Abstract
Rice sheath blight, a major devastating disease caused by Rhizoctonia solani, poses huge threat to global rice production. Despite being the most hostile necrotrophic fungus with a wide host range, the mechanism involved in R. solani pathobiology is poorly understood. The evolutionarily developed nonhost resistance (NHR) provides broad-spectrum disease resistance to food crops. According to previous reports, Arabidopsis PEN1, PEN2 and PEN3 act as key components of cell wall-based pre-haustorial defenses against non-adapted pathogens. However, the involvement of these PEN genes in NHR against R. solani has remained unexplored till date. Therefore, the objective of this work is to study the efficacy of Arabidopsis PEN genes in NHR against R. solani. For this, hyphal colonization and penetration structures were monitored in Arabidopsis wild type Columbia-0 (Col-0) and mutants- pen1, pen2–3 and pen3–1. Comparatively, R. solani colonization on the leaves of Arabidopsis wild types was less than that on the rice leaf surface. Also, among the pen mutants studied, pen2–3 allowed maximum penetration and colonization by R. solani during early hours of infection as evidenced by both microscopic and macroscopic observations. Advanced lesion area, reduced chlorophyll content and increased fungal biomass accumulation also corroborated with the disease severity in pen2–3. However, R. solani resistance was restored in complemented PEN2-GFP comparable to Col-0. Altogether, our results demonstrate major involvement of PEN2 during pre-penetration, and its contribution to NHR against R. solani for enhanced disease resistance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhizoctonia solani (teleomorph, Thanatephorus cucumeris), a multinucleated filamentous necrotroph, causes diseases like sheath blight and banded leaf disease in monocots like rice, maize and sorghum; aerial blight and stem rot in legumes like mung bean and soybean; sheath rot in sugarcane; damping off of cotton; black scurf and sprout canker in potato; heart rot in cabbage and foliar blights in other fruits and plantation crops (Ajayi-Oyetunde and Bradley 2018). Rice sheath blight is the most devastating disease challenging global food security amongst other diseases caused by R. solani and can potentially cause around 50% reduction in rice yield worldwide (Zheng et al. 2013). R. solani is both soilborne as well as waterborne pathogen. This non-sporulating fungus survives in the form of sclerotia during the inactive phases of its infection cycle (Kumar et al. 2016). Under favorable conditions, these sclerotia germinate into mycelia, which further form atypical hyphal aggregates called infection cushions, required for host penetration (Kumar et al. 2011; Lazniewska et al. 2012; Singh et al. 2012; Taheri and Tarighi 2011). It is also reported that the hyphal growth and penetration of R. solani is facilitated by protrusions or openings on the leaves of host surface, such as trichome, stomata or papillae (Basu et al. 2016). R. solani induces programmed cell death with loss of photosynthetic activity and development of necrotic lesions in the host tissues (Mondal et al. 2012; Taheri and Tarighi 2011).
Flentje (1963) reported that the infection cushions develop only in the susceptible host which is suppressed in the resistant host. Although, data regarding the in-depth studies about the mechanism of infection process of R. solani in various host is scarce, it is reported that R. solani produces typical infection structure by forming cluster of hyphae with bulbous end during its pre-penetration stage (Lazniewska et al. 2012). This clustering of T-shaped branched hyphae and formation of infection cushions further leads to penetration by defeating the barriers present in the host tissue, enters via penetration peg and promotes colonization (Pannecoucque and Höfte 2009).
Majority of plant diseases are caused by fungal pathogens (Lazniewska et al. 2012). In order to fend off various pathogenic attacks, plants possess immune system with multilayered continuum of both pre-formed and acquired barriers. The disease resistance is mediated by sequential basal and resistance (R)-gene mediated hypersensitive response that does not always involve recognition of pathogenic cues (Gill et al. 2015). Nonhost resistance demarcates the host range of phytopathogenic microorganisms, representing the hallmarks of basic compatibility. Thus, adapted pathogens always try to suppress or evade the plant’s basal defense mechanism by secreting a repertoire of effector molecules (Speth et al. 2007) against robust nature of innate immunity (Nuernberger and Lipka 2005). Durability of NHR has increased attention to revamp resistance in crop. An elaborated suite of plant defense system induces downstream cell-autonomous responses of PAMP-triggered immunity (PTI) including production of reactive oxygen species (ROS), MAP-kinase signaling, transcriptional induction of pathogenesis-related (PR) genes, and callose deposition (Bittel and Robatzek 2007).
In Arabidopsis, three PENETRATION genes - PEN1 (PENETRATION1), PEN2 (PENETRATION2) and PEN3 (PENETRATION3) are reported to allow limited (entry of non-adapted powdery mildews and provide cell wall based defenses (Lipka et al. 2008). PEN1 encodes a syntaxin (SYP121/PEN1), which belongs to the SNARE superfamily proteins. PEN1 plays a role in the papilla formation (Collins et al. 2003). PEN2 encodes myrosinase, associated with peroxisomes and mitochondria (Fuchs et al. 2016), which is implicated in the production of glucosinolate derivatives as antifungal defense compounds (Bednarek et al. 2009; Lipka et al. 2005). The toxic by-products of PEN2 are transported to the sites of pathogen entry by ABC transporter proteins, which are encoded by PEN3 gene. PEN2 and PEN3 have been documented to confer disease resistance against a biotrophic fungus Erysiphe pisi, hemibiotrophic oomycete Phytophthora infestans as well as a necrotrophic fungus P. cucumerina (Stein et al. 2006). The host specificity and virulence mechanism of R. solani is related and dependent upon its anastomosis group (AG) (For instance, R. solani AG-2-1 and AG-4 can infect Arabidopsis, whereas AG-8 cannot (Foley et al. 2013; Sharon et al. 2011). Also various necrosis-inducing effectors of R. solani have been characterized, of which recently studied effector AGLIP1 in R. solani AG1 IA induces cell death in rice protoplasts and Nicotiana benthamiana and effector RsIA_NP8 induces cell death in N. benthamiana by triggering SGT1 (suppressor of the G2 allele of skp1) and HSP90 (heat shock protein 90) (Li et al. 2019; Wei et al. 2020).
Here, our study is focused on the development of early infection structures of R. solani including cushion formation which is indispensable for penetration in both host (rice) and nonhost (Arabidopsis), and to investigate the NHR mechanisms involved in Arabidopsis pen mutants. To our knowledge, the nonhost interaction between Arabidopsis pen mutants and R. solani has never been reported previously. Our results from the study of infection processes by R. solani in rice and Arabidopsis accessions indicate that PEN2 functions to provide NHR at pre-haustorial level by breaching the development of infection cushion formed by R. solani.
Materials and methods
Growth and maintenance of rice and Arabidopsis
Seeds of Oryza sativa L. cv. Swarna, as susceptible rice cultivar, were obtained from NRRI, Cuttack. The seedlings of Swarna were initially grown on moist tissue paper and kept in a Petri plate under controlled condition in the plant growth chamber. The germinated seedlings were then transplanted onto soil in a greenhouse at ~28 °C in the day time and ~ 25 °C at night. Arabidopsis wild type (WT); Col-0 (N1093), Ler-0 (N24238), La-0 (N24596), Nd-0 (N1390), No-0 (N24239), Kin-0 (N1272), T-DNA insertion line of pen1 (N673657, SALK_004484C), ethyl methanesulfonate (EMS) mutants pen2–3 (N66946) and pen3–1 (N66467) and complement PEN2-GFP (N67162) were obtained from NASC, Europe and were grown in the plant growth chamber. The 10 day old seedlings were transplanted on soil mixture containing agropeat: vermiculite (3:1) and maintained at 14 h photoperiod with ~100 μE/m2/s light intensity, 21 °C temperature and 60% humidity.
R. solani culture conditions
R. solani AG-1 IA (AG: anastomosis group) isolate was collected from National Rice Research Institute (NRRI) and was routinely cultured on freshly prepared potato dextrose agar (PDA) medium supplemented with antibiotic spectinomycin (100 μg/mL). The plates were incubated in dark at 27 °C for 10 days until sclerotia are fully developed which were used as inoculum for detached leaf infection assay.
Genotyping of Arabidopsis mutants
Plant genotype for each Arabidopsis mutant with T-DNA insertion (pen1) or point mutation (pen2–3, pen3–1) was confirmed by PCR using specific primers and the genomic DNA isolated from respective accessions. The gDNA was isolated using the CTAB method (Lukowitz et al. 2000). Polymerase chain reaction (PCR) was performed in a 20 μL reaction volume using specific forward and reverse primers. The point mutation in EMS generated pen2–3, pen3–1 mutants were confirmed by the SBP marker designed as per Sahu et al. (2012) upon digestion with the enzyme BsmAI and HphI respectively. The sequences of the primers used for PCR are provided in Table 1.
Infection assay
For detached leaf infection assay, the fourth leaves of rice at the 4-leaf stage (typically 10-week-old plants) and three upper rosette leaves derived from 4-week old plantlets of Arabidopsis (Mukherjee et al. 2010) were mainly used for inoculation with R. solani. Leaves were inoculated with approximately equal size (0.3–0.4 cm diameter), single sclerotium of R. solani and kept in Petri plates with 100% humidity onto moistened Whatman filter paper. Infected leaves were harvested at different time points for staining with trypan, aniline, and DAB (3,3′-diaminobenzidine) followed by microscopic and macroscopic observations. The experiments were carried out three times, and each contained three biological replicates.
Microscopy
Trypan blue, DAB and aniline blue staining were performed to study cell death, H2O2 accumulation and callose deposition respectively, as described by Park et al. (2009). Briefly, for trypan blue staining the infected rosette leaves were cleared in alcoholic lactophenol (2:1) and stained with 250 μg/mL trypan blue in lactophenol (phenol: glycerol: lactic acid: water – 1:1:1:1, v/v). It was further destained with lactophenol, mounted onto glass slide with 50% glycerol and examined under bright field microscope. For DAB staining, infected leaves were incubated in 1 mg/mL aqueous DAB solution for 8 h in dark following which stain was replaced with water and incubated in similar conditions. Destaining was performed with acetic acid: ethanol (96:4 v/v) and mounted with 50% glycerol and observed under bright field microscope. For aniline blue staining, leaf samples were cleared in alcoholic lactophenol and stained overnight with 0.01% aniline blue in 150 mM K2HPO4, pH 9.5. Stained leaves were equilibrated with 50% glycerol and observed using Olympus IX71 microscope.
Extraction and estimation of chlorophyll content
For chlorophyll estimation, untreated and treated leaves were harvested at 30 h post inoculation (hpi). Leaves were ground with 80% acetone and centrifuged at 5000 rpm for 5 min. The supernatant was collected in a tube and the process was repeated until the residue appears colorless. Final volume was adjusted with 80% acetone as per the weight of the leaves in a concentration of 10 mg/mL. The absorbance of the solution was recorded at 645 nm and 663 nm against the blank solvent (acetone) (Rajalakshmi and Banu 2013). The concentrations of chlorophyll a, chlorophyll b and total chlorophyll was measured as mg/g of the sample and were calculated as described by Azizi et al. (2015). Due to difference in sizes of the leaves, as intact leaf was taken without punching it, the W/V ratio was different. Further, the chlorophyll content was analysed among the WT Col-0 and the three pen mutants.
Scanning electron microscopy
Infected leaves were fixed in 4% paraformaldehyde and subsequently washed with phosphate buffer. Leaves were blotted dry with Kimwipe and mounted on metal stubs for visualization under environmental SEM. Each plant had a number of three detached leaves and data from three biological replicates.
Pathogenicity assays and calculation of disease level using detached leaves
The infected rosette leaves from the WT and pen mutants, harvested at 1 day post inoculation (dpi), 2 dpi and 3 dpi were photographed. The percentage of disease level was calculated based on the area of necrotic lesions formed at different time point in all accessions of Arabidopsis. At 1 dpi, 2 dpi and 3 dpi, disease level was calculated in whole plant as follows:
Each plant had a number of three detached leaves and data from three biological replicates were taken for calculation. The data were expressed as means ± SD of three biological replicates (n = 3) of which one of the 3 representative biological replicates is shown.
Quantification of fungal biomass
A comparative study was conducted in order to quantify the fungal biomass using PCR assay. Genomic DNA was extracted from the infected leaves at 30 hpi and 48 hpi and 100 ng/μl DNA was used as a template. R. solani primers bind to conserved regions of the fungal 28S rDNA as described before (Kouzai et al. 2018) and primers of the housekeeping gene Arabidopsis ACTIN2 was used as an endogenous control. Primers are listed in Table 1.
Statistical analysis
To confirm the authenticity and consistency of the data, the experiments were independently completed in three sets. In addition, the data were analyzed using ImageJ: IJ 1.46r (Schneider et al. 2012), GIMP 2.10.10 (Scalschi et al. 2015) and Origin software. Data presented all through the article represents the mean ± SD of three biological replicates (n = 3) of which one of the 3 representative biological replicates is shown. For all the infection assay 3 leaves per plant and 3 seedlings of each Arabidopsis accessions were used. Significant differences among the WT Col-0, pen mutants and complement PEN2-GFP were determined using one-way ANOVA (P < 0.05). The presence of R. solani mycelia over the infected leaf surface from the micrographs for each replicate was automatically identified using the GNU Image Manipulation Program (GIMP 2.10.10) software in “Select- By colour” tool option and trypan blue intensity were recorded as the area covered by the total number of selected pixels.
Results
Arabidopsis Col-0 is a nonhost to rice sheath blight pathogen R. solani
As reported by Keijer 1996 that adhesion of hyphae on the host plant surface initiates during 10–12 h of infection, R. solani AG-1 IA (hereafter referred to as R. solani) showed enhanced hyphal growth on susceptible rice leaves (Swarna cultivar) from 12 hpi to 72 hpi. We also observed repetitive T-shaped/ perpendicular hyphal branching on the adjacent epidermal cells at 12 hpi (Fig. S1a). The hyphal branching continued to proliferate up to 24 hpi and marked the initiation of small aggregate of compact hyphae termed as infection cushions. We observed prominent dome-shaped infection cushions at 48 hpi and it increased in number by 72 hpi. We also recorded the number of infection cushions per 100 μm2 area observed under microscope at 24 hpi and 72 hpi (Fig. S1b). R. solani developed enhanced hyphal growth on susceptible rice leaves (Swarna cultivar) upon increased time points from 12 hpi to 72 hpi.
In order to study the interaction between Arabidopsis and R. solani we infected six Arabidopsis WT accessions with R. solani. Infection structures were monitored microscopically at 24 and 30 hpi by trypan blue staining which revealed the inability of R. solani to colonize any of the six accessions. Among all the wild accessions screened, Col-0 and Ler-0 were the most resistant as compared to Swarna in terms of hyphal growth (Fig. 1a). R. solani mycelial threads grew all over the leaf surface with prominent runner hyphae or short perpendicular- lateral branching but no complex hyphal branching or specialized infection cushions developed in Col-0 as compared to rice which showed infection cushion at 30 hpi. The findings confirm that Arabidopsis is a nonhost for rice sheath blight pathogen and has therefore been used for further investigations.
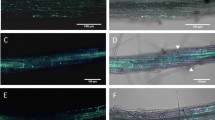
Development of infection structures on Arabidopsis WT accessions and Rice (Swarna) upon infection with R. solani. (a) The intensity of trypan blue stained fungal hyphae in Arabidopsis wild ecotypes was quantified and normalized with the intensity of Swarna obtained from GIMP. Data represent the mean ± SD of three biological replicates (n = 3) of which one of the 3 representative biological replicates is shown. The experiment was carried out three times with similar results. Upper panel (b-e) shows the trypan blue stained infected leaves of Col-0 whereas the lower panel (f-h) is for rice. Leaves inoculated with water were used as control (b, f). The extent of disease progression was observed at 24 hpi (c, g) and 30 hpi (d, e, h). Infection cushions are clearly seen in rice at 30 hpi which was absent in Col-0. Cell wall thickening at the site of infection was observed in Col-0 (e). Stained leaves were observed under bright field microscope. Leaves inoculated with water were used as control. Scale bar = 50 μm
Arabidopsis pen mutants allow differential R. solani growth
We used the Arabidopsis penetration mutants– pen1, pen2–3 and pen3–1 which was verified for homozygosity using PCR based SBP molecular markers (Fig. S2, Table 1). We assessed Arabidopsis - R. solani interaction at cellular level by infecting rosette leaves of four week old plants with R. solani sclerotia and the disease phenotype was monitored at 6, 12, 24, 30, 36 and 48 hpi. No mycelial growth was observed after 6 and 12 hpi in either Col-0 or any of the pen mutants. However, pen mutants showed profuse hyphal branching where each tip formed right-angled or T-shaped branching at both 24 and 30 hpi. Among the pen mutants screened, pen2–3 showed maximum fungal colonization and development of early infection cushions at 30 hpi, which otherwise did not appear in pen1 and pen3–1. Also, Col-0 did not show extensive mycelial colonization (Fig. S3). On the other hand, we observed prominent infection cushions at 36 and 48 hpi in pen2–3, which however initiated in pen3–1 at 48 hpi and were completely absent in Col-0 and pen1 (Fig. S3).
To examine the results obtained from the time course experiment, we focused our observations to 30 hpi, where initiation of infection cushions were observed; and 48 hpi, where well-formed infection cushions were observed (Fig. 2a). Infected leaves of pen2–3 showed several runner hyphae entangled together to form lobate appressoria and initiation of infection cushion at 30 hpi. At 48 hpi, there was more hyphal proliferation in pen2–3, which resulted in the formation of compact bundles of fungal hyphae, entrenched onto the leaves surface and the formation of infection cushions (Fig. 1a). We quantified the hyphal growth in the leaves infected with R. solani (Fig. 2b) by using the GIMP. R. solani colonization was higher in pen2–3 at 30 hpi that progressed further by 48 hpi as compared to other pen mutants, WT Col-0 and Swarna. The extensive hyphal branching and cushion formation in the mutants is a clear indication of compromised resistance in pen2–3 to R. solani when the PEN2 gene is not functional.
Development of infection cushion upon infection by R. solani in Arabidopsis WT (Col-0) and pen mutants. (a) Leaves of WT and pen mutants (pen1, pen2–3 and pen3–1) of Arabidopsis were stained with trypan blue upon infection at 30 hpi and 48 hpi by R. solani. Maximum cushions formed at 48 hpi in pen2–3. Red arrows indicate infection cushions. The experiment was carried out three times, and each contained three biological replicates. Scale bar = 50 μm. (b) The intensity value of Col-0 and pen mutants were normalized with the intensity of Swarna at both 30 and 48 hpi respectively. The intensity of trypan blue stained fungal hyphae was quantified using GIMP 2.10.10 software. Data represent the mean ± SD of three biological replicates (n = 3) of which one of the 3 representative biological replicates is shown. The experiment was carried out three times with similar results
pen2–3 allows formation of extensive infection cushions
In order to observe the surface morphology during the initial stage of infection in WT and pen mutants, we performed SEM analysis of the infected leaf tissue. With increasing time points, all the pen mutants showed repeated dichotomous right-angled branching pattern, of which pen2–3 developed complex hyphal clusters forming infection cushions at 48 hpi (Fig. S4). In pen2–3, profuse mycelial branching and initiation of infection cushions were observed at 30 hpi; while initiation of infection cushions in pen3–1 was observed at 48 hpi. Also, pen3–1 developed swollen hyphal tips at 48 hpi that lead to formation of lobate appressoria. Compact aggregation of hyphal branches in an infection cushion lead to the formation of infection pegs at the base of the cushion which allows the pathogen to colonize and proliferate further (Dodman et al. 1968). Col-0 showed secondary hyphal branching at 30 hpi with no infection cushions (Fig. S4). SEM analysis revealed that the infection cushions were found exclusively in pen2–3.
pen2–3 undergoes oxidative burst and callose accumulation upon infection with R. solani
Generation of ROS and deposition of callose by plants acts as an early defense responses against biotic stress at the site of plant-pathogen interaction (Fauth et al. 1998; Luna et al. 2011). Therefore, we monitored the deposition of H2O2 as yellowish-brown precipitate by DAB staining (Fig. 3a). We observed that pen2–3 exhibited relatively higher level of H2O2 accumulation followed by pen1 and pen3–1 as compared to Col-0. Higher level of H2O2 in pen2–3 can be correlated with increased hypersensitive cell death at the infection site with increasing time point (Fig. 3a). Callose deposition pattern was also examined in the leaves challenged with R. solani using aniline blue stain. The mutants accumulated significantly more callose whereas Col-0 showed reduced callose deposition (Fig. 3b). The epidermal cells of pen2–3 showed higher callose deposition along the cell margin, which directed compromised disease resistance.
Accumulation of H2O2 and callose at the infection sites of Arabidopsis leaves challenged with R. solani. Infected leaves of WT and pen mutants of Arabidopsis were stained with (a) DAB and (b) aniline blue at 30 hpi and 48 hpi. (a) Production of H2O2 was observed at the site of infection as yellowish-brown precipitate. (b) Callose deposition was observed UV excitation of a fluorescence microscope. Each experiment was carried out with three biological replicates of which one of the 3 representative biological replicates is shown. Scale bar = 50 μm
Macroscopic lesion phenotypes induced by R. solani vary between WT and pen mutants of Arabidopsis
Phenotyping of the differential extent of infection in the pen mutants and WT upon challenge with R. solani sclerotia after 3 dpi were studied by observing macroscopic lesion formation. Col-0 and the pen mutants were almost indistinguishable up to 1 dpi (Fig. 4a). Necrotic lesions developed as early as 2 dpi in pen1, pen2–3 and pen3–1, while WT only showed chlorotic patches and slight lesion formation along the leaf margin. Contrastingly, when the infection period was increased to 3 dpi, mutants were clearly distinguished from the WT, especially the infected leaves of pen2–3. Among the pen mutants, pen2–3 showed maximum necrotic lesion and chlorosis at 2 dpi that increased up to 3 dpi. The mycelial colonization was clearly observed in pen1, pen2–3 and pen3–1 at 2 dpi whereas it was seen only at 3 dpi in WT Col-0, with maximum colonization and lesion length in pen2–3 (Fig. 4b). Based on this experiment, percentage of disease severity was calculated (Fig. S5). Infected leaves of pen2–3 showed maximum necrotic lesion with an average of 0.139%, 36.38% and 67.54% at 1 dpi, 2 dpi and 3 dpi respectively.
Macroscopic observation of disease progression in infected Arabidopsis WT and pen mutants. (a) Leaves from four week old seedlings were infected with R. solani sclerotia and photographed at 0 dpi, 1 dpi, 2 dpi and 3 dpi. Leaves inoculated with water were used as control. The coverage of necrotic lesions increased with time in each accession with pen2–3 being the most affected. The experiment was carried out three times, and each contained three biological replicates of which one of the 3 representative biological replicates is shown. (b) Enlarged image of (a) showing the colonization of mycelia on the leaf surface of Col-0 at 3 dpi and on penetration mutants at 2 dpi with maximum lesion in pen2–3
R. solani infection reduces photosynthetic efficiency in pen2–3
Pathogen virulence dampens photosynthetic performance (Okorski et al. 2008). R. solani pathogenesis leads to down-regulation of photosynthesis (Ghosh et al. 2017). Thus, total chlorophyll content was estimated to analyze the effect of R. solani infection in Arabidopsis accessions at 2 dpi. Infected leaves showed reduced chlorophyll content in all the accessions. pen2–3 showed maximum reduction in cellular chlorophyll content as compared to WT, pen1 and pen3–1 (Fig. S6). The reduction in chlorophyll content co-related with both microscopic and macroscopic observations. Higher amount of chlorophyll content was recorded in infected leaves of WT Col-0 as compared to that of pen mutants.
PEN2 provides penetration resistance against R. solani
The disease severity caused by R. solani was at maximum in case of pen2–3. To authenticate and confirm the result that pen2–3 mutant triggers cushion formation, we infected the complemented line of Arabidopsis that expresses green fluorescent protein (GFP)-tagged PEN2 driven by native 5′ regulatory region in pen2–1 background. As observed in previous experiment, pen2–3 allowed formation of infection cushions on the leaf surface (Fig. 2). On the other hand, PEN2-GFP showed similar resistance against R. solani as WT Col-0 with no cushion formation at either 30 hpi or 48 hpi (Fig. 5). This experiment substantiated our hypothesis that PEN2 contributes to penetration resistance against R. solani as it does not allow formation of infection cushions.
Comparison of infection structure of R. solani in PEN2 complemented Arabidopsis. Leaves of WT Col-0, PEN2-GFP and pen2–3 mutants of Arabidopsis were stained with trypan blue after 30 hpi and 48 hpi. Maximum cushions formed at 48 hpi in pen2–3. Red arrows indicate infection cushions. Scale bar = 50 μm
Fugal biomass increases in pen mutants with progress in time
The relative quantification of R. solani DNA was performed by using qPCR of the infected leaves of pen mutants and complemented PEN2-GFP with respect to Col-0 at both 30 hpi as well as 48 hpi (Fig. 6). We observed that pen2–3 accumulated the highest amount of fungal biomass, followed by pen3–1 and pen1. The results obtained revealed that the fungal biomass increased over time in pen mutants (especially in pen2–3), PEN2-GFP and WT and reached to a maximum at 48 hpi. One-way ANOVA (P < 0.05) was performed to study the significant differences at 30 hpi and 48 hpi (Fig. 6). These results were consistent with the disease phenotype and disease severity as observed in previous experiments.
Quantification of fungal biomass in infected Arabidopsis leaves. Genomic DNA extracted from the infected leaves of Col-0, pen1, pen2–3, pen3–1, and PEN2-GFP at (a) 30 hpi and (b) 48 hpi was used in PCR reaction using 100 ng/μl DNA using specific primers of R. solani AG1 (28S rDNA). Primers of the housekeeping gene Arabidopsis ACTIN2 were used as an endogenous control. Data represent the mean ± SD of three biological replicates (n = 3) of which one of the 3 representative biological replicates is shown. Significant differences among the wild type Col-0, pen mutants and complement PEN2-GFP were determined using one-way ANOVA (P < 0.05)
Discussion
In this study, we investigated the pathogenicity and infection process of R. solani in rice, Arabidopsis WTs as well as pen mutants. The interaction between rice and R. solani showed the initiation of infection cushion formation at 24 hpi and progresses up to 72 hpi (Fig. 1a, b). The coherent investigation of our present study revealed that pen2–3 contributes to nonhost resistance in Arabidopsis thaliana to R. solani. Of the three pen mutants studied namely, pen1, pen2–3 and pen3–1, pen2–3 demonstrated severely compromised penetration resistance to R. solani. Absence of PEN2 showed significantly maximum hyphal colonization and number of atypical infection cushions in Arabidopsis, alike to rice challenged with R. solani, which clearly depicts the importance of PEN2 in providing nonhost resistance against sheath blight.
Thus, we found that PEN2 provides disease resistance against rice sheath blight pathogen R. solani for the first time. As per the previous report by Lazniewska et al. (2012), we observed similar infection cushions in pen2–3 mutants upon infection with R. solani. Unlike pen2–3, pen1 and pen3–1 mutants did not allow cushion formation at 30 hpi (Fig. 2a). In pen3–1, the runner hyphae gave rise to swollen hyphal tips instead of forming cushion like structures. This experiment made it quite apparent that there is a clear distinction between hyphal branching formed by WT and infection cushion formed by pen2–3.
The observation from DAB staining clearly indicated the production of H2O2 in pen2–3 at the interaction site of R. solani mycelia and Arabidopsis in the epidermal cells as compared to pen1, pen3–1 and WT Col-0. As it is already reported that the production of H2O2 not only provide direct defense response but also follows downstream signal transduction cascade which in turn lead to activation of hypersensitive response (HR) (Jones 2006; Waetzig et al. 1999; Wojtaszek 1997). The production of H2O2 was not only observed at the site of infection in pen2–3 but also in the neighboring cells of the inoculated leaves which signifies that neighboring cells defense system has already activated and shows up HR response by following defense signaling cascade. Contrastingly, pen1 showed reduced accumulation of H2O2 at the early hour of infection. pen3–1 showed little higher accumulation of H2O2 than pen1 but less than pen2–3 (Fig. 3a). Thus, higher production of H2O2 in pen2–3 at an early stage may indicate the defense pathways activating the early signaling event are intact in the mutant.
Production of callose; composed of β-(1, 3)-glucan polymer, serves as a biomarker against intrusion by pathogens at the site of infection in host (Luna et al. 2011). Callose deposition varied in all the three pen mutants. pen1 failed to produce enhanced callose in response to R. solani infection. In contrast to pen2–3, which showed thick callose deposition at the site of infection, pen1 showed patch like pattern (Fig. 3b). The notable increase in the deposition of callose in pen2–3 mutant may explain the number and extent of attempted sites of fungal penetration. Previously it has been reported that papillae composed of callose are deposited at the sites of penetration into the cell wall upon infection by the fungal pathogen, which results in the cell wall thickening (Ellinger et al. 2013).
Macroscopic infection symptom results clearly showed the elevation in lesion formation. This results correlates with the increased formation of infection cushions in pen2–3 (Fig. 4a). As per the previous study in rice, reported by Ghosh et al. (2017), we also found that Arabidopsis WT and pen mutants did not show any anatomical alterations during the early infection stage i.e., 1 dpi, whereas, upon 3 dpi of pathogenesis it showed severe necrotic lesions (Fig. 4b). Further, the specific events in leaves of pen2–3 leaves viz. chlorosis, decline in chlorophyll content, higher necrotic lesions and increased leaf senescence suggested that pen2–3 was most susceptible among the other two pen mutants. Higher chlorophyll content in WT as compared to the mutants indicated higher physiological stress in mutant plants especially in pen2–3 upon treated with R. solani.
The findings of our study about pen2–3 allowed increased penetration by R. solani, concur with previous findings which reported compromised resistance to Magnaporthe oryzae, a hemibiotrophic pathogen, in a pen2 background (Maeda et al. 2009; Maeda et al. 2010). As confirmed from both macroscopic and microscopic analysis in this study, pen2–3 showed cushion formation, which is the hallmark of pre-penetration stage, and also showed maximum lesion length. We further validated our hypothesis that the PEN2 gene might have some role in providing defense resistance against R solani and the toxic products from PEN2 might be acting on the developing mycelia and not allowing the fungus to grow any further. Confirmation from the infection of complement line of pen2–3 restored the disease resistance as found in Col-0 (Fig. 6). In addition to the phenotypic and microscopic observations, molecular assay was also performed to quantify the total fungal biomass in the leaves of WT, mutants and complement PEN2 line challenged with R. solani also act a plants defense indicator against conquering fungus. We observed an increase in the amount of fungal DNA in WTs and mutants with increasing time intervals. However, pen2–3 showed higher amount of fungal DNA as compared to Col-0 and its complement line (PEN2-GFP) (Fig. 6). Although the microscopic data of pen3–1 didn’t show disease severity microscopically even at 48 hpi (fig. 2b), significant increase in fungal biomass at 48 hpi in pen3–1 (Fig. 6b) compared to Col-0 and pen1–2, revealed it involvement at the later stage of infection. PEN3 independently also plays role in mediating NHR, mutation in this pre-penetration gene leads to breach of plant immunity in Arabidopsis and allow the entry of R. solani (Fig. 2a and 6). The susceptibility in pen3 can also be related to similar phenotype as has been observed against powdery mildew of barley and multiple fungal pathogens of wheat mediated by ABC transporters (Krattinger et al. 2009; Stein et al. 2006). This might be due to the fact that, during infection PEN2 functions together with PEN3 i.e., the hydrolytic enzymes produced by PEN2 are transported by PEN3 at the site of infection (Stein et al. 2006). Possibly, the activation of pre-penetration resistance genes and their by-products do not allow further growth of R. solani. Further characterization of the genes in multiple plant hormone pathway mutants may lead to a clear idea on the involvement of the genes in NHR disease resistance against rice sheath blight.
Conclusion
In summary, our study has uncovered the involvement of PEN2 gene from Arabidopsis at pre-penetration stage as an early infection process in providing disease resistance against broad host range necrotrophic pathogen, R. solani that has not been reported previously. Concurrent with the data obtained from the disease severity as noted in cases of pen2–3 mutant from trypan blue staining, aniline blue staining, DAB staining, chlorophyll content and fungal DNA estimation, it is noteworthy to conclude the potential of the PEN2 that does not allow the R. solani infection cushion formation. Additionally, the role of PEN3 can also be studied to know its possible involvement during pre-invasive stage of infection against R. solani. Further studies can correlate the concurrent function of PEN2 and PEN3 against this necrotrophic sheath blight pathogen of rice and can underpin significant insight into their role in pre and post-invasive resistance.
References
Ajayi-Oyetunde O, Bradley C (2018) Rhizoctonia solani: taxonomy, population biology and management of rhizoctonia seedling disease of soybean. Plant Pathol 67:3–17
Azizi P, Rafii MY, Mahmood M, Abdullah SNA, Hanafi MM, Nejat N, Latif MA, Sahebi M (2015) Differential gene expression reflects morphological characteristics and physiological processes in rice immunity against blast pathogen. Magnaporthe oryzae PloS one 10:e0126188
Basu A, Chowdhury S, Ray Chaudhuri T, Kundu S (2016) Differential behaviour of sheath blight pathogen Rhizoctonia solani in tolerant and susceptible rice varieties before and during infection. Plant Pathol 65:1333–1346
Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106
Bittel P, Robatzek S (2007) Microbe-associated molecular patterns (MAMPs) probe plant immunity. Curr Opin Plant Biol 10:335–341
Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, Schulze-Lefert P (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425:973–977
Dodman R, Barker K, Walker J (1968) A detailed study of the different modes of penetration by Rhizoctonia solani. Phytopathology 58:1271–1276
Ellinger D et al. (2013) Elevated early callose deposition results in complete penetration resistance to powdery mildew in Arabidopsis Plant Physiology:pp 112.211011
Fauth M, Schweizer P, Buchala A, Markstädter C, Riederer M, Kato T, Kauss H (1998) Cutin monomers and surface wax constituents elicit H2O2 in conditioned cucumber hypocotyl segments and enhance the activity of other H2O2Elicitors plant physiology 117:1373-1380
Flentje N, Dodman R, Kerr A (1963) The mechanism of host penetration by Thanatephorus cucumeris. Aust J Biol Sci 16:784–799
Foley RC, Gleason CA, Anderson JP, Hamann T, Singh KB (2013) Genetic and genomic analysis of Rhizoctonia solani interactions with Arabidopsis; evidence of resistance mediated through NADPH oxidases PLoS one 8:e56814
Fuchs R, Kopischke M, Klapprodt C, Hause G, Meyer AJ, Schwarzländer M, Fricker MD, Lipka V (2016) Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28:130–145
Ghosh S, Kanwar P, Jha G (2017) Alterations in rice chloroplast integrity, photosynthesis and metabolome associated with pathogenesis of Rhizoctonia solani. Sci Rep 7:41610
Gill US, Lee S, Mysore KS (2015) Host versus nonhost resistance: distinct wars with similar arsenals. Phytopathology 105:580–587
Jones JD (2006) Dangl JL. The plant immune system Nature 444:323–329. https://doi.org/10.1038/nature05286
Kouzai Y et al. (2018) Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon new phytologist 217:771–783
Krattinger SG et al. (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat science 323:1360-1363
Kumar KVK, Reddy M, Kloepper J, Lawrence K, Groth D, Miller M (2016) Sheath blight disease of rice (Oryza sativa L.)–an overview. Biosciences Biotechnology Research Asia 6:465–480
Kumar KVK et al (2011) Evaluation and selection of elite plant growth-promoting rhizobacteria for suppression of sheath blight of rice caused by Rhizoctonia solani in a detached leaf bio-assay. Int J Appl Biol Pharm Technol 2:488–495
Lazniewska J, Macioszek VK, Kononowicz AK (2012) Plant-fungus interface: the role of surface structures in plant resistance and susceptibility to pathogenic fungi. Physiol Mol Plant Pathol 78:24–30
Li S, Peng X, Wang Y, Hua K, Xing F, Zheng Y, Liu W, Sun W, Wei S (2019) The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Front Microbiol 10:2228
Lipka U, Fuchs R, Lipka V (2008) Arabidopsis non-host resistance to powdery mildews. Curr Opin Plant Biol 11:404–411
Lipka V et al. (2005) Pre-and postinvasion defenses both contribute to nonhost resistance in Arabidopsis science 310:1180-1183
Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol 123(3):795–806
Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J (2011) Callose deposition: a multifaceted plant defense response. Mol Plant-Microbe Interact 24:183–193
Maeda K, Houjyou Y, Komatsu T, Hori H, Kodaira T, Ishikawa A (2009) AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol Plant-Microbe Interact 22:1331–1340
Maeda K, Houjyou Y, Komatsu T, Hori H, Kodaira T, Ishikawa A (2010) Nonhost resistance to Magnaporthe oryzae in Arabidopsis thaliana. Plant Signal Behav 5:755–756
Mondal A, Dutta S, Nandi S, Das S, Chaudhuri S (2012) Changes in defence-related enzymes in rice responding to challenges by Rhizoctonia solani. Archives of phytopathology and plant protection 45:1840–1851
Mukherjee AK, Carp MJ, Zuchman R, Ziv T, Horwitz BA, Gepstein S (2010) Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J Proteom 73(4):709–720
Nuernberger T, Lipka V (2005) Non-host resistance in plants: new insights into an old phenomenon. Mol Plant Pathol 6:335–345
Okorski A, Olszewski J, Pszczółkowska A, Kulik T (2008) Effect of fungal infection and the application of the biological agent EM 1TM on the rate of photosynthesis and transpiration in pea (Pisum sativum L.) leaves. Polish Journal of Natural Sciences 23:35–47
Pannecoucque J, Höfte M (2009) Interactions between cauliflower and Rhizoctonia anastomosis groups with different levels of aggressiveness BMC. Plant Biol 9:95
Park J-Y, Jin J, Lee Y-W, Kang S, Lee Y-H (2009) Rice blast fungus (Magnaporthe oryzae) infects Arabidopsis via a mechanism distinct from that required for the infection of rice. Plant Physiol 149:474–486
Rajalakshmi K, Banu N (2013) Extraction and estimation of chlorophyll from medicinal plants. International Journal of Science and Research:209–012
Sahu, BB, Sumit R, Srivastava SK, Bhattacharyya MK (2012) Sequence based polymorphic (SBP) marker technology for targeted genomic regions: its application in generating a molecular map of the Arabidopsis thaliana genome. BMC Genomics 13(1):20
Scalschi LM et al. (2015) Quantification of callose deposition in plant leaves
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Sharon M, Freeman S, Sneh B (2011) Assessment of resistance pathways induced in Arabidopsis thaliana by hypovirulent Rhizoctonia spp. isolates. Phytopathology 101:828–838
Singh A, Rohila R, WILLOCQUET SSL, SINGH U (2012) Infection process in sheath blight of rice caused by Rhizoctonia solani. Indian Phytopathology
Speth EB, Lee YN, He SY (2007) Pathogen virulence factors as molecular probes of basic plant cellular functions. Curr Opin Plant Biol 10:580–586
Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S (2006) Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18:731–746
Taheri P, Tarighi S (2011) Cytomolecular aspects of rice sheath blight caused by Rhizoctonia solani. Eur J Plant Pathol 129:511–528
Waetzig GH, Sobczak M, Grundler FM (1999) Localization of hydrogen peroxide during the defence response of Arabidopsis thaliana against the plant-parasitic nematode Heterodera glycines. Nematology 1:681–686
Wei M, Wang A, Liu Y, Ma L, Niu X, Zheng A (2020) Identification of the novel effector RsIA_NP8 in Rhizoctonia solani AG1 IA that induces cell death and triggers defense responses in non-host plants. Front Microbiol 11:1115
Wojtaszek P (1997) Oxidative burst: an early plant response to pathogen infection. Biochem J 322:681–692
Zheng A, Lin R, Zhang D, Qin P, Xu L, Ai P, Ding L, Wang Y, Chen Y, Liu Y, Sun Z, Feng H, Liang X, Fu R, Tang C, Li Q, Zhang J, Xie Z, Deng Q, Li S, Wang S, Zhu J, Wang L, Liu H, Li P (2013) The evolution and pathogenic mechanisms of the rice sheath blight pathogen. Nat Commun 4:1424
Acknowledgments
The authors thank Director, NIT Rourkela for providing all facilities at NIT Rourkela, Odisha INDIA. The authors also thank Dr. Arup K. Mukherjee, NRRI, Cuttack for providing the R. solani pure culture.
Financial support
Supported by the S & T Division, Govt. of Odisha (Grant number 27552800232014) and SERB, Govt. of India (YSS/2014/000142).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors read the manuscript and declare that there is no conflict of interest.
Research involving human participants and/or animals
Not applicable. The research involved no human participants or animals.
Informed consent
The research involved no human participants, and no animals so that the statement on the welfare of animals is not required.
Supplementary Information
ESM 1
(PPTX 5868 kb)
Rights and permissions
About this article
Cite this article
Parween, D., Sultan, E., Dalei, K. et al. Arabidopsis nonhost resistance gene PENETRATION 2 is involved in disruption of cushion formation by Rhizoctonia solani during early infection process. Australasian Plant Pathol. 50, 281–292 (2021). https://doi.org/10.1007/s13313-020-00768-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-020-00768-8