Abstract
Genetically transformed root cultures of Rauvolfia serpentina, have been developed following transformation with agropine strains of Agrobacterium rhizogenes. Characterization of Ri transformed root lines on the basis of morphology, karyotype analysis, T-DNA gene integration pattern and ajmalicine content in R. serpentina has been described in the present study. The transformed root lines were morphologically similar and exhibited typical hairy root phenotype. The somatic metaphase plates of the transformed root lines showed 2n = 22 chromosomes which is the diploid chromosome number of the species. The karyotype of both non-transformed and transformed roots showed 20 chromosomes with median to nearly median primary constriction and one pair of chromosomes had two constrictions, one at the median region and the other at the sub terminal region. Based on the pattern of TL and TR gene insertion the Ri transformed root lines can be divided into six types. Complete integration of TL-DNA was noted in 11 root lines induced with strain A. rhizogenes LBA 9402 belonging to Type I and II it is noteworthy that none of the root lines showed presence of aux1 and aux2, which are part of TR-DNA. Variable integration of TL-DNA was characteristic of several R. serpentina root lines. The Ri transformed root lines showed significant variability (p ≤ 0.05) in ajmalicine content (0.004 ± 0.001 to 0.229 ± 0.014 mg g−1 DW) after 4 weeks of culture on solid modified MS medium. Thus, we can conclude that the Ri transformed root lines of R. serpentina are morphologically and cytologically stable however variable pattern of TL and TR-DNA genes integration as well as ajmalicine content was noted among the different root lines. Thus, genetically variant transformed root lines of R. serpentina with high ajmalicine content can be selected for scale up studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rauvolfia serpentina (L). Benth.ex Kurz. (Apocynaceae) is an indigenous medicinal plant, synthesizing nearly 50 indole alkaloids. Its decoction has been traditionally used for psychiatry and central nervous system disorders. Several studies have established the role of the alkaloids as tranquilizers and for treatment of insomnia, insanity, high blood pressure, snake bite, cardiovascular diseases and gastrointestinal diseases [3]. The plant is currently listed as “endangered” by the International Union for Conservation of Nature and Natural Resources (IUCN). Some of the therapeutically important terpenoid indole alkaloids present in the roots of this plant are ajmalicine, ajmaline, reserpine, deserpidine, rescinnamine and yohimbine [38]
Genetically transformed roots obtained following infection with Agrobacterium rhizogenes has long been recognized as an alternative source for production of secondary metabolites [22, 29], due to their fast growth in hormone free media, genetic and biochemical stability and ability to produce natural compounds in levels comparable to field grown plants [18, 32]. Aird et al. [1] reported that hairy root cultures of several species show a stable secondary metabolite production due to their genetic stability at the chromosome level. Hairy root cultures of Datura stramonium have been found to have chromosome number and karyotype typical of the species [4]. The stability of chromosome numbers was noted in transformed root cultures of Swainsona galegifolia [15] and in Ri transformed plants of Tylophora indica [31].
On the other hand, cytological changes were detected in hairy root cultures of Trifolium pratense and Lotus corniculatus [40]. Although transgenic hairy roots regenerated from diploid potato explants also showed genetic stability, hairy root clones arising from monohaploid genotype were either diploid or tetraploid [39]. Variation in chromosome number was observed in the hairy root lines of Artemisia cina originating upon transformation with different strains of A. rhizogenes [16].
The T-DNA of the agropine type Ri plasmid consists of two separate regions designated as the TL-DNA and TR-DNA, which are transferred independently during the infection process [37]. The complete nucleotide sequencing of the TL-DNA revealed the presence of 18 open reading frames (ORFs), four of these ORF10, 11, 12 and 15 correspond to the rolA, rolB, rolC and rolD loci respectively which determine the induction and morphology of the hairy roots. The auxin synthesising genes (aux1 and aux2) and the opine synthesising genes mas1 and mas2 (mannopine) and ags (agropine) are located in the TR-DNA [27, 41]. Single transformation event leads to the formation of single transformed root which can be excised and cultured in vitro as a separate clone. Transgenic root clones are characterized by random T-DNA integration into the host genome and stability in inheritance and expression of T-DNA. Taneja et al. [36] suggested that variations in accumulation of secondary metabolites in hairy root cultures to be due to insertions of T-DNA genes at different positions in the genome, but is unlikely to be the only factor responsible for variations among different hairy root lines. In Catharanthus roseus, role of integration of TL- and/or TR-DNA on growth, morphology, and accumulation of secondary metabolites have been reported [8]. Hänisch ten Cate [19] reported frequent spontaneous deletions of Ri T-DNA in A. rhizogenes transformed potato roots and regenerated plants due to deletion of TL- and TR-DNA insertions during prolonged root culture and during shoot regeneration from root clones.
In course of our studies on genetic transformation in R. serpentina, we have developed root cultures following transformation with agropine strains of A. rhizogenes as reported earlier [9, 10, 23]. In the present paper, we report characterization of Ri transformed root lines on the basis of analysis of morphology, karyotype, T-DNA gene integration pattern and ajmalicine content in R. serpentina.
Materials and methods
Initiation and establishment of Ri transformed root cultures
Excised leaf explants (2–2.5 cm) and excised stem (internode) explants (~2 cm) isolated from 8–10 week old R. serpentina in vitro grown plants were wounded at different sites with a sterile hypodermic needle loaded with 2 ml of bacterial suspension as reported earlier [13]. Wild type supervirulent strain of A. rhizogenes viz. LBA 9402 and A4 obtained from Dr. D. Tepfer, INRA, Versailles France, were used for transformation. Bacterial suspension was prepared by transferring a loopful of bacterium to 10 ml liquid YMB medium [20] and incubated on a rotary shaker at 26 °C at 180 rpm for 24 h in dark. 200 μM acetosyringone (Sigma) was added to bacterial cultures (~1010 cells ml−1) 1 h prior to infection. All the explants were cocultured for 72 h in dark followed by washing with sterile distilled water. The explants were then cultured on 25 ml of 0.75 % agar solidified MS (Murashige and Skoog) [26] medium supplemented with 500 mg l−1 ampicillin at 24 ± 1 ºC in dark and 55–60 % relative humidity. Fifteen explants were used for each experiment and the experiments were repeated twice. Root initiation was noted from the wound sites of stem and leaf explants within 3 weeks of infection. The roots were excised and cultured in dark in 90 mm sterile disposable Petri plates containing 25 ml solid modified MS medium, (i.e., MS medium with 1/5 th total nitrogen, macro and micronutrients, myoinositol, ammonium iron citrate and Gamborg’s vitamin [17]) supplemented with 500 mg l−1 ampicillin. Each excised primary root was designated as a separate root line. The root lines maintained in vitro for over 3 years on solid modified MS medium by regular subculturing at an interval of 4 weeks were used in the present study.
Cytological analysis of Ri transformed root lines
The root tips obtained from A. rhizogenes transformed root lines (five root tips per root line; ten randomly selected root lines) of R. serpentina maintained under dark condition as well as non-transformed roots from in vitro plants were used for cytological analysis. The root tips were pretreated in 0.002 M 8-hydroxyquinoline solution at 18 ºC for 5 h, and then fixed in 3:1 absolute ethanol:glacial acetic acid and stored at -20 ºC. Root tips were transferred to 45 % acetic acid for 15 min, stained overnight in mixture of 2 % aceto-orcein:1(N) HCl (9:1) before squashing in 45 % acetic acid on glass slides to obtain well-scattered metaphase plates.
Cells with good metaphase spreads (two metaphase plates per root tip/ five root tips per root line) were photographed using software ProgRes® CapturePro 2.5, Systeme GmbH, Germany, to take measurements of each chromosome [long arm length (l), short arm length (s), chromosome length (CL) and total chromatin length (TCL)]. The arm ratios (r = l/s) were calculated and the chromosomes were categorized following the nomenclature of Levan et al. [21] [metacentric (m) (1.00–1.70), submetacentric (sm) (1.70–3.00), subtelocentric (st) (3.00–7.00) and telocentric (t) (7.00-∞)]. Chromosomes were first arranged into groups according to arm ratios and then according to the decreasing length in each group. For estimation of karyotype asymmetry Stebbins indices [35] were followed. The intrachromosomal and interchromosomal asymmetry indices were calculated using the formula A1 = 1-(n ∑i = 1qi/pi)/n and A2 = s/x, respectively [43].
Detection of T-DNA gene integration pattern in transformed hairy root genome
Molecular characterisation of the Ri transformed root lines was performed based on the presence and/or absence of different T-DNA genes. Genomic DNA was extracted from Ri transformed root lines as well as non-transformed roots of R. serpentina according to the procedure of Dellaporta et al. [14]. The quality and quantity of the DNA was determined with a UV–vis spectrophotometer (Thermo Scientific) and 0.8 % agarose gel, comparing with a standard lambda DNA marker (Bangalore Genei, India). Plasmid DNA from A. rhizogenes strain A4 to be used as positive control for PCR analysis was isolated following the standard alkali lysis protocol.
For each PCR reaction, 100 ng DNA and 0.25 μM of each primer (forward and reverse) were mixed with 2.5 μl of 10XTaq DNA polymerase Buffer A (GeNeiTM) containing 1.5 mM (final concentration) of MgCl2, 100 μM of each dNTP and 1U Taq DNA polymerase (GeNeiTM) in a final volume of 25 μl. Amplification was done in a programmed thermal cycler (Gene Amp® PCR system 2400; Perkin Elmer, Foster City, Calif) using the following programme: initial denaturation at 95 °C for 3 min, followed by 35 cycles of 30 s at 95 °C (denaturation), 30 s at 55 °C (annealing) and 1 min at 72 °C (extension), with a final extension step at 72 °C for 7 min. The amplified products were detected according to the amplicon size, by 1.2 % agarose gel electrophoresis using ethidium bromide stain and 1X TAE as running buffer. Documentation of amplified products was done with BioRad GelDocTM EZ Imager. The sequences of primers used were as reported by Taneja et al. [36].
Extraction, identification and quantitative analysis of ajmalicine
Alkaloid extraction was performed following the modified method of Panwar et al. [28]. Roots of each line were harvested after 4 weeks of culture on solid modified MS medium and oven-dried at 55 °C for 3 h. Dried root samples (0.1 g DW) were powdered and extracted thrice with 5 ml methanol (AR grade) for 24 h. After filtration with filter paper (Whatman®) the extracts were pooled and evaporated to dryness. The dried extracts were resuspended in 600 μl methanol (HPLC grade; Spectrochem) and filtered using 0.22 μm Milipore filters. HPLC was performed according to Panwar et al. [28] using carbinol:acetonitrile (60:40, v/v) as the mobile phase for isocratic elution at a flow rate of 0.2 ml min−1 and detection at 254 nm. Standard sample of ajmalicine was prepared by dissolving the powder (Sigma-Aldrich) in carbinol (0.5 mg ml−1). A calibrated graph was prepared by plotting peak areas versus amount of ajmalicine injected (0.1 to 0.4 mg ml−1). The relationship was linear over five measurements. Detection of ajmalicine was done on the basis of retention time (6.2 ± 0.05 min) and spiking with the standard compound while quantitative analysis was done based on peak areas. HPLC analysis was performed using Agilent Technologies (1260 Infinity) and reverse phase Agilent C18 column (100x4.6 mm). Extraction and analysis of all samples were done in triplicate with an average variation of ±0.005 mg gm−1 DW in content of ajmalicine observed. The reproducibility of injections expressed as variation (%) of injection was 2 %. Data were examined by a one-way analysis of variance (ANOVA) to detect significant differences (p ≤ 0.05) between the mean [33]. A post hoc mean separation was performed by the Tukey’s multiple comparison test at 5 % probability level using SPSS software (version 16.0). Variability in the data was expressed as the mean ± standard error (SE).
Results and discussion
Morphological analysis of Ri transformed root lines
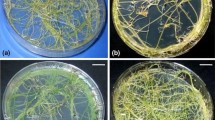
Phenotype of the transformed root lines showed the typical transformed nature including high degree of branching and plagiotropic growth. The primary roots were long (elongated up to 3 to 4 cm after 4 weeks of culturing) with lateral branches and were embedded within the medium surface in solid cultures. The roots were creamish and did not have extensive root hairs. The phenotypes remained stable over 3 years in vitro. The root lines did not differ in their morphology on the basis of branching pattern and lateral density. The non-transformed roots under similar cultural conditions showed poor growth with no branching and necrosed after 2 weeks.
Chromosome analysis of Ri transformed root lines
The chromosome number of ten randomly selected root lines of R. serpentina were studied and also compared with roots of in vitro grown plants. The somatic metaphase plates of the non-transformed roots as well as the transformed root lines showed 2n = 22 chromosomes which is the diploid chromosome number of the species [6]. The chromosomes are small in size (size range: <2.0 to < 4.0 μm). Out of the 22 chromosomes, 20 chromosomes had median to nearly median primary constriction and one pair of chromosomes had two constrictions: one constriction at the median region and the other at the sub terminal region. On the basis of Stebbins chromosome asymmetry indices, the karyotype falls into category 4A which defines a symmetrical karyotype. The karyotype of both non-transformed and transformed roots were similar (Table 1, Fig. 1). No variation was observed in chromosome morphology between transformed and non-transformed roots. These reports are consistent with several species in which hairy root cultures have been found to have chromosome number typical of the species. Aird et al. [1] obtained hairy root lines showing normal chromosome number after 6–18 months culture in the genera C. roseus, D. stramonium, Nicotiana rustica, N. umbratica, N. africana, Phaseolus vulgaris and Beta vulgaris. Similar observations were reported in Artemisia annua [25]. On the other hand variation in chromosome number has been observed in transformed root clones of Vicia faba [30] and Onobrychis viciaefolia [42]. Minor alteration in chromosomal structure was revealed by karyotype analysis of transformed roots of Lycopersicon peruvianum Mill. by Banerjee-Chattopadhyay et al. [7].
a–d Mitotic metaphase plates of Rauvolfia serpentina showing 2n = 22 chromosomes a, c: mitotic metaphase plate of Agrobacterium rhizogenes transformed root tip cell of R. serpentina and corresponding line diagram; b, d: mitotic metaphase plate of non-transformed root tip cell of R. serpentina and corresponding line diagram; bars represent 5 μm. ; (e,f) Idiogram of R. serpentina showing 2n = 22 (20 m + 2 m:st), e: A. rhizogenes transformed root line. f: non-transformed root; bar represents 1 μm
Molecular analysis of Ri transformed root lines
Ri transformed root lines were analyzed for presence of 18 ORFs of TL-DNA (ORF1 to ORF18) and five genes of TR-DNA (mas2, mas1, aux2, aux1, ags). All the amplified PCR products were of the expected size, and were identical to those of the positive control. None of the primers produced any amplification when the genomic DNA of the non-transformed roots (negative control) was used as template. None of the root lines showed complete integration of TL and TR-DNA. However, all transformed root lines showed integration of rolA (ORF10), rolB (ORF11) and rolD (ORF15) genes.
Based on the pattern of TL and TR gene integration the Ri transformed root lines (Fig. 2) can be divided into six types (Type I to VI) as shown in Table 2. Of the 19 Ri transformed root lines analyzed, complete integration of TL-DNA was noted in 11 root lines induced with strain A. rhizogenes LBA 9402 belonging to Type I and II, which differed in presence of mas1 and mas2 in Type I roots but not in Type II roots. It is noteworthy that none of the root lines showed presence of aux1 and aux2, which are part of TR T-DNA. Type III, IV and V did not show insertion of any of the five TR-DNA ORF analysed. Variable integration of TL-DNA was also characteristic of R. serpentina root lines. ORF1 was not detected in four root lines obtained following infection with strain LBA 9402 and two root lines obtained following infection by strain A4 [Type IV]. The root line RsA4 (L9) [Type V] showed absence of rolC (ORF12) gene, whereas in line RsA4 (L10) [Type VI] absence of T-DNA genes spanning ORF1 to ORF7 was noted. The insertion of rolB gene is the only absolute requirement for the production of hairy roots [27]. In some plants, such as tobacco, rolA, rolB and rolC genes are reported to induce roots when inserted alone but with decreased efficiency while, for the establishment of the full ‘hairy root syndrome’, rolA, rolB and rolC genes appear to act synergistically [11, 12, 34]. Alpizar et al. [2] reported the loss of rolA and rolD in many of the 55 hairy root lines studied in coffee.
a–e Agarose [1.2 % (w/v)] gel electrophoresis of PCR products with T-DNA ORF specific primers, a: ORF1, b: ORF12 (rolC), c: ORF 15 (rolD), d: mas1 and e: ags specific primers. Lane 1: molecular marker; Lane 2: positive control (pRiA4); Lane 3 negative control (genomic DNA from non-transformed root); Lanes 4–13: genomic DNA of A. rhizogenes transformed root lines RsIX2, RsIX3, RsIX4, RsIX5, RsIX6, RsIX8, RsIX9, RsIX10, RsIX11, RsIX14
There are very few reports on analysis of T-DNA ORFs in transformed root cultures as done in R. serpentina. Taneja et al. [36] have studied the presence or absence of all 23 of the ORFs of RiT-DNA in ten root lines of C. roseus. They reported the absence of several ORFs of TL-DNA in two root lines, including some ORFs that correspond to rol genes. All five of the ORFs of TR-DNA were found to be either entirely present or absent. However in R. serpentina fragmented insertion of TR-DNA was also detected.
In the present study, transformed root lines of R. serpentina showing similarity in morphology and chromosome characters, differed with respect to the T-DNA genes integrated in their genome. The presence of TR-DNA in addition to TL-DNA did not have any influence on the root morphology. Such observations supports the previous reports of Alpizar et al. [2] in which presence of different rol and aux genes, as established by PCR analysis, did not have any direct effect on root morphology of transformed roots of Coffea arabica. However, the presence of TR-DNA was found to be associated with the formation of rooty calli or other callus morphology in C. roseus [8] and Withania somnifera [5]. Majority of the hairy root lines are reported to be TL +/TR +, and very few are TL +/TR −, as in many plant species such as Duboisia hybrid, tobacco, D. metel [24], C. roseus [8, 36] and W. somnifera [5].
Variability in T-DNA content has been previously correlated with distinct morphological phenotypes in different species. However, in the present study no such correlation could be obtained as the hairy root lines of R. serpentina were morphologically identical irrespective of the A. rhizogenes strain used for transformation.
Ajmalicine content in Ri transformed root lines
The Ri transformed root lines showed significant variability (p ≤ 0.05) in ajmalicine content (0.004 ± 0.001 to 0.229 ± 0.014 mg g−1 DW) (Fig. 3) after 4 weeks of culture in solid modified MS medium. High accumulation of ajmalicine was obtained in the root lines RsIXA (0.229 ± 0.014 mg g−1 DW), RsIX10 (0.228 ± 0.014 mg g−1 DW) and RsIX11 (0.204 ± 0.02 mg g−1 DW). Variations in secondary metabolite content among different transformed root lines and plants regenerated from roots have been reported in many species [8, 13, 23, 24].
Ajmalicine content of roots excised from non-transformed in vitro plant (NTR) and A. rhizogenes transformed root lines after 4 weeks of culture on solid modified MS medium in the dark at 24 ˚C. Means with same letter were not significantly different at p ≤ 0.05 according to ANOVA and Tukey’s multiple comparison test
Thus, we can conclude that the Ri transformed root lines of R. serpentina are morphologically and cytologically stable however variable pattern of TL and TR-DNA genes integration as well as ajmalicine content was noted among the different root lines. Thus, genetically variant transformed root lines of R. serpentina with high ajmalicine content can be selected for scale up studies.
References
Aird ELH, Hamill JD, Rhodes MJC. Cytogenetic analysis of hairy root cultures from a number of plant species transformed by Agrobacterium rhizogenes. Plant Cell Tissue Organ Cult. 1988;15:47–57.
Alpizar E, Dechamp E, Lapeyre-Montes F, Guilhaumon C, Bertrand B, Jourdan C, et al. Agrobacterium rhizogenes-transformed roots of coffee (Coffea arabica): conditions for long-term proliferation and morphological and molecular characterization. Ann Bot. 2008;101:929–40.
Anonymous. The wealth of India: A dictionary of Indian raw materials and industrial products. New Delhi: CSIR; 2003.
Baíza AM, Quiroz-Moreno A, Ruíz JA, Loyola-Vargas VM. Genetic stability of hairy root cultures of Datura stramonium. Plant Cell Tissue Organ Cult. 1999;59:9–17.
Bandyopadhyay M, Jha S, Tepfer D. Changes in morphological phenotypes and withanolide composition of Ri-transformed roots of Withania somnifera. Plant Cell Rep. 2007;26:599–609.
Banerjee N, Sharma AK. Chromosome constitution and alkaloid content in Rauwolfia L. (Apocynaceae). Cytologia. 1989;54:723–8.
Banerjee-Chattopadhyay S, Schwemmin AM, Schwemmin DJ. A study of karyotypes and their alterations in cultured and Agrobacterium transformed roots of Lycopersicon peruvianum Mill. Theor Appl Genet. 1985;71:258–62.
Batra J, Dutta A, Singh D, Kumar S, Sen J. Growth and terpenoid indole alkaloid production in Catharanthus roseus hairy root clones in relation to left- and right termini-linked Ri T-DNA gene integration. Plant Cell Rep. 2004;23:148–54.
Benjamin BD, Roja G, Heble MR. Agrobacterium rhizogens mediated transformation of Rauvolfia serpentina: regeneration and alkaloid synthesis. Plant Cell Tissue Organ Cult. 1993;35:253–7.
Benjamin BD, Roja G, Heble MR. Alkaloid synthesis by root cultures of Rauwolfia serpentina transformed by Agrobacterium rhizogenes. Phytochemistry. 1994;35:381–3.
Blakesley D, Chaldecott MA. The role of endogenous auxin in root initiation Part II. Sensitivity and evidence from studies on transgenic plant tissues. Plant Growth Regul. 1993;13:77–84.
Cardarelli M, Spano L, Mariotti D, Mauro ML, Van Sluys MA, Constantino P. Identification of the genetic locus responsible for non-polar root induction by Agrobacterium rhizogenes. Plant Mol Biol. 1985;5:385–91.
Chaudhuri KN, Ghosh B, Tepfer D, Jha S. Genetic transformation of Tylophora indica with Agrobacterium rhizogenes A4: growth and tylophorine productivity in different transformed root clones. Plant Cell Rep. 2005;24:25–35.
Dellaporta SL, Woods J, Hicks JB. A plant DNA minipreparation: version 2. Plant Mol Biol Rep. 1983;1:19–22.
Ermayanti TM, McComb JA, O'Brien PA. Cytological analysis of seedling roots, transformed root cultures and roots regenerated from callus of Swainsona galegifolia (Andr.) R. Br. J Exp Bot. 1992;44:375–80.
Ermayanti TM, Octavia Y, Hafizh EA. Cytological analysis of root cultures of Artemissia cina. Ann Bogoriensesns. 2004;9:50–8.
Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50:151–8.
Georgiev MI, Pavlov AI, Bley T. Hairy root type plant in vitro systems as sources of bioactive substances. Appl Microbiol Biotechnol. 2007;74:1175–85.
Hänisch ten Cate CH H, Loonen AE, Ottaviani MP, Ennik L, van Eldik G, Stiekema WJ. Frequent spontaneous deletions of Ri T-DNA in Agrobacterium rhizogenes transformed potato roots and regenerated plants. Plant Mol Biol. 1990;14:735–41.
Hooykass PJJ, Klapwjik PM, Nuti MP, Schilperoort RA, Rorsch A. Transfer of the A. tumefaciens Ti plasmid to avirulent Agrobacteria and Rhizobium ex planta. J Gen Microbiol. 1977;98:477–84.
Levan A, Sandberg A, Fredga K. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–20.
Mano Y, Nabeshima S, Matsui C, Ohkawa H. Production of tropane alkaloids by hairy root cultures of Scopolia japonica. Agric Biol Chem. 1986;50:2715–22.
Mehrotra S, Goel MK, Rahman LU, Kukreja AK. Molecular and chemical characterization of plants regenerated from Ri-mediated hairy root cultures of Rauwolfia serpentina. Plant Cell Tissue Organ Cult. 2013;114:31–8.
Moyano E, Fornalé S, Palazòn J, Cusidò RM, Bonfill M, Piñol MC. Effect of Agrobacterium rhizogenes T-DNA on alkaloid production of Solanaceae plants. Phytochem. 1999;52:1287–92.
Mukherjee S, Das S, Jha S. Chromosomal stability in transformed hairy root cultures of Artemissia annua L. Cell Chromos Res. 1994;17:71–6.
Murashige T, Skoog F. A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant. 1962;15:473–97.
Nilsson O, Olsson O. Getting to the root: the role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiol Plant. 1997;100:463–73.
Panwar GS, Attitalla IH, Guru SK. An efficient in vitro clonal propagation and estimation of reserpine content in different plant parts of Rauwolfia serpentina L. Am-Eurasian J Sci Res. 2011;6:217–22.
Payne J, Hamill JD, Robins RJ, Rhodes MJC. Production of hyoscyamine by hairy root cultures of Datura stramonium. Planta Med. 1987;53:474–8.
Ramsay G, Kumar A. Transformation of Vicia faba cotyledon and stem tissues by Agrobacterium rhizogenes: infectivity and cytological studies. J Exp Bot. 1990;41:841–7.
Roychowdhury D, Ghosh B, Chaubey B, Jha S. Genetic and morphological stability of six-year-old transgenic Tylophora indica plants. Nucleus. 2013;56:81–9.
Roychowdhury D, Majumder A, Jha S. Agrobacterium rhizogenes mediated transformation in medicinal plants: prospects and challenges. In: Chandra S, Verma A (eds). Biotechnology for medicinal plants. Springer,2013;29-68.
Sokal RR, Rohlf FJ. Introduction to biostatistics. New York: WH Freeman; 1987.
Spena A, Schmülling T, Koncz C, Schell J. Independent and synergistic activity of RolA, B and C loci in stimulating abnormal growth in plants. Embo J. 1987;6:3891–9.
Stebbins GL. Chromosomal evolution in higher plants. London: Edward Arnold Publ Ltd; 1971. p. 49–113.
Taneja J, Jaggi M, Wankhede DP, Sinha AK. Effect of loss of T-DNA genes on MIA biosynthetic pathway gene regulation and alkaloid accumulation in Catharanthus roseus hairy roots. Plant Cell Rep. 2010;29:1119–29.
Villaine F, Casse-Delbart F. Independant induction of transformed roots by the TL and TR regions of the Ri plasmid of agropine type Agrobacterium rhizogenes. Mol Gen Genet. 1987;206:17–23.
Virmani OP, Popli SP, Misra LN, Gupta MM, Srivastava GN, Abraham Z, Singh AK. Dictionary of Indian Medicinal Plants. CIMAP, Lucknow: India;1992:387.
Vries-Uijtewaal ED, Gilissen LJW, Flipse E, Sree Ramulu K, De Groot B. Characterization of root clones obtained after transformation of monohaploid and diploid potato genotypes with hairy root inducing strains of Agrobacterium. Plant Sci. 1988;58:193–202.
Webb KJ, Jones S, Robbins MP, Minchin FR. Characterization of transgenic root cultures of Trifolium repens, Trifolium pratense and transgenic plants of Lotus corniculatus. Plant Sci. 1990;70:243–54.
White FF, Taylor BH, Huffman GA, Gordon MP, Nester EW. Molecular and genetic analysis of the transferred DNA regions of the root inducing plasmid of Agrobacterium rhizogenes. J Bacteriol. 1985;164:33–44.
Xu Z-Q, Jia J-F. The reduction of chromosome number and the loss of generation ability during subculture of hairy root cultures of Onobrychis viciaefolia transformed by Agrobacterium rhizogenes. Plant Sci. 1996;120:107–12.
Zarco CR. A new method for estimating karyotype asymmetry. Taxon. 1986;35:526–30.
Acknowledgments
Smita Ray thanks University Grants Commission, for the award of the Minor Research Project No. F. PSW-117/12–13 (ERO) S. No. 214374 and Head, Department of Botany, Calcutta University for facilities provided.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ray, S., Samanta, T., Majumder, A. et al. Cytogenetic characterization of Agrobacterium rhizogenes transformed root lines of Rauvolfia serpentina . Nucleus 57, 105–112 (2014). https://doi.org/10.1007/s13237-014-0112-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-014-0112-1