Abstract
Increasing evidence shows that abnormal microRNA (miRNA) expression is involved in tumorigenesis. MiR-25 was previously reported to act as tumor suppressor or oncogene in diverse cancers. However, their expression, function, and mechanism in gastric cancer (GC) are not well known. Here, we show that miR-25 was overexpressed in primary tumor tissues of GC patients and was significantly correlated with a more aggressive phenotype of GC in patients. MiR-25 inhibition significantly decreased the proliferation, invasion, and migration of GC cells in vitro. Furthermore, miR-25 repressed F-box and WD-40 domain protein 7 (FBXW7) expression by directly binding to 3-untranslated region (UTR) of FBXW7, and the inverse correlation was observed between the expressions of miR-25 and FBXW7 mRNA in primary GC tissues. Moreover, the restoration of FBXW7 led to suppressed proliferation, invasion, and migration of GC cells. In vivo, miR-25 promotes tumor growth of GC. Taken together, miR-25 promotes GC progression by directly downregulating FBXW7 expression and may be employed as a novel prognostic marker and therapeutic target of GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is the second leading cause of cancer-related death worldwide [1]. The highest incidence of GC is frequently found in Eastern Asia, Eastern Europe, and South America, and the lowest in North America and most parts of Africa [2]. Although clinical outcome of GC has gradually improved through earlier diagnosis, surgical resection, and chemotherapy, 5-year survival rates of patients with GC are still low [3]. GC is a biologically heterogeneous disease accompanying various genetic and epigenetic alterations. A large number of oncogenes and tumor suppressor genes have been reported to be responsible for the development of GC, but the molecular mechanisms underlying the migration and invasion of advanced GC remains unclear.
MicroRNAs (miRNAs) are short, non-coding RNA molecules that regulate gene expression by directly binding to the 3′-untranslated region (UTR) of their target gene mRNA [4]. MiRNAs have been found to regulate a variety of cellular processes such as cell proliferation, differentiation, invasion, migration, and epithelial–mesenchymal transition [5–8]. An increasing number of miRNAs were shown to be involved in metastasis and invasion of GC, including miR-21 [9], miR-29 [10, 11], miR-30b [12], miR-141 [13], and miR-223 [14]. Although compelling evidences indicated that miR-25 is associated with gastric carcinogenesis, where miR-25 exerted potential proliferative, antiapoptotic, cell cycle-promoting effects, and tumorigenic activity [15, 16], the role of miR-25 in GC metastasis and invasion remains unclear.
F-box and WD-40 domain protein 7 (FBXW7) is a component of SCF (complex of SKP1, CUL1, and F‑box protein)-type ubiquitin ligases and regulates a network of proteins with central roles in cell division, cell growth, and cell differentiation, including cyclin E, MYC, JUN, and Notch [17–20]. FBW7 is also a tumor suppressor, the regulatory network of which is perturbed in many human malignancies. Milne et al. reported that loss of FBXW7 played a role in conventional gastric carcinogenesis, and Li et al. demonstrated that miR-223 functioned as an oncogene in human gastric cancer by targeting FBXW7 [21, 22].
In this study, we validated that miR-25 was substantially increased and downregulated expression of miR-25 significantly inhibited proliferation, invasion, and migration of GC cells. We found that miR-25 directly targeted FBXW7, and FBXW7 overexpression could partially attenuate the effect of miR-25 in GC. Furthermore, our data showed that miR-25 directly downregulated FBXW7 expression through binding to FBXW7-3′-UTR, and the expression of FBXW7 was negatively correlated with miR-25 in GC tissues.
Materials and methods
Patients and samples
A total of 40 GC tissues and matched normal tissues were surgically collected in our department, and informed consent was taken from all subjects. This work was approved by the Ethics Committee of the No. 281 Hospital of the Chinese People’s Liberation Army.
Cell culture
GC cell lines of GES-1, AGS, MKN-45, NUGC-3, HGC-27, and SGC-7901 were purchased from ATCC and cultured with F12 (AGS), DMEM/high glucose (MKN-45 or HGC-27), or RPMI 1640 (GES-1, NUGC-3, or SGC-7901) medium (HyClone) supplemented with 10 % FBS at 37 °C in 5 % CO2, respectively.
RNA extraction and quantitative reverse-transcriptase polymerase chain reaction
Trizol reagent (Invitrogen) was used to isolate total RNAs from frozen tissues and GC cells according to the manufacturer’s protocol. Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) assays for FBXW7 and miR-25 were performed via PrimeScriptTM RT reagent Kits (TaKaRa), TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems), SYBR® Green Realtime PCR Master Mix, and Permix Ex Taq (TaKaRa) according to the manufacturer’s protocol. Primers used were as follows: FBXW7 forward, 5′-CCACTGGGCTTGTACCATGTT-3′ and reverse, 5′-CAGATGTAATTCGGCGTCGTT-3′; and GAPDH forward, 5′-CGGATTTGGTCGTATTG-3′, and reverse, 5′-GAAGATGGTGATGGGATT-3′. Primers for U6 and miR-25 were purchased from GeneCopoeia (RiboBio). FBXW7 level was normalized with GAPDH, and miR-25 level was normalized with U6.
Cell proliferation assay
HGC-27 and SGC-7901 cells were seeded at a density of 104 per well in 96-well plates, respectively. The cells were transfected with anti-miR-NC, anti-miR-25, si-NC, si-FBXW7, empty vector, FBXW7 vector or cotransfected with miR-25 and FBXW7 vector, respectively. After 48 h, cell proliferation was analyzed using Cell Counting kit 8 (CCK8, Beyotime) according to the manufacturer’s protocol.
Cell migration and invasion assays
1 × 106 cells/ml of HGC-27 and SGC-7901 cells were prepared after transfection with anti-miR-NC, anti-miR-25, si-NC, si-FBXW7, empty vector, and FBXW7 vector or cotransfection with miR-25 and FBXW7 vector, for 24 h, respectively. The migration and invasion of the cells were analyzed using QCMTM Laminin Migration Assay (ECM220) and Cell Invasion Assay kit (ECM 550). Cells which had migrated or invaded to the lower membrane were counted using a microscope (Olympus, Tokyo, Japan).
Vector construct
For 3′-UTR of FBXW7, complementary 53-mer DNA oligonucleotides containing the putative miR-25 target site within the 3′-UTR of human FBXW7 mRNA were synthesized with flanking SpeI and HindIII restriction enzyme digestion sites (sense, 5′-GATTCACTTAGAAATTTTATTTTCTTATAACTTAAGTGCAATAAAATGTGTT-3′; antisense, 5′-AACACATTTTATTGCACTTAAGTTATAAGAAAATAAAATTTCTAAGTGAATC-3′). Another construct containing mutant seed region was also generated as a control (GTGCAAT to ATCCGGT). The DNAs and pMIR-REPORTTM Luciferase vectors were used to build the luciferase report vectors. The mutant 3′-UTR of FBXW7 served as a control. For FBXW7 vector, Homo sapiens full open reading frame cDNA clone for FBXW7 was transcribed, and the product was amplified by using primers with flanking SpeI and HindIII restriction enzyme digestion sites, followed by the DNAs were inserted into pcDNA3.1 vector.
Oligonucleotide transfection
Transfection of anti-miR-NC, anti-miR-25, miR-NC, or miR-25 via micrOFFTM anti-miR-25 kit and micrONTM miR-25 kit (RiboBio) according to the manufacturer’s protocol. Transfection of the luciferase report vectors, empty vectors, and miR-25 vectors via Lipofectamine 2000 (Invitrogen).
Luciferase assay
The HEK293T cells were transfected with wide-type pMIR-miR-25-3′-UTR or mutant pMIR- miR-25-3-UTR, and Renilla luciferase control vector (pRL-TK, Promega) using Lipofectamine 2000, and then the cells were transfected with miR-NC or miR-25, respectively. After transfection for 24 h, all of the cells were lysed via dual luciferase reporter assay system, and then the fluorescence activity was detected via GloMax Luminometer. The firefly luciferase activity was normalized to the Renilla luciferase activity.
Lentivirus packaging and transduction
MiR-25 and miR-control precursor sequences were amplified from human genomic DNA and cloned into the lentiviral vector pLVX-shRNA1 (Clontech Laboratories, Inc., Palo Alto, CA, USA). Virus packaging was performed in HEK293T cells. pLV-miR-25 or pLV-miR-control and Lenti-X HTX Packaging Kit were cotransfected using the Xfect transfection reagent. The AGS cells were transduced with pLV-miR-25 or pLV-miR-control. The cell line that stably expressed miRNA-25 was named LV-miR-25-AGS, and the control vector cell line was named LV-miR-control-AGS.
Animal studies
Five-week-old female nude mice (BALB/c,nu/nu) were maintained under specific pathogen-free conditions. Animal experiments were performed to evaluate tumor growth of GC cells in vivo. Briefly, to determine the proliferation capacity of LV-miR-25-AGS and LV-miR-control-AGS in vivo, a total of 1 × 106 cells were injected into the axillary fossae of each anesthetized nude mouse (n = 10 animals per group). Every 7 days post-inoculation, the length and width of individual tumor were measured with calipers, and the volume (mm3) was calculated according to the formula: 1/2× length × width2. The curve of tumor growth was depicted 35 days after inoculation. Mouse tumors were harvested and weighed.
Western blot assay
Cultured cells were lysed in RIPA buffer, and the lysates were analyzed using the standard Western blotting analyses. FBXW7 proteins were detected with anti-FBXW7 mouse monoclonal Abs (Abcam), and GAPDH proteins serving as an internal reference were detected with anti-GAPDH rabbit monoclonal Abs (Abnova). These were followed by incubation with HRP-conjugated secondary Abs (Santa Cruz Biotechnology). Bound proteins were visualized by using SuperSignal® West Dura Extended Duration Substrate kit.
Statistical analysis
One-way ANOVA and two-tail Student’s t test were used to analyze results using SPSS 16.0. Pearson’s correlation was used to analyze the relationship between the expressions of miR-25 and FBXW7 mRNA. P < 0.05 was considered statistically significant.
Results
MiR-25 was increased in GC tissues and cell lines and correlated with metastatic capacity in GC tissues and upregulation of miR-25 conferred poor prognosis in patients with GC
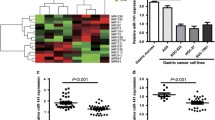
Expression of miR-25 in 40 GC patient tissues and matched normal tissue samples was detected by qRT-PCR. MiR-25 was significantly increased in GC patient tissues compared with matched normal tissues (Fig. 1a). Similarly, expression of miR-25 in five GC cell lines AGS, MKN-45, NUGC-3, HGC-27, and SGC-7901 was significantly increased compared with that in human gastric mucosa cell line GES-1 (Fig. 1d). When the 40 tumors were stratified, based on clinical progression, miR-25 was markedly increased in the GC tissues of patients with lymph node metastasis (Fig. 1b). Moreover, using the Kaplan–Meier method and logrank test, the overall survival of GC patients with high miR-25 expression was significantly shorter than those with low miR-25 expression (Fig. 1c).
MiR-25 was increased in GC tissues and cell lines and correlated with metastatic capacity in GC tissues, and upregulation of miR-25 conferred poor prognosis in patients with GC. a Expression of miR-25 in 40 GC tissues and matched normal tissues was detected by qRT-PCR. b qPCR data of miR-25 levels in primary GC (with or without metastasis). c Overall survival curves for two groups defined by low and high expression of miR-25 in patients with GC. d Expression of miR-25 in five GC cell lines, AGS, MKN-45, NUGC-3, HGC-27, and SGC-7901, and human gastric mucosa cell line (GES-1) was detected by qRT-PCR. Expression of miR-25 was normalized with U6. All assays were performed in duplicates.*P < 0.05, **P < 0.01 compared with the control
MiR-25 regulates the proliferation, invasion, and migration of GC cells in vitro
To determine whether miR-25 promotes the proliferation, migration, and invasion potential of GC cells, we carried out the experiments: HGC-27 and SGC-7901 cells were transfected with anti-miR-25 or anti-miR-NC for 24 h, followed by the proliferation, invasion, and migration of those cells were analyzed, respectively. As expected, the results showed that expression of miR-25 in HGC-27 and SGC-7901 cells transfected with anti-miR-25 was significantly decreased by qRT-PCR (Fig. 2a, b) and anti-miR-25 significantly suppressed the proliferation (Fig. 2c, d), invasion (Fig. 2e, f), and migration (Fig. 2g, h) of HGC-27 and SGC-7901 cells (P < 0.01). Consistent with the previous study [16, 23], our data showed that anti-miR-25 significantly reduced the proliferation of HGC-27 and SGC-7901 cells. Together, miR-25 promotes the proliferation, invasion, and migration of GC cells.
MiR-25 inhibition decreased the proliferation, migration, and invasion of GC cells in vitro. HGC-27 and SGC-7901 cells were transfected with anti-miR-NC or anti-miR-25 for 24 h, respectively. a, b Expression of miR-25 in HGC-27 and SGC-7901 cells transfected with anti-miR-NC or anti-miR-25 was detected by qRT-PCR. After transfection, 1 × 106 cells were used to perform the proliferation (c, d), invasion (e, f), and migration (g, h) assays. All assays were repeated in duplicates. *P < 0.05, **P < 0.01 compared with the control
FBXW7 was a direct target of miR-25
The function of miRNAs in tumor development is dependent on targeting their key target genes, so it is crucially important to identify the targets of miR-25. We used TargetScan 6.2 to screen the target gene of miR-25. Interestingly, we found that FBXW7 as a tumor suppressor is associated with cell division, growth, and differentiation. To identify whether FBXW7 is a target of miR-25, we constructed vectors containing the wild-type 3′-UTR or mutant 3′-UTR of FBXW7 mRNA, which was individually fused directly downstream of the firefly luciferase gene (Fig. 3a). For the luciferase assays, the wild-type or mutant vector was cotransfected into HEK293T cells with miR-NC or miR-25. As shown in Fig. 3b, miR-25 significantly reduced the relative luciferase activity of the wild-type 3′-UTR of FBXW7 (P < 0.01), while the luciferase activity of the mutant 3′-UTR was not significantly changed. We also confirmed that miR-25 significantly decreased the mRNA and protein expression of FBXW7 (P < 0.01) (Fig. 3c, d). Together, miR-25 downregulated FBXW7 expression through direct binding to the 3′-UTR of FBXW7.
FBXW7 was a direct target of miR-25. a The graph shows the wild-type or mutant binding site of FBXW7-3′-UTR for miR-25 and the construction model of wild-type or mutant pLUC- FBXW7-3′-UTR vectors. b Luciferase activity assays of luciferase vectors containing wild-type or mutant FBXW7-3′-UTR were performed after transfection with miR-NC or miR-25. The luciferase activity was normalized to Renilla luciferase activity. The assays were repeated in six wells. Western blot and qRT-PCR assays show the expressions of FBXW7 mRNA (c) and FBXW7 protein (d) in HGC-27 and SGC-7901 cells after transfection with miR-NC or miR-25. For Western blot assays, GAPDH served as an internal control. For qRT-PCR assays repeated in duplicates, GAPDH served as an internal control for FBXW7 and RNU6B for miR-25. *P < 0.05, **P < 0.01 compared with the control
MiR-25 promoted GC cell growth and motility by targeting FBXW7
To further investigate whether the effect of miR-25 on promoting proliferation, invasion, and migration of GC cells was via targeting FBXW7, we addressed the experiments that the GC cells were transfected with si-NC, si-FBXW7, empty vector or FBXW7 vector, and miR-25 or cotransfected with miR-25 and FBXW7 vector. The results showed that suppression of FBXW7 expression by siRNA promoted cell proliferation, invasion, and migration and overexpression of FBXW7 protein with pcDNA3.1-FBXW7 inhibited cell proliferation, invasion, and migration (Fig. 4a–d). Function investigation revealed that the proliferation, invasion, and migration of GC cells were significantly enhanced after transfection with miR-25, whereas the restoration of FBXW7 markedly led to the migration, invasion, and proliferation suppression (Fig. 4e–h). We also evaluated the expression of FBXW7 in 40 GC tissues and the corresponding normal tissues by qRT-PCR. Data showed that the average level of FBXW7 mRNA was significantly decreased in GC tissues compared with that in the corresponding normal tissues (Fig. 5a). Moreover, FBXW7 mRNA level was inversely correlated with miR-25 level (Fig. 5b). Together, miR-25 promotes the metastasis, invasion, and proliferation potential of GC cells by targeting FBXW7.
MiR-25 promoted GC cell growth and motility by targeting FBXW7. a–d Cell proliferation, invasion, and migration ability assays after HGC-27 and SGC-7901 cells were transfected with si-NC, si-FBXW7, empty vector, or FBXW7 vector, respectively. e, f HGC-27 and SGC-7901 cells were cotransfected with miR-25 and pcDNA3.1-FBXW7 or the vector. CCK8 assay was used to measure proliferation. g In vitro invasion assay of HGC-27 and SGC-7901 cells with miR-25 and pcDNA3.1-FBXW7 or the vector. h In vitro migration assay of HGC-27 and SGC-7901 cells cotransfected with miR-25 and pcDNA3.1-FBXW7 or the vector. Data were drawn from three independent experiments.*P < 0.05, **P < 0.01, #P < 0.05, ##P < 0.01 compared with miR-25 group
MiR-25 was inversely correlated with FBXW7 expression in GC tissues. a Expression of FBXW7 in 40 GC tissues and that in the corresponding nontumor tissues was detected by qRT-PCR. b Correlation analysis between miR-25 and FBXW7 mRNA level in GC tissues (Spearman’s correlation analysis, r = −0.4536; P < 0.01)
MiR-25 promotes tumor growth of GC in vivo
To further investigate the effect of miR-25 in vivo, a lentivirus vector was constructed to mediate the expression of miR-25, and two stable cell lines were established named LV-miR-25-AGS and LV-miR-control-AGS. These cell lines were injected into the axillary fossae of the female nude mice. To evaluate tumor growth, the length (L) and width (W) of tumor were measured every 7 days post-inoculation. The volumes of the tumors resulting from LV-miR-control-AGS injection were significantly smaller than those resulting from LV-miR-25-AGS injection (Fig. 6a). The mice were sacrificed 35 days post-inoculation. In agreement with the tumor volumes, the weight of tumors from LV-miR-control-AGS group were significantly lower than LV-miR-25-AGS (Fig. 6a). The data suggest that miR-25 promote the growth of AGS-engrafted tumors in vivo.
Discussion
Although a number of molecular drivers of gastric cancer have been described over the years, only very recently, miRNAs have emerged as key players in the pathogenesis of this disease [24, 25]. In this study, we determined that miR-25 was overexpressed in tumor tissues of GC patients compared with matched adjacent normal tissues and miR-25 was significantly correlated with a more aggressive phenotype of GC in patients. MiR-25 inhibition repressed the proliferation, invasion, and migration of GC cells in vitro. In addition, we identified FBXW7 as a direct miR-25-target gene and confirmed its direct interaction with the miR-25. In vivo, miR-25 promotes tumor growth of GC. Furthermore, our observation for an inverse correlation between miR-25 expression and FBXW7 expression in GC tissues fills this important void in literature for the missing experimental evidence for the function of miR-25 and FBXW7 in gastric pathogenesis.
Mature miR-25 was believed to be associated with tumor carcinogenesis, such as miR-25 promoted tumor growth, migration, and invasion in gastric cancer by targeting tob1 [16]. Another two studies showed that miR-25 promoted apoptosis resistance in cholangiocarcinoma by targeting TRAL death receptor-4 [26] and miR-25 promotes adult neural stem/progenitor cell proliferation and neuronal differentiation [27]. In our data, we confirmed that miR-25 was overexpressed in primary tumor tissues of GC patients compared with matched non-tumor tissues. These data implied that miR-25 overexpression was involved in GC carcinogenesis. Moreover, we showed that miR-25 inhibition repressed the proliferation, invasion, and migration of GC cells in vitro, suggesting that miR-25 can partially control the metastasis, invasion, and proliferation potential of GC. Apart from miR-25 overexpression in a large variety of human tumors, it is downregulated in the other types of human tumors, such as in colon cancer and anaplastic thyroid carcinomas [28, 29]. Therefore, miR-25 expression may be oppositely changed in the development of different tumors.
FBXW7 regulates a network of proteins with central roles in cell division, cell growth, and cell differentiation [24–27]. Most FBXW7 substrates are proto-oncogenes that are broadly implicated in the pathogenesis of human cancers. FBXW7 is a tumor suppressor, and loss of FBXW7 function leads to chromosomal instability, probably owing to hyperactivation of its many oncogenic substrates [30]. Therefore, the altered expression of FBXW7 is recognized to be one of the major causes of carcinogenesis or cancer development. FBXW7 expression is repressed in tumors. For example, its expression is suppressed in gliomas and correlates with patient survival [31, 32]. Mori et al. reported that FBXW7 mRNA expression was significantly lower in colorectal cancer tissues than the corresponding normal tissues and the low FBXW7 expression group showed a significantly poorer prognosis than patients in the high expression group [33]. Lu et al. reported that miR-25 regulated Wwp2 and Fbxw7 and promoted reprogramming of mouse fibroblast cells to iPSCs [34]. Consistent with these evidences, our data also indicated that miR-25-induced loss of FBXW7 enhanced the proliferation, invasion, and metastasis of GC cells and restoration of FBXW7 led to the proliferation, invasion, and migration suppression. Together, we highlighted the regulatory mechanism that miR-25-induced loss of FBXW7 promotes the metastasis, invasion, and proliferation potential of GC cells through direct binding to the 3′-UTR of FBXW7.
In conclusion, the present study highlighted the regulatory mechanism that miR-25-induced loss of FBXW7 enhanced the proliferation, invasion, and migration of GC cells and indicated that miR-25 may be a biomarker for the prognosis of GC patients.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–90.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66.
Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–55.
Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–81.
Hur K, Toiyama Y, Takahashi M, Balaguer F, Nagasaka T, Koike J, et al. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–26.
Wang X, Chen X, Wang R, Xiao P, Xu Z, Chen L, et al. MicroRNA-200c modulates the epithelial-to-mesenchymal transition in human renal cell carcinoma metastasis. Oncol Rep. 2013;30:643–50.
Wang Z, Cai Q, Jiang Z, Liu B, Zhu Z, Li C. Prognostic role of MicroRNA-21 in gastric cancer: a meta-analysis. Med Sci Monit. 2014;20:1668–74.
Han TS, Hur K, Xu G, Choi B, Okugawa Y, Toiyama Y, Oshima H, Oshima M, Lee HJ, Kim VN, Chang AN, Goel A, Yang HK. MicroRNA-29c mediates initiation of gastric carcinogenesis by directly targeting ITGB1. Gut. 2015;64:203–14.
Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, et al. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497–506.
Qiao F, Zhang K, Gong P, Wang L, Hu J, Lu S, et al. Decreased miR-30b-5p expression by DNMT1 methylation regulation involved in gastric cancer metastasis. Mol Biol Rep. 2014;41:5693–700.
Zhou X, Xia Y, Su J, Zhang G. Down-regulation of miR-141 induced by helicobacter pylori promotes the invasion of gastric cancer by targeting STAT4. Cell Physiol Biochem. 2014;33:1003–12.
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. MiRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–33.
Kan T, Sato F, Ito T, Matsumura N, David S, Cheng Y, et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology. 2009;136:1689–700.
Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y, Mao XH, Wu C, Yang SM, Zeng H, Zou QM, Guo G: MicroRNA-25 promotes gastric cancer migration, invasion and proliferation by directly targeting transducer of ERBB2, 1 and correlates with poor survival. Oncogene. 2014;34:2474–83.
Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93.
Cheng Y, Li G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012;31:75–87.
Brandt Y, Mitchell T, Wu Y, Hartley RS. Developmental downregulation of Xenopus cyclin E is phosphorylation and nuclear import dependent and is mediated by ubiquitination. Dev Biol. 2011;355:65–76.
Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–7.
Milne AN, Leguit R, Corver WE, Morsink FH, Polak M, de Leng WW, et al. Loss of CDC4/FBXW7 in gastric carcinoma. Cell Oncol. 2010;32:347–59.
Li J, Guo Y, Liang X, Sun M, Wang G, De W, et al. MicroRNA-223 functions as an oncogene in human gastric cancer by targeting FBXW7/hCdc4. J Cancer Res Clin Oncol. 2012;138:763–74.
Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86.
Matuszcak C, Haier J, Hummel R, Lindner K. MicroRNAs: promising chemoresistance biomarkers in gastric cancer with diagnostic and therapeutic potential. World J Gastroenterol. 2014;20:13658–66.
Liu HS, Xiao HS. MicroRNAs as potential biomarkers for gastric cancer. World J Gastroenterol. 2014;20:12007–17.
Razumilava N, Bronk SF, Smoot RL, Fingas CD, Werneburg NW, Roberts LR, et al. MiR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–75.
Brett JO, Renault VM, Rafalski VA, Webb AE, Brunet A. The microRNA cluster miR-106b~25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging (Albany NY). 2011;3:108–24.
Li Q, Zou C, Zou C, Han Z, Xiao H, Wei H, et al. MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett. 2013;335:168–74.
Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, et al. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab. 2012;97:E710–8.
Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–9.
Hagedorn M, Delugin M, Abraldes I, Allain N, Belaud-Rotureau MA, Turmo M, et al. FBXW7/hCDC4 controls glioma cell proliferation in vitro and is a prognostic marker for survival in glioblastoma patients. Cell Div. 2007;2:9.
Gu Z, Inomata K, Ishizawa K, Horii A. The FBXW7 beta-form is suppressed in human glioma cells. Biochem Biophys Res Commun. 2007;354:992–8.
Iwatsuki M, Mimori K, Ishii H, Yokobori T, Takatsuno Y, Sato T, et al. Loss of FBXW7, a cell cycle regulating gene, in colorectal cancer: clinical significance. Int J Cancer. 2010;126:1828–37.
Lu D, Davis MP, Abreu-Goodger C, Wang W, Campos LS, Siede J, et al. MiR-25 regulates Wwp2 and Fbxw7 and promotes reprogramming of mouse fibroblast cells to iPSCs. PLoS One. 2012;7:e40938.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gong, J., Cui, Z., Li, L. et al. MicroRNA-25 promotes gastric cancer proliferation, invasion, and migration by directly targeting F-box and WD-40 Domain Protein 7, FBXW7. Tumor Biol. 36, 7831–7840 (2015). https://doi.org/10.1007/s13277-015-3510-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3510-3