Abstract
Overlapping communities are pervasive in real-world networks. Therefore, overlapping community detection is an important task in network analysis. Recently, many overlapping community detection methods are proposed to achieve different goals. However, how to detect communities effectively and efficiently is still an open problem. In this paper, we use our previously proposed method LinkSHRINK to detect overlapping community detection, which is based on density structure and modularity optimization. It successfully solves the excessive overlapping problem. Moreover, it can detect both overlapping communities of multi-granularity and outliers. To deal with very large networks, we choose to sample on the large graph and then parallelize LinkSHRINK by distributed computing frameworks. Experiments are conducted on benchmark networks and some real-world networks with known ground-truth communities. The experimental results demonstrate that LinkSHRINK outperforms most of the baseline methods and its parallel versions PLinkSHRINK and MLinkSHRINK can process large networks efficiently.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
At present, many complex systems are in the form of complex networks or can be modeled by complex network such as interpersonal relationship in human society and academic collaboration networks. Complex networks usually present community structure in which nodes connect densely and connections between them are sparse. However, in real-world networks, communities tend to have overlapping parts, and nodes in the network can be classified into more than one community. Thus, it is more meaningful to find the overlapping community structure in complex networks. Current overlapping community detection methods are mainly divided into two categories:
- 1.
Node-based methods This kind of methods is directly processed on the node graph. Well-known methods in this kind include spectral clustering method (Li et al. 2018), Clique Percolation Method (CPM) (Palla et al. 2005), label propagation methods COPRA (Gregory 2010), SLPA (Xie and Szymanski 2012), hierarchical clustering method SHRINKO (Huang et al. 2011) and density-based methods SCAN (Xu et al. 2007) and OCDDP (Bai et al. 2017).
- 2.
Link-based methods One kind of link-based methods calculates the similarity between links and then partitions the links in the network by using link similarity. Well-known method in this kind is LINK (Ahn et al. 2010). The other kind of link-based methods cannot directly be processed on the node graph, but on the edge graph or link graph induced from node graph. After the transformation from node graph to link graph has been done, it uses the non-overlapping community detection method to detect communities based on link graph. After the non-overlapping community detection methods finished on the link structure, each link induced by two nodes will be classified into a determined community. Thus, nodes in the network can finally belong to multiple communities. Well-known method of this kind is proposed by Evans and Lambiotte (2009).
Besides, some novel methods based on stochastic models such as Brain Ball et al. (2011), Gopalan and Blei (2013) and Sun et al. (2014) are proposed. Recently, there are also some novel methods based on nonnegative matrix factorization (Zhang et al. 2018) and two-step (Sarswat et al. 2017). According to the existing two types of methods, we found there are some shortcomings as follows:
- 1.
Nondeterministic results This situation usually happens in the methods using label propagation, such as COPRA. The community label of the nodes is determined by the random selection of the node label when more than one pair has the same maximum belonging coefficient, which means running the method many times in the same network may find different communities every time.
- 2.
Inaccurate communities For the aforementioned methods, most methods tend to make each node in the network belonging to one community. However, in the real-world network, not all of the nodes belong to any community, called isolated nodes. These nodes have very few connections with others. In Fig. 1, node 6 is an isolated node which cannot be the member of the community which contains other five nodes.
- 3.
Excessive overlapping problem There are too many highly overlapping nodes in the result of community detection. This phenomenon usually occurs in overlapping community detection based on the link partition. We will introduce in detail in Sect. 2.
- 4.
Complex parameters Recently, methods (Lim et al. 2014; Zhu et al. 2013) can solve the excessive overlapping problem by improving density-based clustering method SCAN (Xu et al. 2007). But it comes to the problem that the experimental results highly depend on more than two complex parameters. How to reduce the influence of complex parameters on the algorithm is also a big challenge.
- 5.
Longer running time For overlapping community detection based on link partition, it always takes quite long running time, because the link graph transformed from node graph can be very large.
Based on the above problems, in our previous work (Yin et al. 2016),Footnote 1 we take advantage of both density-based clustering and modularity-based methods to discover deterministic overlapping community structure in networks by using link graph, called LinkSHRINK. However, LinkSHRINK cannot deal with large-scale network. In this paper, we extend LinkSHRINK by using Spark, GraphX and Hadoop to find overlapping communities in large-scale networks. Our contributions are as follows:
- 1.
We can successfully find not only the communities but also outliers (Ester et al. 1996). Compared with SCAN (Xu et al. 2007) requiring parameters \(\epsilon \) and \(\mu \), we have the advantage of only needing one parameter easily tuned.
- 2.
We introduce the notion of the community overlap degree \(\vartheta \), which is essential in analyzing overlapping communities in complex network. Our algorithm can also find the communities with various overlapping granularity.
- 3.
We choose to sample on the large networks and implement our algorithm by distributed computing frameworks Hadoop and Spark GraphX, called MLinkSHRINK and PLinkSHRINK, respectively.
- 4.
We conduct sufficient experiments to show the advantage of PLinkSHRINK. Experimental results show that PLinkSHRINK runs faster on large networks than MLinkSHRINK and LinkSHRINK do. The effectiveness of PLinkSHRINK is about 1% worse than that of LinkSHRINK.
The rest of the paper is organized as follows. We first review the related work in Sect. 2 and introduce some backgrounds in Sect. 3. Then we elaborate LinkSHRINK algorithm in short and propose its parallel versions with detailed implementation in Sect. 4. We also show experimental results in comparison with existing methods in Sect. 5. Finally, we summarize our work and suggest future research directions in Sect. 6.
2 Related work
2.1 Structural clustering algorithm with parameter-free
Newman and Girvan (2004) proposed a metric, called modularity, to measure the quality of the detected communities. The better result of community detection leads to the higher value of modularity. However, it is demonstrated that the community detection methods by maximizing modularity can hardly find small communities (Lancichinetti and Fortunato 2011), called modularity limit. SCAN (Xu et al. 2007) is a density-based method to find communities in network. It introduces two parameters to determine the core vertex. It can find small communities in large network. Moreover, SHRINK (Huang et al. 2010) is proposed to find hierarchical communities requiring no parameter. It not only finds communities, but also identifies hubs and outliers by combining density-based clustering and modularity optimization. Modularity Q (Newman and Girvan 2004) is a metric for evaluating the quality of community discovery which is proposed by Newman and Girvan. Modularity \(Q=\sum _{s=1}^k{[\frac{l_s}{L}-(\frac{d_s}{2L})^2]}\), where L is the number of edges in the network, \(l_s\) is the number of edges in the community s, \(d_s\) is the sum of degree of the nodes in community s. It is suggested that the higher modularity Q is, the better network partition result the method gets. However, recent research shows that network partition with higher modularity Q cannot reveal relatively smaller community structure. It usually tends to identify larger communities. In order to get rid of the restrictions, compared with the methods based on modularity optimization, the SHRINK takes advantage of the density theory trying to get the relatively high modularity Q. In this paper, we are motivated to find overlapping communities in the similar idea with SHRINK, which breaks the limit of modularity (Fortunato and Barthelemy 2007).
2.2 Clustering based on link partition
LINK (Ahn et al. 2010) was firstly proposed to find overlapping community by using link partition. The basic idea of LINK is shown as follows.
Link graph is firstly constructed, and then LINK hierarchically clusters the nodes in link graph to detect the communities. Initially, each link community only contains one link. Then LINK maximizes the value of objective function as shown in formula (1) by merging two link communities with the highest similarity.
where M denotes the number of the links in the network, c denotes a link community, \(m_c\) represents the number of the links (edges) in the link community c, \(n_c\) represents the number of the nodes in the link community c, the numerator \(m_c-(n_c-1)\) denotes the number of edges linking the other \(n_c-1\) edges induced by \(n_c\) nodes, the denominator denotes the maximal number of edges linking the other \(n_c-1\) edges, \(\frac{m_c-(n_c-1)}{(n_c-2)(n_c-1)}\) denotes the link density in the community c and D denotes the average link density of all the communities. After obtaining the link communities, LINK transforms the link communities into the node communities. The node community consists of the nodes contained in the edges of the link community. The edges induced by the common node belong to different link communities, and the common node will belong to different node communities. And the common node will be the overlapping node.
However, LINK partitions each edge into a determined link community, which may lead to excessive overlapping problem. As shown in Fig. 2, LINK finds three link communities colored by yellow, blue and green, and their corresponding node communities are {1,2,3,4,5,6},{6,7},{0,7,8,9}. Node 6 and 7 are considered as overlapping nodes in the network which is actually not in agreement with the reality.
2.3 Community detection in large networks
The traditional stand-alone algorithms have been unable to deal with large networks very well. Currently, there are mainly two ways to solve this problem. One way is to reduce the size of the large network by sampling which leads to lose a little accuracy to exchange for time efficiency. The well-known method in this field includes Lim et al. (2014). The other way is the parallelization of existing methods based on the distributed computing frameworks, such as Hadoop and Spark. Well-known methods in this field include Qiao et al. 2017 and Jin et al. 2015. Wang et al. (2015) parallelized the overlapping communities detection algorithm in parallel framework GraphLab. Zeng and Yu (2015) presented a parallel hierarchical graph clustering algorithm that uses modularity as clustering criteria to effectively extract community structures in large graphs of different types. Zhang et al. (2016) proposed a parallel LPA to detect community in social network. Li et al. (2015) proposed a parallel multi-label propagation method to detect overlapping communities. Wickramaarachchi et al. (2014) and Cheong et al. (2013) presented efficient approaches to detect communities in large-scale networks by parallelizing Louvain algorithm for community detection. Moon et al. (2016) developed two parallel versions of the GN algorithm to support large-scale networks based on MapReduce and GraphChi. Kuzmin et al. (2013) presented highly scalable variants of a community detection algorithm called SLPA to detect overlapping communities of social networks. Thang (2017) parallelized BigClam (Yang and Leskovec 2013) to detect overlapping communities.
3 Background
In this section, we introduce method and platforms which are used in this paper.
- 1.
SHRINK (Huang et al. 2010) detects community based on density. Due to SHRINK combined modularity optimization-based method with heuristic strategy, it solves the problem of SCAN (Xu et al. 2007) which detects community depend on two sensitive parameters, \(\epsilon \) and \(\mu \). And SHRINK keeps the advantage of finding both hub nodes and outliers. It defines the structural similarity between two nodes. Let \(\sigma (u,v)\) be the structural similarity of nodes u and v. If \(\sigma (u,v)\) is the largest similarity between nodes u, v and their adjacent neighbor nodes, then {u, v} is a dense pair. A micro-community is a maximal connected component linked by edges induced by dense pairs. Greedy SHRINK clusters the network via greedy shrinkage of the dense pairs. Thus, each dense pair in a micro-community is considered separately. Starting with an arbitrary node u in a network G, it finds the dense pair containing u. If there is a node v adjacent to u that forms a dense pair {u, v} and its modularity gain is positive, it merges node v and u to form a super-node \(u'\). Then it checks whether there exists a dense pair containing \(u'\) and tries to shrink it. The above process is repeated until there does not exist a shrinkable dense pair containing current node. Then the algorithm continues with next unvisited node. The clustering is accomplished when all the nodes in the network G are visited.
- 2.
Hadoop is a popular open-source software framework for distributed storage and processing of very large data deployed on computer clusters. The core of Hadoop is composed of two parts. The one is the storage part known as Hadoop distributed file system (HDFS). The other one is the processing part called MapReduce engine.
- 3.
Spark is a distributed computing framework which is designed for low latency and iterative computation on historical data and streaming data. Compared with Hadoop, Spark is more suitable for iterative and interactive operations for the reason that Spark has an advanced DAG execution engine that supports cyclic data flow and in-memory computing.
- 4.
GraphX is Apache Spark’s API for graphs and graph-parallel computation. It is a distributed graph processing framework and supplies rich easy interfaces for graph computing and mining. GraphX unifies ETL, exploratory analysis, and iterative graph computation within a single system. It also competes on performance with the fastest graph systems while retaining Spark’s flexibility, fault tolerance and ease of use.
4 Method
4.1 LinkSHRINK
In this paper, considering drawbacks mentioned above, we introduce LinkSHRINK which discovers different types of the communities with diverse levels of overlapping granularity via parameter \(\omega \). Some definitions and basic concepts are shown as follows:
Definition 1
(outliers) Given a network \(G = (V,E)\), where V and E denote the node and edge set in the network G, respectively. There usually are some independent nodes, which cannot be grouped into any communities. We define them as outliers: \(Outliers = \{v|v\in V,\not \exists V_i' \in CR \wedge v \in V_i' \}=V-\cup _{i=1}^kV_i'\), where CR means all the communities detected in the network G.
Definition 2
(Structural similarity) Let \(G=(V,E)\) be a unweighted undirected network. The structure neighborhood of a node u is the \(\varGamma (u)\) containing u and its adjacent nodes: \(\varGamma (u)=\{v\in V|\{u,v\}\in E\}\cup \{u\}\). The structural similarity between two adjacent nodes u and v is then
Definition 3
(Dense Pair) Give a network \(G=(V,E)\), \(\sigma (u,v)\) is the structural similarity of nodes u and v. If \(\sigma (u,v)\) is the largest similarity between nodes u, v and their adjacent neighbor nodes: \(\sigma (u,v)=max\{\sigma (x,y)|(x=u,y\in \varGamma (u)-\{u\})\vee (x=v,y\in \varGamma (v)-\{v\})\}\), then \(\{u,v\}\) is called a dense pair in G, denoted by \(u\leftrightarrow _{\varepsilon }v\), where \(\varepsilon =\sigma (u,v)\) is the density of pair \(\{u,v\}\).
Definition 4
(micro-community) Given a network \(G = (V,E)\), \(C(a) = (V',E',\varepsilon )\) is a connected sub-graph of G represented by node a. C(a) is a local micro-community iff (1) \(a\in V'\); (2) for all \( u\in V'\),\(\exists v \in V' (u \leftrightarrow _{\varepsilon } v)\); (3) \(\not \exists u\in V (u \leftrightarrow _{\varepsilon } v \wedge u\in V'\wedge v \not \in V' )\), which \(\varepsilon \) is density of the micro-community C(a) and \(u \leftrightarrow _{\varepsilon } v\) means similarity \(\varepsilon \) is the largest similarity between nodes u, v and their adjacent neighbor nodes.
Definition 5
(community overlap degree) Let set of clusters \(CR = \{C_1,C_2,\ldots ,C_k\}\) be the communities found from the network, where \(C_i\) is a set of nodes classified into the same community i. The community overlap degree \(\vartheta \) between community \(C_i\) and community \(C_j\) can be computed by formula (3), which represents the degree of overlap between two communities.
where \(C_i\cap C_j\) denotes the length of the intersection between community \(C_i\) and community \(C_j\), and \({\mathrm{min}}(|C_i|,|C_j|)\) denotes the smaller length between community \(C_i\) and community \(C_j\).
Definition 6
(community connection) Given a network \(G = (V,E)\), let \(C_1\) connect to \(C_2\) iff \(\exists e(u,v)\in E \wedge u \in C_1 \wedge v \in C_2\), where \(C_1,C_2 \in CR\).
The overall framework of LinkSHRINK is shown as in Algorithm 1. It consists of four steps, (1) generating the link graph (line 1); (2) detecting link communities (line 3); (3) transforming the link community to the node community (lines 6–12); and (4) merging communities (line 13).

4.1.1 Link-Graph transformation
Given a graph \(G = (V,E)\), where \(V = \{v_1,v_2,\ldots ,v_n\}\) represents the node set and \(E = \{e_1,e_2,\ldots ,e_m\}\) represents the edge set. Among edges, \(e = (u,v)\) represents an edge induced by two nodes u and v. Edge e(u, v) in the original graph is corresponding to node \(v_e(u,v)\) in the link graph. There is an edge between two nodes in link graph only if their corresponding two edges in original graph are induced by a common node. Figure 3 shows the transformation from an original graph to a link graph. In Fig. 3, edge (1,2) in original graph is transformed to node 1–2 in link graph. Nodes 1–2 and 2–4 in link graph are linked because edges (1,2) and (2,4) in original graph are induced by the common node 2.
4.1.2 Algorithm based on modularity and hierarchical clustering
Algorithm 2 shows the procedure of the clustering on the link graph. This procedure is based on the framework of method SHRINK (Huang et al. 2010), and we make some changes.

Initially, each node is considered as an independent micro-community. Then in lines 4–20, the candidate micro-community C(i) is obtained, where similarity \(\sigma \) between them is the largest similarity for them and their adjacent neighbors. After that, in lines 22–27, we use the similarity-based modularity function \(Q_s\) proposed by Feng et al. (2007) to determine whether those candidate micro-communities can be merged. \(\varDelta Q_s\) can be calculated by formulas (4) and (5) where \(US_{i,j}=\sum _{u\in C_i,v\in C_j}\sigma (u,v)\) is the total similarities of the links between two communities, \(DS_i=\sum _{u\in C_i,v\in V}\sigma (u,v)\) is the total similarities between nodes in cluster \(C_i\) and any node in the network, and \(TS=\sum _{u,v\in V} \sigma (u,v)\) is the total similarities between any two nodes in the network.
If \(\varDelta Q_s(C) >0\), nodes in set C will be merged into a new micro-community MC represented by any node in set C, called super-node v. All other nodes in set C will be ignored, and edges induced by them link to the representative super-node v. Continue this procedure until there are no micro-communities available to be merged. The procedure of Algorithm 2 is shown in Fig. 4.
4.1.3 Merge community
After the procedure of clustering on the link graph, we obtain the link partitions which will be transformed into node communities. Actually, node communities obtained at this step always tend to have high overlapping degree. To find communities with different overlapping granularity, we need to merge communities based on the community overlap degree \(\vartheta \). The main idea of this step is combining two connected communities \(C_x\), \(C_y\) with \(\vartheta (C_x,C_y )\ge \omega \) where \(\omega \) denotes threshold value. It is noticed that the set of clusters \(NC = \{ C_1,C_2,\ldots ,C_k \}\) actually has links between communities. There exists an edge between cluster \(C_i\) and \(C_j\) if there exists edges or an edge between nodes in \(C_i\) and \(C_j\). Therefore, after a round of the merging, we need adjust the edge structure in clusters NC.
With varying the parameter \(\omega \), we can find different kinds of the overlapping communities with different overlapping granularity. For example in Fig. 5, our method can find one overlapping node 7 in the toy network which LINK can also do. However, LinkSHRINK also finds the outlier node 14 which belongs to none of the clusters and LINK cannot do. In Fig. 5b, we find four clusters with the overlapping nodes 1, 4, 5, 7, 8, 11, 12. It can be seen that connections between nodes in cluster in Fig. 5a are relatively more sparse than connection in cluster in Fig. 5b. Hence, Fig. 5b shows the overlapping granularity of the community partition is more precise compared with Fig. 5a. Two types of the overlapping community results shown in Fig. 5 seem to be reasonable both. However, most overlapping community detection methods can only find the result in Fig. 5a. In addition, we also show our algorithm LinkSHRINK can find reasonable clusters on other toy networks in Fig. 6.
For the network in Fig. 2, edge e(6, 7) is considered as a outlier which cannot belong to any cluster in link graph by our method. Accordingly, we find the overlapping communities solving the excessive overlapping problem which is shown in Fig. 7.
Community detection result of the network in Fig. 2 by LinkSHRINK
4.1.4 Running time complexity
Finally, we analyze the computational complexity of LinkSHRINK. Let n, \(n'\), m and \(m'\) denote the number of nodes and edges in the original graph and link graph, respectively. Let k denote the number of communities. Transforming original graph to link graph requires \(O(mm')\). After forming the link graph, finding micro-communities using Algorithm 2 requires \(O(m'\log n')\), where \(\log n'\) denotes the number of iteration in Algorithm 2. Transforming link communities to node communities needs \(O(n')\) computations. Merging communities requires \(O(k^2)\) which k is far smaller than n. Overall, LinkSHRINK requires \(O(mm'+m'\log n'+n'+k^2)\) computations.
4.2 LinkSHRINK based on parallel computing framework
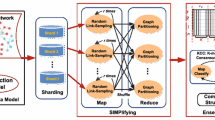
For a node x in original graph having l edges with other nodes, it will generate \((l* (l - 1))/2\) new edges in Link-Graph. Therefore, the size of the Link-Graph is far larger than the original graph. LinkSHRINK is hard to handle the large networks efficiently due to its time complexity. To solve this problem, we first sample on the generated Link-Graph. Then we parallelize the LinkSHRINK based on Hadoop and Spark, called MLinkSHRINK and PLinkSHRINK, respectively.
4.2.1 Sample
Sampling can reduce the running time. Let \(LG=(V',E')\) be a Link-Graph, for each node \(v \in V'\), we take a random sample of size \(n_v\) from the set of incident links of v. It is important to choose size \(n_v\). Formula (6) shows the calculation of \(n_v\):
where \(d_v\) is the degree of node v. For parameter \(\alpha \) and \(\beta \), they are set to 2\(<d_v>\) or \(<d_v>\) and 1 respectively which perform well on all data sets in Sect. 5.
4.2.2 PLinkSHRINK
For spark, we implement the PLinkSHRINK by using GraphX. There are two main structures in GraphX: NodeRDD[VD], EdgeRDD[ED], where VD and ED are the attribute of Node and edge, respectively. Then a graph G can be created by NodeRDD and EdgeRDD just like Graph[VD,ED].

The main process of PLinkSHRINK as shown in Algorithm 3 is on the whole the same as LinkSHRINK. The main difference between them is that LinkSHRINK merges one micro-community candidate at each iteration, while PLinkSHRINK combines all micro-community candidates at each iteration by two leftOuterJoin Spark RDD actions which saves a lot of time. Figure 8 shows how the two leftOuterJoin actions work, where nodes 1 and 2 need to be merged and so are the nodes 4 and 5.
Note that, when we calculate similarity for Link-Graph (lines 3–10), the original graph \(G'\) is the input graph with new edges which connecting two nodes having at lest one common neighbor. In lines 27–29, new Link-Graph is generated with initialing node IS value. Here, we split \(DS_i\) value into two parts, one is \(IS_i\) and the other is \(OS_i\) where \(IS_i\) is total similarities of nodes in micro-community \(S_i\), \(OS_i\) is total similarities of nodes in micro-community \(S_i\) and any other nodes out of the micro-community \(S_i\). The relationship between them shows as formula (7).
For large networks, such as network with millions nodes, because of the limit of the leftOuterJoin action, the time of each iteration increases. However, in the last several iterations, there are only very few nodes that need to be merged. For saving running time, we ignore these left nodes which will not have a significant impact on the results. Therefore, we set a threshold \(\partial \). PLinkSHRINK stops when the proportion of nodes to be merged in Link-Graph is smaller than threshold \(\partial \). Here, the set of \(\partial \) is a trade-off between the effectiveness and efficiency. Given a small \(\partial \) such as \(10^{-4}\), PLinkSHRINK cannot early stop in a small network which means PLinkSHRINK stops only if there is no node left to be merged. However, PLinkSHRINK early stops when the proportion of nodes to be merged in Link-Graph is smaller than \(\partial \) in a large network.
4.2.3 MLinkSHRINK
Next, we will introduce the implementation of the parallel LinkSHRINK algorithm, MLinkSHRINK. MLinkSHRINK consists of 6 jobs based on MapReduce.
Job1: Calculation of Link-Graph’s Similarity
Map Phase The mapper takes a pair of (key, value) as input, where key is the node input and value is its adjacency node list. The node in the Link-Graph corresponds to an edge in the original graph. The input node in the Link-Graph consists of the id of the adjacent nodes. To illustrate MLinkSHRINK, we take the node and its adjacency node list \(< v : v_1,v_2,\ldots ,v_s>\) as an example. Note that one edge is represented by using the node pair in an increasing order. For each neighbor node \(v_i (i = 1,2,\ldots ,s)\) of v, the mapper emits a (key, value) pair, in which key is the edge(\(v, v_i\)) and value is the adjacency node list of v.
Reduce Phase In the reducer, for the key \((v,v_i)\), the corresponding values will include the adjacency node lists of v and \(v_i\). The two adjacency node lists consist of all the information needed to calculate the similarity of the edge, including the adjacency information of two adjacent nodes in original graph. Therefore, we can calculate the similarity of \((v, v_i)\) in the reducer.
Job2: Adding the Similarity of the Edge to the Adjacency Nodes List of the Two Adjacent Nodes.
Map phase The mapper takes a pair of (key, value) as input, where the key is the input edge and the value is its similarity. For each adjacent node of the edge, the mapper emits a (key, value) pair, in which the key is node and the value is the joint of the other node and the similarity.
Reduce phase The reducer combines all the adjacent information of every node.
Job3: Calculation of the Sum of the Similarity
Map phase The mapper calculates the sum of the similarity of every node.
Reduce phase The reducer calculates the sum of all the similarity.
After the above preparation, MLinkSHRINK steps into the stage of iterations. In the process of each iteration, MLinkSHRINK needs three jobs which are running in sequence until the algorithm stops.
Job4: Finding the nodes to be merged
Map phase The mapper takes a pair of (key, value) as input, where the key is input node and the value is the adjacent node list which contains the information of adjacent nodes and the similarity of the corresponding edge. The mapper emits a (key, value) pair, in which the key is the edge with the largest similarity and the value consists of the set of the cohesion of node, the overall similarity of the node and the similarity of the edge.
Reduce phase If the size of the received values of the edge is two, two adjacent nodes, respectively, select the edge in the map phase. Then the edge can be regarded as a candidate edge to be cut. The reducer gets the information of two nodes and the edge. After the calculation of \(\varDelta Q\). If \(\varDelta Q\) is positive, reducer outputs the edge. Otherwise, the edge is cut in the next job.
Job5: Reconstruction of the Graph
In this step, MLinkSHRINK needs to get the information of the edge to be cut, which can be obtained by job 4 in advance. Then, the information is saved in hash table for quickly query.
Map phase The mapper takes a pair of (key, value) as input, where the key is the input node and the value is the adjacent node list. Firstly, mapper checks whether this node is in the hash table. If not, we make this node as the key to be emitted. Otherwise, some adjacent edges of this node are to be cut. The mapper gets the information of the adjacent node from the hash table. If this node id is smaller than the adjacent node, then we make this node as the key. And smaller one will be the representative node. This edge can be cut off. After that, for each adjacent node in the adjacent list, the reducer checks whether the adjacent node is in the hash table. If not, mapper emits a (key, value) pair, in which the value is the joint of the adjacent node and the similarity of the edge. Otherwise, we get the adjacent node of the adjacent node from hash table. Choose the smaller one as the value and combine with the similarity of the edge as the value to be emitted.
Reduce phase The reducer combines all the information of adjacent nodes for this node and updates the similarity of the edges. The reducer emits a (key, value) pair, in which the key is node and the value is the combined adjacent list. After this phase, we achieved the goal of reconstruction of the graph.
Job6: Updating the Partition of the Community in the Link-Graph
Map Phase First communities are obtained in previous iteration. The mapper takes a pair of (key, value) as input, where the key is the representative node of the community and the value is the members of the community. Then we check whether the representative node is in hash table. If not, the mapper emits a (key, value) pair, in which the key is the representative node and the value is the members of the community. Otherwise, some edges of the representative node need to be removed. If the representative node id is smaller than the adjacent nodes obtained from hash table, the mapper emits a (key, value) pair, in which the key is the representative node and the value is the members of the community. Otherwise, the mapper emits a (key, value) pair, in which the key is adjacent node and the value is joint of the representative node and the members of the community of the representative node.
Reduce Phase The reducer takes a pair of (key, values) as input. The key is set to be the new representative node and the combination of information in values to be the members of the community.
Note that jobs 4, 5 and 6 are repeated until the output of the job 4 is empty, which means none of the edges are to be cut. Finally, another job transforms the community structure of link partition to the node communities.
Both PLinkSHRINK and MLinkSHRINK can find the same communities as LinkSHRINK does in the toy network in Fig. 4.
5 Experiment
In this section, we evaluate the performance of our proposed algorithm LinkSHRINK using both synthetic benchmarks and real-world networks. LinkSHRINK is implemented in C++. We compare LinkSHRINK with some other state-of-the-art overlapping community detection algorithms which are listed as follows:
- 1.
The CPM (Clique Percolation Method) (Palla et al. 2005)Footnote 2
- 2.
The COPRA (Community Overlap Propagation Algorithm) (Gregory 2010)Footnote 3
- 3.
The link partition method: LINK (Ahn et al. 2010)Footnote 4
- 4.
The link partition method: LINK1. Here, we do a little change on the LINK algorithm. We simply delete the link community which only contains a single edge founded by LINK.
- 5.
The SHRINKO (Huang et al. 2011): SHRINKO transformed from SHRINK simply by recognizing the hubs in the network as the overlapping nodes.
- 6.
The OCDDP (Bai et al. 2017): OCDDP detects overlapping communities based on density peaks. It adopts a similarity-based method to set distances among nodes, a three-step process to select cores of communities and membership vectors to represent belongings of nodes.
- 7.
The GraphSAGE (Hamilton et al. 2017): GraphSAGE is a network embedding approach which maps the nodes in network into low-dimension vector, and then we adopt fuzzy C-means method (Bezdek et al. 1984) to detect overlapping communities.
- 8.
The PBigClam (Thang 2017): PBigClam is a parallel version of BigClam (Yang and Leskovec 2013) which detects overlapping communities by using nonnegative matrix factorization.
All stand-alone experiments are conducted on a 2.66-GHz and 8-GB RAM Pentium IV computer.
For PLinkSHRINK and MLinkSHRINK, we conduct experiments on some small networks to verify the correctness of the results compared with LinkSHRINK. Then we also conduct experiments on some larger networks to compare time performance among them.
We deploy PLinkSHRINK and MLinkSHRINK on a cluster of five nodes as DataNodes and NodeManagers, within which one node acts as both NameNode and ResourceManager. The hardware specific of each node is Intel Xeon E5-2620 v2 CPU, 64 GB main memory. Hadoop version is 2.6.0, while Spark version is 1.5.1. Spark is operating in conjunction with Hadoop in the scheme of ON YARN.
5.1 Synthetic networks
-
1.
Data Sets For synthetic networks, we adopt the LFR benchmark networks (Lancichinetti and Fortunato 2009). Some important parameters of the benchmark network are shown in Table 1.
-
2.
Evaluation Metric The ground-truth community is given in the synthetic network. Hence, we use an extended version of the normalized mutual information (NMI) for overlapping community detection proposed by Lancichinetti et al. (2009). So far, NMI is the most famous and widely used to evaluate the result of the community detection on the networks with the ground-truth community. The larger NMI is, the better cluster result is. The value of NMI is from 0 to 1. NMI equal to 1 means two partitions of the network are identical and equal to 0 on the contrary.
$$\begin{aligned} {\rm NMI}(X|Y)=1-[H(X|Y)+H(Y|X)]/2 \end{aligned}$$(8)NMI can be computed by formula (8), where X and Y denote the cluster partitions and H(X|Y) represents the normalized conditional entropy of a cover X with respect to Y which is shown in formula (9). Here, C represents the real community partition of a network.
$$\begin{aligned} H(X|Y)=\frac{1}{|C|}\sum _k\frac{H(X_k|Y)}{H(X_k)} \end{aligned}$$(9) -
3.
Parameters Set The main parameters of LFR benchmark networks are shown in Table 2.
Here, for S1, S2 and S4, S5, parameter on is set to 100 and 300 which represent networks with less and more overlapping nodes, respectively. For S1 and S3, parameter mu is set to 0.1 and 0.3 which represent networks with low and high mixing degree, respectively. S6 represents networks with varying degree k .
The parameters of each algorithm are set as follows: k in CPM is set to 3–8, v in COPRA is set to 2–10, \(\omega \) in LinkSHRINK is set to 0.5–1. We always adopt the best experimental results of every method. Here LinkSHRINK does not consider the useless small communities, and we can simply delete the community in which the number of nodes is smaller than the \({\mathrm{min}}_c\). Figure 9 shows the community detection results on six groups of LFR benchmark networks by using eight algorithms mentioned above, respectively. It is noticed that LS, CPM, GS, L, L1, COPRA, SO, OCDDP represent LinkSHRINK, CPM, GraphSAGE, LINK, LINK1, COPRA, SHRINKO, OCDDP methods, respectively.
- 1.
Compared with LINK and LINK1 On every LFR benchmark network, our algorithm performs better than LINK and LINK1 do. This is because our algorithm is more reasonable to deal with the isolated edges in networks.
- 2.
Compared with COPRA It can be seen, the NMI of the community detection result by our algorithm is better than COPRA almost on all the LFR benchmark networks except some networks such as S6 with the \(k=12, 15\) and S1 with the \({\rm om}=6\). With the increase in om, finding overlapping community becomes more difficult and the NMI of these networks by our algorithm showing a downward trend which is reasonable. However, results of COPRA have a vibration trend for the reason that the result of COPRA is nondeterministic.
- 3.
Compared with CPM The NMI of our algorithm is better than that of CPM on all of the LFR benchmark networks.
- 4.
Compared with SHRINKO As the COPRA, our algorithm performs better than SHRINKO on most of the LFR benchmark networks except the networks S1 with the \({\rm om}=6\) and S3 with the \({\rm om}=6\). However, SHRINKO comes to the problem of the having vibration trend obviously what we mentioned in Sect. 2.
- 5.
Compared with OCDDP Overall, OCDDP almost has the same performance with LinkSHRINK. LinkSHRINK only obtains 15 of 30 better results than OCDDP does.
- 6.
Compared with GraphSAGE The NMI of our algorithm is better than that of GraphSAGE on most of the LFR benchmark networks except the networks S2 with \({\rm om}=5,6\).
As shown above, LinkSHRINK can find better overlapping community structure on synthetic networks compared with some others mentioned above.
To evaluate sampling on Link-Graph, we conduct experiments on some larger synthetic networks whose detail is shown in Table 3. Here, we process LinkSHRINK on every data set which has two versions: one is generating Link-Graph with sampling, and the other is generating Link-Graph without sampling. The results are shown in Figs. 10 and 11.
It can be seen that sampling on the data set has nonsignificant impact on the performance. However, its running time reduces by about 30 percents and the level of the time reduction is proportional to the size of the data.
5.2 Real-world networks
-
1.
Data Sets
For real-world networks, we choose four very classic and commonly used community evaluation data sets whose details are shown in Table 4:
-
2.
Evaluation Metrics
Due to the unknown ground truth in real-world networks, we cannot adopt NMI to evaluate the effectiveness of the community detection methods. Accordingly, we use extended modularity \(Q_{ov}^E\) (Shen et al. 2009) for overlapping community detection which uses the number of communities to which a node belongs as a weight for Q as shown in formula (10).
$$\begin{aligned} Q_{ov}^E= \frac{1}{2m}\sum _c{\sum _{i,j\in c}{\left[ A_{ij}-\frac{k_ik_j}{2m}\right] \frac{1}{O_iO_j}}} \end{aligned}$$(10)where \(O_i\) denotes the number of communities to which node i belongs. \(A_{ij}\) is equal to 1 if there is an edge between nodes i and j, otherwise 0. \(k_i\) denotes the degree of the node i. m denotes the number of all edges in the network and c denotes a community.
Figure 12 shows a comparison between our three algorithms and other six methods: COPRA, SHRINKO, LINK, OCDDP, GraphSAGE and PBigClam. All the parameters set are the same as the parameters set in the LFR benchmark. From the results, we can see that the performances of our three approaches are better than SHRINKO and much better than the other five methods on most networks. Besides, we can see our methods LinkSHRINK, PLinkSHRINK and MLinkSHRINK have almost the same performance on these data sets.
-
3.
Example: PDZBase Network
PDZBase network is a network of protein–protein interactions from PDZBase. The community structures in this network majorly present radiation and star-like structure. Overlapping community detection algorithms based on node-structure is more difficult to find the star-like community structure. As illustrated in Fig. 13, the LinkSHRINK algorithm can achieve good result on this network, where 19 communities as well as 1 outlier are found. Almost all communities present the shape of star which seems very reasonable. However, COPRA is difficult to find communities in this network and achieves the lowest value of \(Q_{ov}^E\) observed in Fig. 12.
-
4.
Large Network
For large real-world networks, we choose DBLP collaboration network with 317,080 nodes and 1,049,866 edges. After transforming original graph into link graph, there are about 1,049,866 nodes and 21,780,889 edges in link graph which becomes a huge network. After sampling on the original network with the parameters \(\alpha \) and \(\beta \) being equal to 1\(\times \langle d_v \rangle \) and 1, the edges in link graph descend about 67 percent which looks very effective in Fig. 14.
We run algorithms LinkSHRINK, PLinkSHRINK and MLinkSHRINK on the link graph transformed from DBLP network. Here parameter \(\partial \) for PLinkSHRINK is set to 0.0001 which is quite small. Note that LinkSHRINK cannot finish for the reason that computation resources such as CPU and memory are overflowed, while PLinkSHRINK, MLinkSHRINK and PBigClam finish in 3, 6.3 and 5.4 h, respectively, shown in Fig. 15. PLinkSHRINK finds more than 76,400 author communities whose size distribution is shown in Fig. 16.
For Spark, the running time of PLinkSHRINK correlates with the executer cores. Performance improving by increasing number of cores is shown in Fig. 17. For cases of executer cores are at least and most, the running time difference between them is close to 1 h.
Besides, we use LFR benchmark to generate five large networks from 100,000 nodes to 300,000 nodes, the detail of data set is shown as in Table 5.
Note that LinkSHRINK cannot finish on these large data sets. However, by sampling on the data sets and the distributed computing framework, we can complete overlapping community detection in a reasonable period of time, especially PLinkSHRINK. As shown in Fig. 18, the running time of MLinkSHRINK increases more strongly. PBigClam needs longer running time than PLinkSHRINK does. On the other hand, the running time of PLinkSHRINK is 80 percent lower than that of MLinkSHRINK. The reason is that LinkSHRINK is iterative which is better by Spark. For MLinkSHRINK, each iteration needs to start three jobs need more time. When there are too many iterations, that will be a big expense.
6 Conclusions
LinkSHRINK is overlapping community detection method combining density-based clustering with modularity optimization. It maximizes modularity by using density-based clustering in link graph. Finally, it finds overlapping communities by merging reductant nodes with parameter \(\omega \). LinkSHRINK not only finds overlapping communities, but also identifies the hubs and outliers. It avoids the problem of excessive overlapping problem and reveals the overlapping community structure with different overlap degrees by using parameter \(\omega \). To make LinkSHRINK handle large network, we sample the Link-Graph to improve the efficiency with losing little accuracy. Meanwhile, we implement LinkSHRINK based on Spark and Hadoop. LinkSHRINK outperforms state-of-the-art methods on the synthetic and real-world networks. PLinkSHRINK and MLinkSHRINK can also find communities in large network with millions of edges efficiently without losing accuracy much. In the future, we would like to extend LinkSHRINK to detect overlapping communities in dynamic networks and in larger networks, such as networks with millions of nodes.
Notes
This manuscript is an extended version of a previous conference publication (Yin et al. 2016).
References
Ahn YY, Bagrow JP, Lehmann S (2010) Link communities reveal multiscale complexity in networks. Nature 466(7307):761
Bai X, Yang P, Shi X (2017) An overlapping community detection algorithm based on density peaks. Neurocomputing 226:7–15
Ball B, Karrer B, Newman ME (2011) Efficient and principled method for detecting communities in networks. Phys Rev E 84(3):036103
Bezdek JC, Ehrlich R, Full W (1984) Fcm: the fuzzy c-means clustering algorithm. Comput Geosci 10(2–3):191–203
Cheong CY, Huynh HP, Lo D, Goh RSM (2013) Hierarchical parallel algorithm for modularity-based community detection using gpus. In: European conference on parallel processing, Springer, pp 775–787
Ester M, Kriegel H, Sander J, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. In: International conference on knowledge discovery and data mining, AAAI Press, pp 226–231
Evans T, Lambiotte R (2009) Line graphs, link partitions, and overlapping communities. Phys Rev E 80(1):016105
Feng Z, Xu X, Yuruk N, Schweiger TA (2007) A novel similarity-based modularity function for graph partitioning. In: International conference on data warehousing and knowledge discovery, Springer, pp 385–396
Fortunato S, Barthelemy M (2007) Resolution limit in community detection. Proc Natl Acad Sci 104(1):36–41
Gopalan PK, Blei DM (2013) Efficient discovery of overlapping communities in massive networks. Proc Natl Acad Sci 110(36):14534–14539
Gregory S (2010) Finding overlapping communities in networks by label propagation. New J Phys 12(10):103018
Hamilton W, Ying Z, Leskovec J (2017) Inductive representation learning on large graphs. In: Advances in neural information processing systems, pp 1024–1034
Huang J, Sun H, Han J, Deng H, Sun Y, Liu Y (2010) Shrink: a structural clustering algorithm for detecting hierarchical communities in networks. In: ACM international conference on information and knowledge management, ACM, pp 219–228
Huang J, Sun H, Han J, Feng B (2011) Density-based shrinkage for revealing hierarchical and overlapping community structure in networks. Phys A Stat Mech Appl 390(11):2160–2171
Jin S, Yu PS, Li S, Yang S (2015) A parallel community structure mining method in big social networks. Math Probl Eng 2015:934301
Kuzmin K, Shah SY, Szymanski BK (2013) Parallel overlapping community detection with slpa. In: 2013 international conference on social computing, IEEE, pp 204–212
Lancichinetti A, Fortunato S (2009) Benchmarks for testing community detection algorithms on directed and weighted graphs with overlapping communities. Phys Rev E 80(1):016118
Lancichinetti A, Fortunato S (2011) Limits of modularity maximization in community detection. Phys Rev E 84(6):066122
Lancichinetti A, Fortunato S, Kertész J (2009) Detecting the overlapping and hierarchical community structure in complex networks. New J Phys 11(3):033015
Li R, Guo W, Guo K, Qiu Q (2015) Parallel multi-label propagation for overlapping community detection in large-scale networks. In: International workshop on multi-disciplinary trends in artificial intelligence, Springer, pp 351–362
Li Y, He K, Kloster K, Bindel D, Hopcroft J (2018) Local spectral clustering for overlapping community detecion. ACM Trans Knowl Discov Data 12(2):17
Lim S, Ryu S, Kwon S, Jung K, Lee JG (2014) Linkscan*: overlapping community detection using the link-space transformation. In: IEEE international conference on data engineering, IEEE, pp 292–303
Moon S, Lee JG, Kang M, Choy M, Jw Lee (2016) Parallel community detection on large graphs with mapreduce and graphchi. Data Knowl Eng 104:17–31
Newman ME, Girvan M (2004) Finding and evaluating community structure in networks. Phys Rev E 69(2):026113
Palla G, Derényi I, Farkas I, Vicsek T (2005) Uncovering the overlapping community structure of complex networks in nature and society. Nature 435(7043):814
Qiao S, Guo J, Han N, Zhang X, Yuan C, Tang C (2017) Parallel algorithm for discovering communities in large-scale complex networks. Chin J Comput
Sarswat A, Jami V, Guddeti RMR (2017) A novel two-step approach for overlapping community detection in social networks. Soc Netw Anal Min 7(1):47
Shen H, Cheng X, Guo J (2009) Quantifying and identifying the overlapping community structure in networks. J Stat Mech Theory Exp 2009(07):P07042
Sun B, Shen H, Cheng X (2014) Detecting overlapping communities in massive networks. Europhys Lett 108(6):68001
Thang ND (2017) Community detection in large-scale networks. Master’s thesis, ThangLong University
Wang S, Dong Y, Li Z, Chen H, Qian J (2015) The identification of overlapping communities in large-scale complex networks. Acta Electron Sin 43(8):1575–1581
Wickramaarachchi C, Frincu M, Small P, Prasanna VK (2014) Fast parallel algorithm for unfolding of communities in large graphs. In: 2014 IEEE high performance extreme computing conference, IEEE, pp 1–6
Xie J, Szymanski BK (2012) Towards linear time overlapping community detection in social networks. In: Pacific-Asia conference on knowledge discovery and data mining, Springer, pp 25–36
Xu X, Yuruk N, Feng Z, Schweiger TA (2007) Scan: a structural clustering algorithm for networks. In: ACM SIGKDD international conference on Knowledge discovery and data mining, ACM, pp 824–833
Yang J, Leskovec J (2013) Overlapping community detection at scale: a nonnegative matrix factorization approach. In: Proceedings of the sixth ACM international conference on Web search and data mining, ACM, pp 587–596
Yin D, Wu B, Zhang Y (2016) Linkshrink: overlapping community detection with link-graph. In: IEEE international conference on data science in cyberspace, IEEE, pp 44–53
Zeng J, Yu H (2015) Parallel modularity-based community detection on large-scale graphs. In: 2015 IEEE international conference on cluster computing, IEEE, pp 1–10
Zhang H, Niu X, King I, Lyu MR (2018) Overlapping community detection with preference and locality information: a non-negative matrix factorization approach. Soc Netw Anal Min 8(1):43
Zhang Q, Qiu Q, Guo W, Guo K, Xiong N (2016) A social community detection algorithm based on parallel grey label propagation. Comput Netw 107:133–143
Zhu M, Meng F, Zhou Y (2013) Density-based link clustering algorithm for overlapping community detection. J Comput Res Dev 12(006)
Acknowledgements
This work is supported by the National Key R&D Program of China under Grant 2018YFC0831500. We are grateful to the anonymous reviewers for their careful reading and valuable suggestions.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, Y., Yin, D., Wu, B. et al. PLinkSHRINK: a parallel overlapping community detection algorithm with Link-Graph for large networks. Soc. Netw. Anal. Min. 9, 66 (2019). https://doi.org/10.1007/s13278-019-0609-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13278-019-0609-3