Abstract
Due to the application of low-dose computed tomography screening, more and more early-stage lung cancers have been diagnosed. Thus, it is essential to characterize the gene expression profile of early-stage lung cancer to develop potential biomarkers for early diagnosis and therapeutic targets. Here, we analyzed microarray data of 181 early-stage lung cancer patients. By comparing gene expression between different tumor and lymph node metastasis stages, we identified various differentially expressed protein-coding genes and long noncoding RNA (lncRNA) in the comparisons of T2 vs. T2 and N1- vs. N0-stage lung cancer. Functional analyses revealed that these differentially expressed genes were enriched in various tumorigenesis or metastasis-related pathways. Survival analysis indicated that two protein-coding genes, C7 and SCN7A, were significantly associated survival of lung cancer. Notably, a novel lncRNA, LINC00313, was highly expressed in both T2- and N1-stage lung cancers. On the other hand, LINC00313 was also upregulated in lung cancer and metastasized lung cancer tissues, compared with adjacent lung tissues and primary lung cancer tissues. Additionally, higher expression level of LINC00313 indicated poor prognosis of lung cancer (hazard ratio = 0.658). Overall, we characterized the expression profiles of protein-coding genes and lncRNA in early-stage lung cancer and found that LINC00313 could be a biomarker for lung cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lung cancer is the leading cause of cancer-related death [1]. According to the randomized controlled National Lung Screening Trial (NLST), the lung cancer-related mortality and overall mortality have decreased due to the application of low-dose computed tomography (CT) instead of chest radiography [2]. With the application of low-dose CT screening, more and more lung cancer cases are diagnosed at early stage. For patients with early-stage lung cancer detected by screening, concerns have raised about the appropriate treatment for these patients [3–5]. Therefore, it is important to characterize the molecular basis and understand the altered gene expression in early-stage lung cancer. Additionally, prognostic biomarkers and makers that are predictive of metastasis and benefit from chemotherapy are needed for early-stage lung cancer [6, 7]. However, few studies have focused on these issues.
Gene expression microarray is a feasible and effective approach to characterize gene expression profile and searching messenger RNA (mRNA)-based biomarkers. For lung cancer, microarrays have been widely used, which provide abundant resource for data mining [8–10]. Due to the advance of high-throughput technology, evidence has demonstrated that long noncoding RNA (lncRNA) is actively transcribed from human genome and plays an important role in all aspects of tumor biology [11–13]. Compared with protein-coding genes, lncRNA exhibits stronger cell-type specific expression manner [14], suggesting that lncRNA could be a potential biomarker [15]. Given that a number of probe sets were matched with lncRNA, reannotation of published microarray data and analyzed lncRNA expression profile is a feasible and widely used method [16–18].
In this study, we performed data mining of the GSE50081 dataset [9], which includes gene expression data of 181 early-stage lung cancer patients. We compared the protein-coding gene and lncRNA expression profiles between different tumor and lymph node stages and performed functional annotation of the differentially expressed protein-coding genes. SCN7A, C7, and LINC00313 were associated survival of lung cancer.
Methods and materials
Dataset and calculation of differentially expressed genes
Gene Expression Omnibus is a public online database that has various high-throughput data, including microarray. In the present study, we selected the GSE50081 dataset for further data mining [9]. The GSE50081 dataset consists of microarray data of 181 lung cancer patients, including TNM stages and survival data. The Affymetrix Human Genome U133 Plus 2.0 Array, which is widely used in various research areas, was utilized in the GSE50081 data set. For microarray data analysis, the processed series matrix file was first downloaded. Since the series matrix data has already been background subtracted and normalized by RMA method, the data was subjected to differentially expressed gene detection. The differentially expressed genes were calculated by the Limma algorithm [19], and P value < 0.05 was considered as significant. RNA sequencing data of lung cancer from The Cancer Genome Atlas (TCGA) were accessed through the website lncRNAtor (http://lncrnator.ewha.ac.kr/).
Probe set annotation
Sequences of lncRNA were downloaded from the LNCipedia (http://www.lncipedia.org/) and 79,586 lncRNA larger than 200 nt were downloaded. Sequences of Affymetrix Human Genome U133 Plus 2.0 Array probe set were downloaded from the Affymetrix website. The probe sets were reannotated by Blast software, and 12,156 lncRNA completely matched with probe sets were identified.
Gene Ontology and KEGG pathway analysis
Gene Ontology (GO) analysis was applied to analyze the main function of the differential expression genes according to the Gene Ontology (www.geneontology.org), which can organize genes into hierarchical categories and uncover the gene regulatory network on the basis of biological process and molecular function [20, 21]. Specifically, two-side Fisher’s exact test and X 2 test were used to classify the GO category, and the false discovery rate (FDR) [22] was calculated to correct the P value; the smaller the FDR, the smaller the error in judging the P value. The FDR was defined as \( FDR=1-\frac{N_k}{T} \) where N k refers to the number of Fisher’s test P values less than X 2 test P values. We computed P values for the GOs of all the differential genes. Enrichment provides a measure of the significance of the function: as the enrichment increases, the corresponding function is more specific, which helps us to find those GOs with more concrete function description in the experiment. Within the significant category, the enrichment Re was given by Re = (n f /n)/(N f /N) where “n f ” is the number of flagged genes within the particular category, “n” is the total number of genes within the same category, “N f ” is the number of flagged genes in the entire microarray, and “N” is the total number of genes in the microarray [23].
Pathway analysis was used to find out the significant pathway of the differential genes according to Kyoto Encyclopedia of Genes and Genomes (KEGG). Still, we turn to Fisher’s exact test and X 2 test to select the significant pathway, and the threshold of significance was defined by P value and FDR. The enrichment Re was calculated like the equation above [24–26]. KEGG pathway analysis allowed us to determine the biological pathways for which a significant enrichment of differentially expressed mRNAs existed (P < 0.05 was considered statistically significant).
Statistical analysis
Kaplan-Meier survival and univariate Cox proportional hazards regression analyses were conducted to explore the prognostic value of differentially expressed coding genes or lncRNA. According to the median expression value of a specific target gene, patients were classified as “high expression” or “low expression”, and survival analysis was conducted between the two groups. LINC00313 expression level between lung cancer tissues and adjacent lung tissues, primary lung cancer tissues, and metastasized lung cancer tissues were calculated by Student’s t test, and P < 0.05 was statistically significant. All statistical analyses were performed with SPSS software (version 18.0, SPSS Inc.).
Results
Differential expression profile between T2 and T1 stages
We first analyzed the gene expression among different tumor stages. Compared with T1-stage lung cancer, there were 94 protein-coding genes upregulated and 228 genes downregulated in T2-stage lung cancer. KEGG analysis showed that the differentially expressed genes were significantly enriched in the pathways of “cell cycle”, “p53 signaling pathway”, “pathways in cancer”, and other cancer-related pathways (Fig. 1a). For lncRNA, we found that 238 lncRNAs were upregulated and 217 were downregulated in T2-stage lung cancer (with the threshold P < 0.01). The top differentially expressed genes were shown in Tables 1 and 2, and the full lists were provided in the supplementary materials.
Functional analysis of differentially expressed genes between different tumor and lymph node stages. KEGG analysis for differentially expressed protein-coding genes between T2- and T1-stage lung cancer (a). Gene Ontology (biological process) analysis of differentially expressed protein-coding genes between N1- and N0-stage lung cancer (b). The items with P < 0.05 were considered as significantly enriched. The top enriched items and enrichment score were shown
Differential expression profile between N1 and N0 stages
Lymph node metastasis is an important prognostic factor for NSCLC. By comparing patients with and without lymph node metastasis, we found that 47 protein-coding genes were upregulated and 163 were downregulated in N1-stage patients compared with N0-stage patients. On the other hand, 210 lncRNA were upregulated and 81 were downregulated in N1-stage patients. Functional GO annotation analysis showed that the altered genes were associated with “cell migration”, “localization of cell”, “cell motility”, “cell motion”, and other metastasis-related biological processes (Fig. 1b) while there were 210 lncRNAs upregulated and 81 downregulated in N1-stage lung cancer (with the threshold P < 0.01). The top differentially expressed genes were shown in Tables 3 and 4, and the full list was provided in the supplementary materials.
Survival analysis
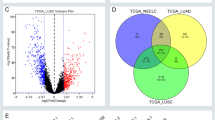
Given that all samples included in the GSE50081 dataset were lung cancer tissues, it is unknown whether these genes show a differential expression pattern between lung cancer tissues and normal lung tissues. Thus, we used a list of differentially expressed genes as validation set, which were differentially expressed between lung cancer tissues and corresponding adjacent tissues (these genes were identified by microarrays in our unpublished work, GSE66654). Using the differentially expressed protein-coding genes between T and N stages as two independent training sets, Venny plot revealed that 11 genes were common in the three groups (Table 5 and Supplementary Figure S5). As shown in Table 3, the 11 genes were not overlapped with the gene signature in the original study. To test the potentially prognostic role of these 11 genes, we analyzed whether they were associated with the survival of lung cancer patients. Kaplan-Meier curve and Cox regression were performed for the 11 genes, and results indicated that the expression level of two genes, SCN7A and C7, were significantly associated with the survival of lung cancer patients (Fig. 2). In addition, the predictive efficacy was improved with the combination of C7 and SCN7A (Fig. 2).
Given the specific expression nature of lncRNA, we assessed whether the differentially expressed lncRNAs could be predictive biomarkers of survival or metastasis. First, the top differentially expressed lncRNAs between different T and N stages were validated with TCGA RNA-seq data sets (tumor vs. normal and metastasis tumor vs. primary tumor by the lncRNAtor website). Notably, we found a novel lncRNA; LINC00313 was highly expressed both in lung cancer tissues and metastasized lung cancer tissues (Fig. 3). Additionally, LINC00313 expression was predictive of lung cancer survival, namely lung cancer patients with higher expression level of LINC00313 would have a shorter overall survival (hazard ratio = 0.658, Fig. 3).
Discussion
Due to the wide application of low-dose CT screening, more and more lung cancer patients are diagnosed at early stage. However, there are many debates about the primary treatment options for early-stage lung cancer [4, 27]. Nevertheless, it is paramount important to characterize the altered gene expression profile and identify biomarkers predictive of survival or chemotherapy, which will help understand molecular feature of early-stage lung cancer. To date, researchers have developed several mRNA-based biomarkers by microarray, and several gene signatures have been confirmed effective as prognostic biomarkers [7, 9, 28]. In these studies, the large amount of microarray data offer valuable source for data mining.
In the present study, we utilized a microarray series of 181 lung cancer patients and compared gene expression profiles between different tumor and lymph node stages. Comparing the expression data of patients with and without lymph node metastasis, we found 210 differentially expressed genes, and functional GO enrichment suggest that the differentially expressed genes were enriched in the biological processes of “cell adhererion cell motily”, which were closely associated with invasion and metastasis of cancer. By comparing data of T2- and T1-stage patient, we found 322 differentially expressed genes. Functional annotation analysis revealed that many cancer-related pathways were enriched among the differentially expressed genes, such as “Cell adhesion molecules (CAMs)”, “Cell cycle”, “p53 signaling pathway”, “Pathways in cancer”, and “Tight junction”. These results suggested that gene expression profiles were different among patients with different tumor and lymph node stages. KEGG pathway analysis and GO analysis are mostly used and powerful data mining tools. By KEGG and GO analyses, we found and revealed the altered pathways in different stages of lung cancer. Our work may help understand the molecular basis of lung cancer.
Since the dataset analyzed included only lung cancer patients, the expression profile of these genes between normal lung tissues and lung cancer tissues is unknown. Thus, we used a gene list of differentially expressed genes between lung cancer and normal lung tissues (the data were from our unpublished work) as a validate gene set. Using Venny plot to select genes which were common in three groups, 11 genes were identified. By cox regression, we found that the two genes (SCN7A and C7) were significantly associated with survival of lung cancer patients. The original data set was designed to validate the prognostic value and predictive efficacy of a 15-gene signature. But, C7 and SCN7A were not included in the 15-gene signature [9]. Additional literature review was performed for C7 and SCN7A, and reports about SCN7A and C7 in the paradigm of cancer research were few. This indicated that the C7 and SCN7A are potential novel prognostic biomarkers of lung cancer, and they may play an important role in lung cancer. However, further studies are warranted to validate our results.
Due to rapid development of high-throughput transcriptome, accumulating evidence suggests that at least 90% of the total mammalian genome is actively transcribed while only less than 2% of the genome sequence is protein-coding genes [29]. And numerous noncoding RNAs are transcribed from genome, of which microRNAs (miRNA) and lncRNA are mostly investigated [30, 31]. It is widely known that lncRNAs play an important role in cancer, such as the process of carcinogenesis, invasion, and metastasis of cancer [13]. Dysregulation of lncRNA has been found in many types of cancer, like breast cancer [32], prostate cancer [33], and lung cancer [34]. Although several genome-wide transcriptome studies have identified a lot of lncRNAs, only a small proportion of lncRNAs has been well characterized. The functional role and molecular mechanism of several cancer-associated lncRNAs have been well characterized. Additionally, it was also found that these cancer-associated lncRNAs could be potential biomarkers, as the dysregulated expression was associated with clinicopathological characteristics, even prognosis.
In current study, we re-annotated the probe set of Human Genome U133 Plus 2.0 microarray using the Lincpedia database. Among the differentially expressed lncRNAs, we noted that a novel lncRNA, LINC00313, which was upregulated both in T2- and N1-stage lung cancer and could be a prognostic biomarker of lung cancer. In addition, expression level of LINC00313 was also analyzed using TCGA RNA sequencing data. In consistence, LINC00313 was highly expressed in lung cancer tissues compared with normal tissues. Intriguingly, compared with primary lung cancer, the expression level of LINC00313 was higher in metastasized lung cancer tissues, which was in accordance with the high expression level in N1 stage. These findings confirmed that LINC00313 could be a potential biomarker for lung cancer while further in vitro studies are warranted to clarify the underlying molecular mechanism. Many functional lncRNAs have been characterized in lung cancer, and several of them were associated with prognosis or other clinical characteristics. By data mining of the dataset, we identified a set of differentially expressed lncRNAs between different stages of lung cancer while further studies are warranted to identify the functional roles and clinical value of these lncRNAs.
To summarize, we performed data mining of a data set of 181 microarrays and found that a set of protein-coding genes and lncRNAs was differentially expressed between different stages. Additionally, SCN7A, C7, and LINC00313 were significantly associated with the survival of lung cancer.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90.
National Lung Screening Trial Research T, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med. 2011;365(5):395–409.
Senthi S, Lagerwaard FJ, Haasbeek CJ, Slotman BJ, Senan S. Patterns of disease recurrence after stereotactic ablative radiotherapy for early stage non-small-cell lung cancer: a retrospective analysis. Lancet Oncol. 2012;13(8):802–9.
Senan S, Paul MA, Lagerwaard FJ. Treatment of early-stage lung cancer detected by screening: surgery or stereotactic ablative radiotherapy? Lancet Oncol. 2013;14(7):e270–4.
Chang JY, Senan S, Paul MA, Mehran RJ, Louie AV, Balter P, Groen HJ, McRae SE, Widder J, Feng L, van den Borne BE, Munsell MF, Hurkmans C, Berry DA, van Werkhoven E, Kresl JJ, Dingemans AM, Dawood O, Haasbeek CJ, Carpenter LS, De Jaeger K, Komaki R, Slotman BJ, Smit EF, Roth JA. Stereotactic ablative radiotherapy versus lobectomy for operable stage I non-small-cell lung cancer: a pooled analysis of two randomised trials. Lancet Oncol. 2015.
Zhu CQ, Pintilie M, John T, Strumpf D, Shepherd FA, Der SD, et al. Understanding prognostic gene expression signatures in lung cancer. Clin Lung Cancer. 2009;10(5):331–40.
Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102(7):464–74.
Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28(29):4417–24.
Der SD, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, et al. Validation of a histology-independent prognostic gene signature for early-stage, non-small-cell lung cancer including stage IA patients. J Thorac Oncol. 2014;9(1):59–64.
Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona FJ, Sayols S, et al. A prognostic DNA methylation signature for stage I non-small-cell lung cancer. J Clin Oncol. 2013;31(32):4140–7.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34(2):613–20.
Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7.
Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–45.
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–59.
Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5(20):9864–76.
Yang J, Lin J, Liu T, Chen T, Pan S, Huang W, et al. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85(2):110–5.
Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, et al. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48(1):1–8.
Wettenhall JM, Smyth GK. limmaGUI: a graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20(18):3705–6.
The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34(Database issue):D322-326.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9.
Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, et al. Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nat Biotechnol. 2007;25(6):663–8.
Schlitt T, Palin K, Rung J, Dietmann S, Lappe M, Ukkonen E, et al. From gene networks to gene function. Genome Res. 2003;13(12):2568–76.
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–80.
Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30.
Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17(10):1537–45.
Louie AV, Senthi S, Palma DA. Surgery versus SABR for NSCLC. Lancet Oncol. 2013;14(12), e491.
Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, et al. A 12-gene set predicts survival benefits from adjuvant chemotherapy in non-small cell lung cancer patients. Clin Cancer Res. 2013;19(6):1577–86.
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–504.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–14.
Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–55.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–6.
Srikantan V, Zou Z, Petrovics G, Xu L, Augustus M, Davis L, et al. PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci U S A. 2000;97(22):12216–21.
Schmidt LH, Spieker T, Koschmieder S, Schaffers S, Humberg J, Jungen D, et al. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6(12):1984–92.
Acknowledgments
This study is founded by the Natural Science Foundation of China (81372321 to Lin Xu; 81201830 and 81472200 to Rong Yin), Natural Science Foundation for High Education of Jiangsu Province (13KJB320010 to Rong Yin), Jiangsu Provincial Special Program of Medical Science (BL2012030 to Lin Xu), and Jiangsu Province Ordinary University Graduate Student Research Innovation Project for 2013 (CXLX13_571 to Mantang Qiu).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ming Li and Mantang Qiu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Li, M., Qiu, M., Xu, Y. et al. Differentially expressed protein-coding genes and long noncoding RNA in early-stage lung cancer. Tumor Biol. 36, 9969–9978 (2015). https://doi.org/10.1007/s13277-015-3714-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3714-6