Abstract
RING box protein1 (RBX1), an essential component of SCF E3 ubiquitin ligases, plays an important role in gastric cancer. In the study, miR-194 and RBX1 expression was evaluated in 76 pairs of gastric tumor and non-tumor tissue samples by qRT-PCR, and clinicopathological characteristics were analyzed. CCK8, transwell assay, wound healing assay, and flow cytometry assay were performed to evaluate the effect of miR-194 on gastric cancer (GC) cellular proliferation, invasion, migration, apoptosis, and cell cycle, respectively. Luciferase reporter assays and Western blotting were used to evaluate whether RBX1 is a direct target of miR-194. The Kaplan-Meier method and log-rank test were used to evaluate the correlation between miR-194 or RBX1 expression and patient survival. Then, we found that miR-194 was significantly downregulated and RBX1 upregulated in GC tissues; both of which showed significant association with tumor size, location, invasion, and tumor node metastasis. Cell proliferation, invasion, and migration were significantly restricted with miR-194 overexpression. miR-194 downregulated RBX1 protein expression, and luciferase assays showed that binding sites in the RBX1 3′UTR were required for miR-194-mediated repression of RBX1, indicating that RBX1 was a direct target of miR-194. Transfection of RBX1 without the 3′UTR restored the miR-194-inhibiting migration function. miR-194 overexpression or RBX1 lowexpression was associated with prolonged survival of GC patients. In conclusion, upregulation of miR-194 can inhibit proliferation, migration, and invasion of GC cells, possibly by targeting RBX1. Aberrant expression of miR-194 and RBX1 is correlated to GC patient survival time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is one with the highest incidence and mortality of tumors and has the second mortality rate of cancer worldwide [1–3]. Many exogenous and genetic transformations have been demonstrated to contribute to the multistep process of GC development [4–6]. However, the intricate mechanisms underlying gastric carcinogenesis are not yet clear, and greater efforts are needed to understand the process. Moreover, the identification of biomarkers to predict the prognosis of patients is crucial for treatment planning.
MicroRNAs (miRNAs) are a class of well-conserved, endogenously expressed, noncoding RNA molecules. miRNAs are short single-stranded RNAs, 19–22 nucleotides in length, that regulate the translation of many genes [7, 8]. miRNAs are involved in multiple biological functions, like proliferation, differentiation, apoptosis, and metabolism [9, 10]. In addition, several miRNAs have been identified to be involved in various aspects of human GC pathogenesis, such as metastasis, invasion, self-renewal, and so on [11, 12].
Protein modification by ubiquitin or ubiquitin-like proteins is one of the primary posttranslational regulatory mechanisms of critical factors involved in cell physiology [13]. Skp1/Cullins/F-box (SCF) complexes are the largest multiunit E3 ubiquitin ligases and ubiquitinate a broad range of proteins [14]. RING box protein1 (RBX1) is an essential element of SCF E3 ubiquitin ligases. Notably, the upregulation or downregulation of components of SCF complexes has been reported in different kinds of cancers [15–19].
We recently reported that miR-194 inhibited the proliferation and migration of GC cells [20]. And a previous study showed that RBX1 expression was upregulated in GC tissues [21]. However, no report has examined the interaction between miR-194 and RBX1 in GC. In this study, we examined the expression of miR-194 and RBX1 in GC tissues and analyzed clinicopathological characteristics. We evaluated the migration and proliferation of GC cells with modulated expression of miR-194. We also verified RBX1 as a direct functional target of miR-194. Our study helps further the understanding of the mechanism underlying GC pathogenesis and may contribute to the development of novel, targeted therapies.
Materials and methods
Clinical sample collection
This study was approved by the Human Research Ethics Committee of Zhengzhou University. We asked every patient to sign the informed consent. A total of 76 pairs of gastric tumor and non-tumor tissue samples were obtained from patients who underwent gastrectomy in the First Affiliated Hospital and Cancer Hospital Affiliated of Zhengzhou University (Zhengzhou, China) between 2006 and 2008. Both tumor tissue and non-tumor tissue samples were based on histopathological evaluation. The biopsies were immediately frozen after resection and stored in liquid nitrogen until use. The tumor histological grade was staged using the seventh TNM staging of the International Union against Cancer/American Joint Committee on Cancer system.
Cell lines and cell culture
The human GC cell lines NCI-N87 and BGC-823 were offered from the Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with 10 % autoclaving fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin in a humidified condition of 5 % CO2 at 37 °C in RPMI 1640.
RNA isolation and qRT-PCR
Total RNA was purified from tissue samples and cell lines using the Qiagen RNeasy kit (Valencia, CA, USA). Complementary DNA (cDNA) was taken place with the high-capacity cDNA reverse transcription kit (F. Hoffmann-La Roche Ltd.). qRT-PCR was used to quantify miR-194 expression, according to the TaqMan MicroRNA Assays protocol (Applied Biosystems), and normalized using U6 small nuclear RNA (RNU6B). The 2−ΔΔCT method was used to calculate the relative expression levels of miR-194 in tumor tissues compared with matched normal adjacent tissues [22]. Primer sequences are as follows: has miR-194, RT 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACATTGTC 3′, Forward 5′ TCCGAAGGTGTACCTCAAC 3′, Reverse 5′ GTGCAGGGTCCGAGGT 3′; U6, RT 5′ GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA 3′, Forward 5′ TCCGATCGTGAAGCGTTC 3′, Reverse 5′ GTGCAGGGTCCGAGGT 3′; RBX1, Forward 5′ TGCAGGAACCACATTATGGAT 3′, Reverse 5′ GAATTCCCACTCTCTGTTGTCC 3′; GAPDH, Forward 5′ AGAGGCAGGGATGATGTTCTG 3′, Reverse 5′ GACTCATGACCACAGTCCATGC 3′.
Western blotting
Protein was extracted from tissue samples and cell lines using RIPA buffer containing PMSF. A BCA protein assay kit (Beyotime, Haimen, China) was used to confirm the total protein concentration. Protein samples were separated by SDS-PAGE and transferred to polyvinyl membranes. Membranes were incubated overnight at 4 °C with primary antibody (rabbit anti-RBX1 antibody, 1:1000 dilution; Invitrogen, Carlsbad, CA, USA) followed by the secondary antibody (HRP-conjugated goat anti-rabbit IgG, 1:3000 dilution; Santa Cruz Biotechnology). Signals were observed by using a chemiluminescence detection kit (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Antibody against GAPDH (Santa Cruz Biotechnology) functioned as an internal reference [23, 24]. Using Typhoon PhosphorImager (GE Healthcare) measures the quality of the protein. Experiments were performed in triplicate.
RNA oligoribonucleotides and cell transfection
The miR-194 (GMR-miR microRNA-194) and scrambled miRNA-negative control used in this study were synthesized by Shanghai GenePharma Co. Ltd. BGC-823 and NCI-N87 cells were planted into six-well plates (2 × 105 cells/well) and settled overnight until they were 50–80 % confluent. Cells were transfected using Lipofectamine™ 2000 (Invitrogen, Carlsbad, CA, USA) in the following experimental groups: non-transfected group (blank control), scrambled miRNA-transfected group (negative control, NC), and transfected group (miR-194). At 24–48 h after transfection, the cells were lysed for analyses.
Cell proliferation assay
Cellular proliferation was measured by water-soluble tetrazolium salt (WST) assay using microplate computer software (Bio-Rad Laboratories, Hercules, CA, USA) according to the protocol of the Cell Counting Kit-8 (CCK8) assay kit (Dojindo, Japan). Reading the absorbance at 450 nm (A450) on a microplate reader (168–1000 Model 680, Bio-Rad) draw the proliferation curves.
Transwell invasion assay
The invasive potential of cells was tested using a Matrigel transmembrane invasion assay. Cells (2 × 105) were resuspended in 200 μL of serum-free medium and planted onto the upper chamber of the transwell filters, and medium (500 μL) containing 10 % FBS was added to the bottom chamber. The cells were to migrate for 24 h at 37 °C in an incubator with 5 % CO2. The membranes were cut out and mounted onto coverslips with the cells on the upper surface, and cell morphology was observed by hematoxylin and eosin (HE) staining. The cells invading the membrane were counted in three randomly selected visual fields from the central to the surrounding parts of the filter using an inverted microscope at ×200 magnification. Each test was done in triplicate.
Wound healing assay
The wound healing assay was used for detection of cell migration. The two groups of cells, BGC-823 and NCI-N87, were cultured to confluence (>90 %) in six-well plates. Then, a straight scratch wound was emerged with a 200-μL sterile pipette tip. The medium was changed with a new one regularly. After 48 h, the cells were photographed. Assays were repeated three times.
Flow cytometric analysis of apoptosis
The flow cytometry apoptosis assay was used for detection of apoptosis. The BGC-823 and NCI-N87 cell lines (transfected with NC and miR-194) were cultured into culture flasks. After 24 h, the cells were collected and washed twice by phosphate-buffered saline (PBS). Then, the cells were centrifugated and resuspended at a density of 1 × 106 cells/mL in 1× binding buffer. After staining with FITC Annexin V and propidium iodide (PI), the cells were measured with a FACSCalibur flow cytometer (BD Biosciences, USA), and the data were analyzed by Cell Quest software (BD Biosciences, USA).
Flow cytometric analysis of cell cycle
The BGC-823 and NCI-N87 cell lines (transfected with NC and miR-194) were cultured into culture flasks. After 24 h, the cells were collected, washed twice by PBS, and fixed with 70 % ethanol storage overnight at 4 °C. Then, the cells were incubated with 100 μL ribonuclease A (RNase A) at 37 °C for 30 min. After that, in dark conditions, cells were stained with 400 μL PI for 30 min at 4 °C. The percentage of cells in G0/G1, S, and G2/M phases of cell cycle was measured with a FACSCalibur flow cytometer (BD Biosciences, USA), and the data were analyzed by Cell Quest software (BD Biosciences, USA).
Dual-luciferase reporter assays
The RBX1 was amplified from human genomic DNA by PCR, containing putative binding sites for miR-194, and cloned into the pmirGLO control vector (Promega, Madison, WI, USA). The 3′UTR of RBX1 was confirmed by sequencing and named pmirGLO-Wt. Site-directed mutagenesis of the miR-194 target site in the RBX1 3′UTR was performed using a QuikChange site-directed mutagenesis kit (Promega, Madison, WI, USA) and pmirGLO-Wt conducted the template to occur the pmirGLO-Mut plasmid. BGC-823 and NCI-N87 cells were cultured in 96-well plates and cotransfected with wild-type or mutant reporter plasmid (100 nM) and miRNA (100 nM) using Lipofectamine™ 2000 (Invitrogen). Luciferase activity was measured by the dual-luciferase assay system (Promega, Madison, WI, USA) 48 h post transfection.
Statistical analysis
Statistical analysis was performed using the SPSS software version 21.0 (SPSS, Chicago, IL). All data are presented as the mean ± SD where applicable. Differences were analyzed with the paired t test. The survival time was defined as the time from the operation until 84 months. The survival curves were calculated by the Kaplan-Meier method. The Kaplan-Meier method and log-rank test were used to evaluate the correlation between miR-194 or RBX1 expression and patient survival. The correlation between miR-194 or RBX1 expression and clinicopathological characteristics was analyzed by Pearson correlation analysis. Differences were considered significant when P < 0.05.
Results
miR-194 is downregulated and RBX1 upregulated in GC tissues
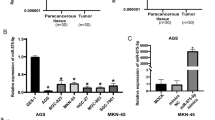
We analyzed the expression of RBX1 and miR-194 in gastric tumor and non-tumor tissues. Significantly higher levels of RBX1 mRNA were observed in GC tissues compared with non-tumor tissues (Fig. 1a). Furthermore, we found that miR-194 was expressed at low levels in tumor tissues and expressed at high levels in non-tumor tissues (Fig. 1b). To elucidate the correlation between miR-194 and RBX1 expression levels and clinicopathological factors in human GC, 76 GC cases were analyzed. As shown in Table 1, no correlation between miR-194 or RBX1 expression level and patients’ age, gender, and histologic type was observed (P > 0.05). However, miR-194 was downregulated in tumor tissues with smaller tumor size (P < 0.01), deeper local invasion (P < 0.01), more advanced TNM stage (P < 0.01), and positive lymph node metastasis (P < 0.01). RBX1 was upregulated in tumor tissues with larger tumor size (P < 0.01), deeper local invasion (P < 0.01), more advanced TNM stage (P < 0.01), and positive lymph node metastasis (P < 0.01). Correlation analysis was carried out to evaluate the relationship between miR-194 and RBX1 expression levels, and results indicated that miR-194 levels were negatively correlated with RBX1 expression levels: the correlation coefficient was −0.379 (P < 0.01). These results indicate a significant correlation with miR-194 upregulation or RBX1 downregulation in GC.
Expression level of miR-194 and RBX1 mRNA in gastric tumor and non-tumor tissues. Relative expression of miR-194 was measured by qRT-PCR; U6 snRNA served as an endogenous control. RBX1 protein expression levels were measured by Western blotting; GAPDH served as an endogenous reference. a RBX1 expression differed significantly between cancer tissues and corresponding distant non-tumor tissues (P < 0.01). b miR-194 expression in gastric tumor tissues was significantly lower than in non-tumor tissues (P < 0.01)
miR-194 overexpression or RBX1 mRNA lowexpression is associated with prolonged survival time in GC
To investigate the relationship between survival time and the expression level of miR-194 or RBX1 mRNA, we divided cases as overexpression (with levels greater than or equal to the mean) or lowexpression (with levels less than the mean). GC patients with miR-194 overexpression had longer survival time (mean, 67.87; 95 % CI, 63.26, 72.49) than GC patients with lowexpression (mean, 49.80; 95 % CI, 42.16, 57.45) (χ 2 = 8.0379, P = 0.004) (Fig. 2b). Furthermore, GC patients with RBX1 lowexpression had longer survival time (mean, 65.05; 95 % CI, 58.88, 71.23) than GC patients with overexpression (mean, 52.62; 95 % CI, 45.92, 59.33) (χ 2 = 6.798, P = 0.009) (Fig. 2a). Together, this data indicates that miR-194 overexpression or RBX1 mRNA lowexpression is correlated with prolonged survival time of GC patients.
Upregulation of miR-194 inhibits proliferation, promotes apoptosis, and arrests cell cycles of BGC-823 and NCI-N87 cells
We next performed CCK8 assays in GC cells to further evaluate the effect of miR-194 on cell proliferation and used the flow cytometry assay for detection of apoptosis and cell cycle. After transfection of BGC-823 and NCI-N87 cells with miR-194, the expression of miR-194 was significantly increased compared with the untransfected (blank) and miR-194-negative control (NC) groups. In CCK8 assays, we observed that the viability of the miR-194 group was decreased compared with the blank and NC groups from 24 h onwards in BGC-823 (Fig. 3a) and NCI-N87 (Fig. 3b) cells (P < 0.05). In contrast, we found no significant difference between the blank and NC groups (P > 0.05). From flow cytometric analysis of apoptosis, we can observed that compared with NC groups, the percentage of apoptotic cells of the miR-194 groups was much higher in both BGC-823 and NCI-N87 cells (P < 0.01) (Fig. 3c, d). In the flow cytometric analysis of cell cycle, we can know that in both BGC-823 and NCI-N87 cells, the percentage of G0/G1 of miR-194 groups was higher than that of NC groups(P < 0.01), while the percentage of S of miR-194 groups was lower than that of NC groups (P < 0.01). There was no significant difference between the two groups about the percentage of G2/M (Fig. 3e, f). These results suggest that upregulation of miR-194 could slow the proliferation of BGC-823 and NCI-N87 and also can promote the apoptosis and arrest cell cycles.
CCK8 assay and flow cytometry assay were performed in BGC-823 and NCI-N87 cells. Significant decreases in BGC-823 (a) and NCI-N87 (b) cell proliferation were observed in the miR-194 group compared with the NC and blank groups (P < 0.05). No difference was found between the blank and NC groups. Higher percentage of apoptotic cells were observed in the miR-194 group compared with the NC groups both in BGC-823 and NCI-N87 cells (P < 0.01) (c, d). In both BGC-823 and NCI-N87, the percentage of G0/G1 of miR-194 groups was significant higher than NC groups, but the percentage of S of miR-194 was lower than NC groups. No difference was found between the miR-194 groups and NC groups of the percentage of G2/M (e, f). Data were presented as the mean of duplicate experiments. NC cells transfected with miR-194 negative control, miR-194 cells transfected with miR-194
Upregulation of miR-194 restricts cell invasion and migration of BGC-823 and NCI-N87 cells
We next conducted a transwell assay and a wound healing assay to evaluate the role of miR-194 in regulating the invasion and migratory activity of GC cells (Fig. 4). The average number of migrating cells penetrating the transwell membrane was significantly lower in the miR-194 group (P < 0.01) compared with the NC group (Fig. 4a, b). The NC group had reached higher cell density at 48 h post wounding compared with the miR-194 group in both cell types (Fig. 4c, d). These data indicate that upregulation of miR-194 restricted the migratory and invasive capacity of BGC-823 and NCI-N87 cells.
Invasive and migratory abilities of GC cells (BGC-823 and NCI-N87) after transfection with miR-194 were assessed by transwell assay and wound healing assay. a Hematoxylin and eosin staining. b The numbers of BGC-823 and NCI-N87 cells passing through the Matrigel were lower in the miR-194 group than in the negative control (NC) groups (P < 0.01). c, d For BGC-823 and NCI-N87, both the NC groups had a higher density of cells at 48 h after wounding compared with the miR-194 group
RBX1 is a direct target of miR-194
Bioinformatic analysis using TargetScan and miRanda indicated that the 3′UTR of RBX1 contains a predicted seed region for miR-194 (Fig. 5a). To determine whether RBX1 is regulated by miR-194, we conducted Western blot analysis and luciferase reporter assays. Western blot analysis showed that RBX1 expression was downregulated in both BGC-823 and NCI-N87 cells after transfection with miR-194 (Fig. 5b, c). To verify whether RBX1 is directly targeted by miR-194, we used a dual-luciferase reporter system with luciferase reporter vectors containing either the wild-type or the mutant 3′UTR of RBX1. Cotransfection with miR-194 significantly decreased the luciferase activity of the reporter containing wild-type 3′UTR in both BGC-823 and NCI-N87 cells, but did not affect the luciferase reporter with the mutant 3′UTR (Fig. 5d, e). Taken together, these data strongly suggest that miR-194 negatively regulates RBX1 expression by directly binding to the putative binding site in the 3′UTR sequence.
RBX1 is a target gene of miR-194 in both human GC BGC-823 and cells. a The wild-type RBX1 3′UTR and RBX1 3′UTR putative binding sequences in miR-194. b, c Western blot analysis of RBX1 expression in cells transfected as indicated. GAPDH was used as the endogenous reference. miR-194 significantly decreased expression of RBX1 in both BGC-823 and NCI-N87 cells. d, e Luciferase reporter vectors with wild-type or mutant-type 3′UTR were cotransfected into BGC-823 or NCI-N87 cells together with NC or miR-194. Cotransfection of miR-194 significantly decreased the luciferase activity of the pmirGLO-Wt 3′UTR reporter (*P < 0.05), but did not decrease that of the pmirGLO-Mut 3′UTR reporter. NC cells transfected with miR-194 negative control, miR-194 cells transfected with miR-194
Expression of RBX1 restores miR-194-inhibiting migration function
We next performed Western blotting and transwell assays to evaluate whether RBX1 is a functional target of miR-194. As demonstrated previously, transfection with miR-194 resulted in decreased endogenous RBX1 levels in BGC-823 cells. RBX1 levels were higher after transfection with vector containing RBX1 lacking the 3′UTR (pcDNA3.1-RBX1). Cotransfection of miR-194 and pcDNA3.1-RBX1 did not impact the increased expression of RBX1 and abrogated the RBX1 expression-reducing effect of miR-194 (Fig. 6a).
Expression of RBX1 abrogates miR-194-inhibiting migration function. a RBX1 levels in BGC-823 cells were lower after transfection with miR-194. However, transfection with RBX1 vector lacking 3′UTR (pcDNA3.1-RBX1) together with miR-194 increased RBX1 expression, abrogating the inhibitory effect of miR-194 on RBX1 expression. b The average numbers of cells penetrating the transwell membrane decreased after transfection with miR-194. Transfection of the RBX1 vector lacking 3′UTR (pcDNA3.1-RBX1) resulted in increased invasive ability. Cotransfection of miR-194 with the mutant RBX1 had no effect on invasion. These results suggest that RBX1 is a major target of miR-194
In transwell assays, the mean number of cells penetrating the transwell membrane was decreased after transfection with miR-194. We observed increased invasive ability of cells transfected with pcDNA3.1-RBX1 (lacking the 3′UTR) compared with controls. However, cotransfection with pcDNA3.1-RBX1 and miR-194 increased the average number of transwell cells, abrogating the effect of miR-194 on decreased cell numbers (Fig. 6b). These results suggest that RBX1 is a major target of miR-194.
Discussion
RBX1 is a meaningful conserved protein from yeast to human and acts a major role in embryogenesis [25, 26]. Abnormal expression of RBX1 resulted in the dysfunction of SCF ligases and caused embryo damage [27]. Lately, some studies indicated that RBX1 was not only vital in regulating the function of multiple physiological cellular but was also related to tumor progression [28, 29]. Other studies showed that altered expression of RBX1 protein played a key role in cancer occurrence and development. Our research indicated that RBX1 was overexpressed in GC tissues. But today, the underlying mechanism remained unsolved. Analysis of RBX1 expression level could help us to better understand its functions and mechanisms in GC and help to understand the development of biomarkers for early diagnosis and prognosis.
Recently, some studies have indicated that miR-194 acted a part in the development of a variety of diseases. In the human digestive tract, miR-194 is specifically expressed and induces the differentiation of intestinal epithelial cells [30]. In GC, the downregulation of miR-194 was correlated to more advanced stages and larger tumor [20]. In liver mesenchymal-like cancer, upregulation of miR-194 expression reduces N-cadherin expression level and induces the changes of cellular biological behavior of this cancer, like the inhibition of cell proliferation, migration, and metastasis [31]. However, the function of miR-194 in GC is still unknown. Thus, our findings of the biological functions of miR-194 in GC are critical to elucidating the role for miR-194 in tumor development. Notably, our study showed that miR-194 expression was significantly lower in human gastric tumor tissues compared with non-tumor tissues.
The results of our study also show that upregulating miR-194 expression level in GC tissues can inhibit the proliferation, migration, and invasion of tumor cell lines. These data suggest that miR-194 could be used in the treatment of GC. Moreover, the expression level of miR-194 has a high correlation with the survival of GC patients and thus could serve as a biomarker for GC prognosis. Similarly, we found that RBX1 is upregulated in GC cells and tissues, suggesting that RBX1 may be a potential therapeutic target in GC.
In summary, our data suggest that upregulation of miR-194 can inhibit proliferation, migration, and invasion of GC cells, possibly by targeting RBX1. Aberrant expression of miR-194 and RBX1 is correlated to GC patient survival time.
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917.
Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66.
Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354–62.
Iravani O, Tay BW, Chua PJ, Yip GW, Bay BH. Claudins and gastric carcinogenesis. Exp Biol Med. 2013;238:344–9.
Jang BG, Kim WH. Molecular pathology of gastric carcinoma. Pathobiology. 2011;78:302–10.
Maurel M, Jalvy S, Ladeiro Y, Combe C, Vachet L, Sagliocco F, et al. A functional screening identifies five microRNAs controlling glypican-3: role of miR-1271 down-regulation in hepatocellular carcinoma. Hepatology. 2013;57:195–204.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–79.
Zhang S-J, Feng J-F, Wang L, Guo W, Du Y-W, Ming L, et al. miR-1303 targets claudin-18 gene to modulate proliferation and invasion of gastric cancer cells. Dig Dis Sci. 2014;59:1754–63.
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, et al. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963–70.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–80.
Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, et al. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284:662–5.
Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49.
Masuda TA, Inoue H, Sonoda H, Mine S, Yoshikawa Y, Nakayama K, et al. Clinical and biological significance of S-phase kinase-associated protein 2 (Skp2) gene expression in gastric carcinoma: modulation of malignant phenotype by Skp2 overexpression, possibly via p27 proteolysis. Cancer Res. 2002;62:3819–25.
Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93.
Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan Y, et al. Overexpression of Cullin1 is associated with poor prognosis of patients with gastric cancer. Hum Pathol. 2011;42:375–83.
Birner P, Schoppmann A, Schindl M, Dinhof C, Jesch B, Berghoff AS, et al. Human homologue for Caenorhabditis elegans CUL-4 protein overexpression is associated with malignant potential of epithelial ovarian tumours and poor outcome in carcinoma. J Clin Pathol. 2012;65:507–11.
Song Y, Zhao F, Wang Z, Liu Z, Chiang Y, Xu Y, et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann Surg Oncol. 2012;19:S509–17.
Migita K, Takayama T, Matsumoto S, Wakatsuki K, Tanaka T, Ito M, et al. Prognostic impact of RING box protein-1 (RBX1) expression in gastric cancer. Gastric Cancer. 2014;17:601–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2 (delta delta c (T)) method. Methods. 2001;25:402–8.
Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26:27–34.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60.
Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20.
Yi Sun MT, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–50.
Tan M, Davis SW, Saunders TL, Zhu Y, Sun Y. RBX1/ROC1 disruption results in early embryonic lethality due to proliferation failure, partially rescued by simultaneous loss of p27. Proc Natl Acad Sci. 2009;106:6203–8.
Wei D, Sun Y. Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: the role in cancer and as cancer targets. Gene Cancer. 2010;1:700–7.
Jia L, Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Curr Cancer Drug Targets. 2011;11:347–56.
Hino K, Tsuchiya K, Fukao T, Kiga K, Okamoto R, Kanai T, et al. Inducible expression of microRNA-194 is regulated by HNF-1alpha during intestinal epithelial cell differentiation. RNA. 2008;14:1433–42.
Meng Z, Fu X, Chen X, Zeng S, Tian Y, Jove R, et al. MiR-194 is a marker of hepatic epithelial cells and suppresses metastasis of liver cancer cells in mice. Hepatology. 2010;52:2148–57.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81272188 and No. 81301726) and the Education Department of Henan province science and technology research key projects (14A310002).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, X., Wang, Y., Zang, W. et al. miR-194 targets RBX1 gene to modulate proliferation and migration of gastric cancer cells. Tumor Biol. 36, 2393–2401 (2015). https://doi.org/10.1007/s13277-014-2849-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2849-1