spectrometry method for the determination of ibrutinib in human plasma has been validated. The procedure involved liquid extraction of the drug and its internal standards (ibrutinib-d5).The chromatographic separation employed a C18 column with isocratic elution, while the mobile phase consisted of ammonium acetate buffer (10 mM)/formic acid (98%)/methanol (15%/0.1%/84.9%). Detection was carried out using Quattro premier mass spectrometer in multiple reaction monitoring mode using Ion Spray with positive ionization. The investigated transition of ibrutinib was at m/z 441.25 (parent)→138.00 (daughter). The method was validated considering different parameters, such as linearity, accuracy, precision, and stability. The method had a total run time of about 6 min and showed acceptable linearity (r = 0.9982) over the working range of 0.100–40.000 ng/ml.
2.3 Population Pharmacokinetic Model
Population pharmacokinetic modeling was performed using non-linear mixed-effect modeling software (Monolix v.2019R2; Antony, France: Lixoft SAS, 2019).
2.3.1 Basic Model Building
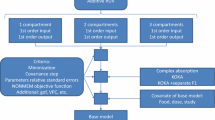
Several structural models were initially compared with variations in the number of compartments (one, two, or three compartments) and/or absorption kinetics (first order, zero order, or dual absorption). Several residual error models that include constant, proportional, and combination were also examined. Following the determination of the basic structural model, the influence of the inclusion of inter-occasion variability was also explored. Model selection was based on the value on Bayesian information criterion (BIC), goodness of fit diagnostic plots, and relative standard errors (RSE) of the estimated parameters [11].
2.3.2 Distribution of Individual Parameters
A log-normal distribution was assumed for individual parameters according to the following equation:
where \({\theta }_{I}\) is the individual pharmacokinetic parameter, \(\theta\) is the population pharmacokinetic parameter, \(\beta\) is the covariate regression term, \({\mathrm{Cov}}_{\mathrm{i}}\) is the covariate for the Ith individual, \({\eta }_{I}\) is the random effect for the ith individual, and \({\upeta }_{\mathrm{ki}}\) is the random effect for the Ith individual at the kth occasion. Those random effects, \({\eta }_{\mathrm{i}}\) and \({\eta }_{ki}\), were assumed to follow normal distributions:\({\upeta }_{\mathrm{i}}\sim N(0,\omega )\) and \({\upeta }_{\mathrm{ki}}\sim N(0,\upgamma )\) where \(\omega\) and \(\gamma\) are standard deviations of the inter-individual and inter-occasion variability terms.
2.3.3 Covariate Analysis
Several characteristics were explored as potential covariates associated with ibrutinib pharmacokinetic parameters. These potential covariates were recorded at the screening phase that included the following:
-
Demographic information: height, weight, body mass index (BMI), and age.
-
Smoking status: subjects were classified into smokers (a subjects who smokes between 1 and 10 cigarettes/day) and non-smokers (a subject who does not smoke).
-
Blood tests: plasma glucose, total bilirubin, creatinine, blood urea nitrogen, alkaline phosphatase, aspartate transaminase, and alanine aminotransferase.
-
Hematological indices: white blood cell count, red blood cell count, hemoglobin concentration, packed cell volume, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, and platelets count.
-
Urine tests: urine specific gravity, urine pH
A stepwise covariate screening process based on the reduction in the value of corrected BIC was implemented [12]. Initially, a forward selection process was applied. In the first step, all potential covariates were individually added to the population model. The covariate that resulted in the lowest BICc was included in the model. At the next step, the remaining covariates were then individually added to the selected model from the first step. The covariate that resulted in the lowest BICc was then selected. This step was repeated with the addition of a covariate each time until no further reduction in the BICc was observed upon the addition of each of the remaining covariates.
A backward deletion was then applied to the model selected from forward addition. Each covariate was individually excluded from the model. The covariate with the lowest BICc upon its elimination was selected. The remaining covariates were then individually eliminated from the model and the BICc value was recorded. Similarly, the model with the lowest BICc value was selected and carried forward. This step was repeated until no further reduction was observed upon the removal of any of the remaining covariates.
2.3.4 Monte Carlo Simulations
Monte Carlo simulations were performed using Simulx (mlxR: R package v.4.0.2; Inria, Paris, France, [13]) based on the final pharmacokinetic model to generate steady-state concentrations for 2000 patients. Two simulation experiments were conducted. The first simulation trial was based on a once-daily dose of 560 mg. a dose selected based on the recommended dose for mantle cell lymphoma. The second simulation experiment assumed a once-daily dose of 420 mg based on the recommended dose for chronic lymphocytic leukemia and Waldenström’s macroglobulinemia. Concentrations were simulated for one dosing interval (24-h). AUC was calculated using the trapezoidal method with the concentrations simulated by the model. Mean steady-state concentration was calculated by dividing the AUC by 24 h.
2.3.5 Model Selection and Evaluation Using Visual Predictive Check
The population pharmacokinetic model was used to generate 1000 simulated datasets for the population. The simulated data set was used to plot visual predictive check (VPC). The 5th, 50th, and 95th percentiles of the simulated concentrations were constructed and compared against the observed concentrations. VPC assesses the predictive performance of the model.
3 Results
3.1 Subject Characteristics
All the subjects were males (n = 39) with a mean age of 29.5 years (SD = 9.3 years). The age ranged from 18 to 48 years. As per protocol requirements, subjects included in the study must have a BMI between 18.5 and 30 kg/m2. The actual BMI mean was 23 kg/m2 (SD = 3.3 kg/m2) with a maximum of 29.4 kg/m2 and a minimum of 18.8 kg/m2. About 59% (n = 23) of subjects were smokers (less than 10 cigarettes/day). More detailed demographic and clinical characteristics of the study population are provided in Table 1.
3.2 Basic Model Building
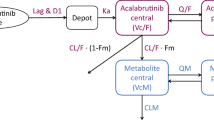
A two-compartment model with zero order absorption with lag time was selected as the best structural model that describes ibrutinib concentrations. The selected error model was proportional. The use of other error models did not reduce the values of BIC. The use of one or three compartments was associated with an increase in the value of BIC. The use of first-order absorption was also associated with increases in the BIC value with no improvement in the predictive performance of the model. Several combinations of dual absorption scenarios were explored. Zero- and first-order absorption were individually tested for each of the first and the second absorption process. The inclusion of dual absorption in the model reduced the BIC value; however, it resulted in a high RSE (> 50%) of one of the absorption parameters. The inclusion of inter-occasion variability (IOV) terms for all the parameters resulted in a reduction in the values of BIC. Hence, the selected basic structural model is a two-compartment model with zero-order absorption with a proportional error model with the inclusion of IOV terms.
3.3 Covariate Screening
Population pharmacokinetic parameters, inter-individual parameters, and inter-occasion variability were successfully estimated, as presented in Table 2. The values of %RSE were less than 50% for all the estimated parameters. We identified smoking status as a significant covariate associated with ibrutinib clearance. Ibrutinib clearance was described according to the following equation:
Thus, clearance for non-smokers is 56.8 L/h and clearance for smokers is 90.6 L/h.
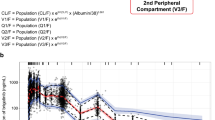
Several plots were prepared to evaluate the performance of the pharmacokinetic model (Figs. 1, 2, 3, and 4). The accuracy of predictions using individual parameters in describing observed concentrations is presented in Fig. 1. Predictions using population parameters (Fig. 2) did not perform very well in terms of describing observed values. This is probably due to the large inter-individual and inter-occasion variability. Distributions of residual errors is presented in Figure 3. Residuals were uniformly distributed around zero. Additionally, there was no apparent pattern over time.
Visual predictive check for ibrutinib plasma concentrations; solid black lines represent the 5th, median, and 95th empirical percentiles of the observed data; the theoretical percentiles of simulated data (n = 1000) were computed from the final model, where dashed black lines represent the 5th, median, and 95th theoretical percentiles of the simulated data; 90% prediction intervals are displayed as shaded gray
To explore whether the observed variability can be reproduced by the model, VPC was applied. A total of 1000 datasets were generated using model parameters. The 10th, 50th, and 90th percentiles from simulated observations were compared to the observed percentiles. Figure 4 demonstrates good agreement between the simulated percentiles and the observed concentrations. This demonstrates the predictive performance of the model.
3.4 Simulation
According to the simulation results (Table 3), a daily dose of 560 mg is expected to result in a mean steady-state concentration of 12.0 ng/ml in smokers and 18.9 ng/ml in non-smokers. This represents a 37% reduction in steady-state concentrations in smokers compared to non-smokers. A 40% reduction in steady-state concentration was also predicted for smokers (8.87 ng/ml) compared to non-smokers (14.9 ng/ml) when a simulated daily dose of 420 mg was given. The observed mean concentrations (over 24 h) following a single dose of 140 mg were 2.69 ng/ml in smokers and 4.37 ng/ml in non-smokers (a difference of 38%).
4 Discussion
This is the first report describing the population pharmacokinetics of ibrutinib in healthy subjects with rich data. A two-compartment open model was adequate to fit the plasma concentration–time data, and proved useful to evaluate the potential contribution of covariates to differences between individuals in pharmacokinetic parameters.
Two previous publications have explored ibrutinib population pharmacokinetics in patients with B-cell malignancies [9] and patients with lymphoid malignancies [10]. Similar to the present model, both publications used a two-compartment model with first-order elimination. However, they used a sequential zero-first-order model to describe the absorption process in contrast to the zero-order absorption implemented currently. The use of sequential absorption resulted in a %RSE of more than 50%. Hence, a sequential absorption model was not used. The estimated value of lag time of 0.315 h is consistent with previous estimates of 0.283 h [9] and 0.238 h [10]. The estimated clearance value (56.8 L/h) was comparable to the clearance estimated in patients with B-cell malignancies (41.3 L/h) [9]. However, a value of 242 L/h was reported in patients with lymphoid malignancies [10]. There is a large discrepancy in the values of the volume of distribution (for both central and peripheral compartments) and inter-compartmental clearance. For example, the volume of distribution of the central compartment was reported previously as 9.59 L in patients with B-cell malignancies [9] and 1010 L in patients with lymphoid malignancies [10] compared to the current estimate of 309 L in healthy adults. It is not clear whether this discrepancy is attributable to physiological differences between healthy and ill patients or due to the use of different models.
The simulation results (Table 3) highlight the impact of smoking on ibrutinib concentrations. Smokers are expected to have around a 40% reduction in steady-state concentrations compared to non-smokers. In fact, a daily dose of 420 mg in non-smokers is expected to result in steady-state concentrations of more than a daily dose of 560 mg in non-smokers. The simulation results are in line with the observed results upon the administration of a single dose of 140 mg. The similarity between the observed and the simulated results confirms the validity of the simulation results. Smoking can potentially modify the clinical effects of ibrutinib. Higher ibrutinib dosing intensity has been found to increase the progression-free survival for patients with chronic lymphocytic leukaemia/small lymphocytic lymphoma [14]. The reduction in ibrutinib concentrations observed in smokers is expected to result in reduced therapeutic outcomes [14]. Reduced ibrutinib levels are also expected to result in reduced toxicities such as neutropenia and hemorrhage [15].
Smoking induction of many metabolizing enzymes including CYP3A4 [16, 17] is a possible explanation of the effect of smoking status on ibrutinib clearance. Ibrutinib undergoes extensive hepatic metabolism mainly by CYP3A4 [7]. Induction of CYP3A4 by tobacco smoking is expected to increase ibrutinib clearance which will in turn reduce plasma concentration. This explains the observed reduction in ibrutinib concentrations in smokers compared to non-smokers (Table 3). This interaction was implanted in the present model through the inclusion of smoking as a covariate for the estimation of ibrutinib clearance.
This study has potential limitations. The inclusion of healthy adults limits the ability to identify important covariates. According to protocol requirements, subjects with abnormal laboratory results were excluded from the study. Hence, the effect of disease states or abnormal laboratory results on ibrutinib pharmacokinetics could not be explored. Additionally, the inclusion of other potential covariates such as CYP3A4 and alcohol consumption could decrease the inter-individual variability observed in the pharmacokinetic parameters [18].
In contrast to the model developed by Marostica et al. [9], our findings indicate that body weight was not significantly associated with ibrutinib pharmacokinetic parameters. The inclusion of weight in the pharmacokinetic model by Marostica et al. was based on allometric scaling principles and on statistical significance. Indeed, it was stated that the inclusion of weight did not result in a significant drop in objective function value and had a minimal effect on ibrutinib volume of distribution with no effect on other pharmacokinetic parameters [9].
5 Conclusion
In summary, a population pharmacokinetic model was developed to describe ibrutinib population pharmacokinetics in healthy adults. Smoking was identified as a covariate that affects the observed ibrutinib concentrations. Inter-individual and inter-occasion variability were quantified. The model can be used to simulate clinical trials or various dosing scenarios. Further studies are needed to examine the effect of smoking on ibrutinib dosing in patients. This project can be extended by inclusion of patients with various degrees of abnormalities, such as various levels of hematological indices and various degrees of liver function. The inclusion of genetic data such as the CYP3A4 genotype is another important aspect that need to be considered in future projects.
References
Wellstein A, et al. Pathway-targeted therapies: monoclonal antibodies, protein kinase inhibitors, and various small molecules. In: Brunton LL, Hilal-Dandan R, Knollmann BC, editors., et al., Goodman & Gilman’s: The Pharmacological Basis of Therapeutics. New York: McGraw-Hill Education; 2017.
Byrd JC, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42.
Wang ML, et al. Targeting BTK with ibrutinib in relapsed or refractory mantle-cell lymphoma. N Engl J Med. 2013;369(6):507–16.
Imbruvica, Highlights of Prescribing Information. 2020. https://imbruvica.com/files/prescribing-information.pdf. Accessed 7 Feb 2021.
de Jong J, et al. Single-dose pharmacokinetics of ibrutinib in subjects with varying degrees of hepatic impairment. Leuk Lymphoma. 2017;58(1):185–94.
de Vries R, et al. Stable isotope-labelled intravenous microdose for absolute bioavailability and effect of grapefruit juice on ibrutinib in healthy adults. Br J Clin Pharmacol. 2016;81(2):235–45.
Scheers E, et al. Absorption, metabolism, and excretion of oral 14C radiolabeled ibrutinib: an open-label, phase I, single-dose study in healthy men. Drug Metab Dispos. 2015;43(2):289–97.
de Jong J, et al. The effect of food on the pharmacokinetics of oral ibrutinib in healthy participants and patients with chronic lymphocytic leukemia. Cancer Chemother Pharmacol. 2015;75(5):907–16.
Marostica E, et al. Population pharmacokinetic model of ibrutinib, a Bruton tyrosine kinase inhibitor, in patients with B cell malignancies. Cancer Chemother Pharmacol. 2015;75(1):111–21.
Gallais F, et al. Population Pharmacokinetics of Ibrutinib and Its Dihydrodiol Metabolite in Patients with Lymphoid Malignancies. Clin Pharmacokinet. 2020;59(9):1171–1183.
Neath AA, Cavanaugh JE. The Bayesian information criterion: background, derivation, and applications. WIREs Comp Stat. 2012;4(2):199–203.
Delattre M, Lavielle M, Poursat M-A. A note on BIC in mixed-effects models. Electron J statist. 2014;8(1):456–75.
Lavielle M, et al. mlxR: simulation of longitudinal data. 2015. https://cran.r-project.org/web/packages/mlxR/index.html. Accessed 28 Oct 2020.
Barr PM, et al. Impact of ibrutinib dose adherence on therapeutic efficacy in patients with previously treated CLL/SLL. Blood. 2017;129(19):2612–5.
Stephens DM, Byrd JC. How I manage ibrutinib intolerance and complications in patients with chronic lymphocytic leukemia. Blood. 2019;133(12):1298–307.
Rahmioglu N, et al. Genetic epidemiology of induced CYP3A4 activity. Pharmacogenet Genomics. 2011;21(10):642–51.
Zevin S, Benowitz NL. Drug interactions with tobacco smoking. Clin Pharmacokinet. 1999;36(6):425–38.
Xu RA, et al. Functional Characterization of 22 CYP3A4 Protein Variants to Metabolize Ibrutinib In Vitro. Basic Clin Pharmacol Toxicol. 2018;122(4):383–7.
Acknowledgements
This work has been carried out during sabbatical leave granted to the corresponding author (Mutasim Al-Ghazawi) from the University of Jordan during the academic year 2019-2020.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Authors’ Contribution
MA and MS wrote the manuscript and analyzed the data. ON and NN designed the study and supervised the conduction of the study. IS designed the study and supervised bioanalysis of the study samples. All authors contributed towards the critical revision and approval of the manuscript.
Funding
Sabbatical leave granted by University of Jordan to Mutasim Al-Ghazawi.
Data Availability
Data are available on request from the corresponding author.
Conflict of Interest
Mutasim Al-Ghazawi, Mohammad I. Saleh, Omaima Najib, Isam Salem, Naji Najib have no conflict of interest to declare.
Ethics Approval
The study protocol was approved by the institutional review board of the International Pharmaceutical Research Center and the clinical trials Committee at the Jordan Food and Drug Administration.
Consent to Participate
Participants included in the study gave written informed consent before entering the study.
Consent to Publish
Not applicable.
Rights and permissions
About this article
Cite this article
Al-Ghazawi, M., Saleh, M.I., Najib, O. et al. Population Pharmacokinetics of Ibrutinib in Healthy Adults. Eur J Drug Metab Pharmacokinet 46, 405–413 (2021). https://doi.org/10.1007/s13318-021-00679-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13318-021-00679-z