Abstract
A total of 2257 lactic acid bacteria were preliminarily screened for antagonistic activity against Lactobacillus sakei subsp. sakei JCM 1157. Strain SKI19 was selected and identified at the subspecies level as Lactobacillus plantarum subsp. plantarum SKI19, using 16S rRNA gene sequence analysis combined with recA and dnaK genes’ amplification. Antibacterial activity of SKI19 was completely lost after treatment of neutralized cell free culture supernatant with proteolytic enzymes, suggesting that SKI19 produced a bacteriocin-like substance that inhibited not only closely related species, but was also effective against Listeria monocytogenes DMST 17303. Viewed under scanning electron microscope, cell membranes of the indicator strain appeared to collapse after exposure to the bacteriocin-like substance. In vitro tests concerning probiotic properties, SKI19 survived under simulated gastrointestinal tract conditions, and adhesion of its cell surface to xylene and chloroform was 90.14 and 89.85%, respectively. Complete inhibition by SKI19 against pathogenic bacteria (Escherichia coli DMST 4212, L. monocytogenes DMST 17303, and Staphylococcus aureus DMST 8840) was observed in co-cultivation under anaerobic conditions. A safety assessment showed that SKI19 was susceptible to several antibiotics and had no haemolytic activity. PCR amplification of virulence factors with the specific primers for ace, asa1, cylLS, efaAfs, hyl, and gelE genes were negative for SKI19. Also, SKI19 did not harbor any hdc, tdc, odc or ldc genes involved in biogenic amine production. The results reveal that SKI19 has probiotic potential and antibacterial activity, and is safe for further application in certain food products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactic acid bacteria (LAB), used in fermenting food, have the “generally regarded as safe” (GRAS) status, and are found in natural environments (Belicova et al. 2013). Physiological characteristics principally associated with them are substrate utilization, metabolic capabilities, and probiotic properties. Besides producing lactic acid during fermentation, LAB also release antimicrobial substances including bacteriocins. These have gained much attention as safe biopreservatives, since they are degraded by protease in the gastrointestinal tract (GI). These antimicrobial compounds are ribosomally synthesized and extracellularly released low molecular mass peptides or peptide complexes (usually 30–60 amino acids) having bactericidal or biostatic activity. Some bacteriocins appear to inhibit different indicator bacteria, including food poisoning microorganisms, foodborne pathogenic bacteria, and closely related species (Swetwiwathana and Visessanguan 2015). Furthermore, LAB have been qualified as potential probiotics, which is of interest because of health and safety concerns that require development of food products that promote healthy well-being beyond basic nutrition. Probiotics are defined as “Live microorganisms which when administered in adequate amount confer a health benefit on the host” (FAO/WHO 2002). In order to be qualified as a candidate probiotic, a strain should be screened both for its essential functional properties (resistance to gastric acidity and bile salt, antagonistic effect against pathogenic bacteria, and adhesion to gut tissues), and for its safety properties (antibiotic resistance, biogenic amine production, virulence factors, and haemolytic activity) (Belicova et al. 2013). Use as probiotics in fermented dairy or sausage products has been documented for Lactobacillus acidophilus, Lactobacillus casei, Lactobacillus reuteri, Lactobacillus plantarum, Pediococcus pentosaceus, and Bifidobacterium longum (Vuyst et al. 2008).
Sai krok e-san mu is a traditional Thai fermented pork sausage, basically made of pork, cooked rice, garlic, salt, and sugar. Various microorganisms, predominantly LAB, are involved in the fermentation. Visessanguan et al. (2015) found L. plantarum, Lactobacillus pentosus, Lactobacillus sakei, and P. pentosaceus, as predominant in meat fermentation. However, meat products fermented by naturally occurring LAB can potentially be contaminated by pathogenic bacteria such as Salmonella spp., Staphylococcus aureus, Bacillus cereus, Escherichia coli, and Listeria monocytogenes (Paukatong and Kunawasen 2001). Chemical preservatives inhibit harmful bacteria, but long-term exposure to artificial preservatives may adversely impact the consumer’s health. This suggests that bacteriocin-producing strains could be used as natural preservatives that inhibit the growth of and the spoilage by pathogenic microorganisms, and the strains’ probiotic properties could allow their use as starter cultures.
The objectives of the present study were to screen LAB isolated from sai krok e-san mu. A chosen strain was further identified and preliminarily characterized for antibacterial compounds and for inhibitory spectrum. Its probiotic potential (survivability in the gut environment, bacterial adhesion to hydrocarbons, and co-cultivation) was also evaluated. Haemolysis, susceptibility to antibiotics, presence of virulence factors, and biogenic amines were also examined, to determine its safety for probiotic use.
Materials and methods
Bacterial strains and growth media
The genetically closely related species used as indicator strains, namely L. sakei subsp. sakei JCM 1157, L. plantarum TISTR 875, and P. pentosaceus TISTR 413, were purchased from the Japan Collection of Microorganisms (JCM) and the Thailand Institute of Scientific and Technological Research (TISTR), respectively. Enterococcus faecium CKF12 and E. lactis CKF16 were from our own cultured stocks, whereas E. faecalis CM6CR07 and E. faecalis CF1GI14 were kindly provided by Dr. Noraphat Hwanhlem (Hwanhlem et al. 2017). These indicator strains were cultured in de Man, Rogosa and Sharpe (MRS) medium under aerobic conditions at 37 °C. The food spoilage and foodborne pathogenic bacteria used in this research were based on the Thai Community Product Standards. All of them, including Gram-positive and Gram-negative bacteria, were purchased from the Department of Medical Science, Ministry of Public Health, Thailand (DMST). E. coli DMST 4212 was cultured in Luria–Bertani (LB) medium. B. cereus DMST 5040 and S. aureus DMST 8840 were cultured in nutrient broth (NB). Clostridium perfringens DMST 16637 was cultured in Thioglycolate broth (TB) under anaerobic conditions (HiMedia, India). L. monocytogenes DMST 17303 and Salmonella were cultured in trypticase soy broth (TSB). The strains were cultured for 18–24 h at 37 °C and sub-cultured at least twice prior to the experiments. All strains used in this study were maintained as stock cultures at − 80 °C in 30% glycerol.
Isolation and screening of bacteriocin-producing LAB
Ten freshly prepared sai krok e-san mu samples were bought from local markets in Songkhla province. Samples (25 g) of sai krok e-san mu were added to 225 ml of normal saline solution (0.85% NaCl). Appropriate decimal dilutions (105–106 CFU/ml) were prepared in normal saline solution and poured onto molten MRS agar (45 °C) containing 1% CaCO3. The plates were incubated at 37 °C for 2–3 days under microaerophilic conditions. The LAB strains that showed clear zones were picked for re-streaking three times to obtain single colonies. Antagonistic activity detection was performed by the agar spot assay. Briefly, 5 µl of overnight cultures in MRS broth were spotted onto bacteriocin screening medium and incubated at 37 °C for 24 h under anaerobic conditions. LAB colonies were overlaid with 10 ml of semi-soft MRS (0.75% agar) containing L. sakei subsp. sakei JCM 1157 (106 CFU/ml). Those LAB colonies that showed clear zones of growth inhibition were then examined by microscopy. Isolates that exhibited Gram-positive, catalase-negative, and non-motile cells were selected for further investigation. Production of antibacterial substances by the selected LAB strains was determined by the drop plate method. Overnight culture of LAB was centrifuged at 10,000×g for 10 min at 4 °C and cell-free culture supernatant was filtered through a 0.22 µm filter. Inhibition was observed by spotting 10 µl of the supernatant onto semi-soft agar seeded with L. sakei subsp. sakei JCM 1157, and then incubating anaerobically overnight at 37 °C. That LAB strain showing the maximal inhibition zone of the indicator strain was selected for further identification and characterization.
Identification of selected LAB strain SKI19
The bacterial cells were extracted using the TIANamp Genomic DNA kit (TIANGEN, China), following the manufacturer’s instructions. The primers and the PCR conditions used for the amplification of 16S rRNA, recA, and dnaK genes are summarized in Table 1. The PCR products were analyzed by electrophoresis in 1% agarose gel and were directly visualized under ultraviolet light. The amplified PCR products were purified by using a PCR purification kit (Vivantis, Malaysia), following the manufacturer’s instructions prior to sequencing (1st BASE, Malaysia). The obtained sequences were edited by BioEdit Sequence Alignment Editor V 7.2.1 program, and were compared to sequences in the GenBank database using the Basic Local Alignment Search Tool (BLAST). Phylogenetic trees were constructed using the neighbor-joining method in the Molecular evolutionary genetics analysis (MEGA 7.0) software.
Characterization of antibacterial compounds and inhibitory spectrum
SKI19 was cultured in MRS broth (100 ml) and incubated at 37 °C for 18–24 h. Then, the entire culture broth was centrifuged at 10,000×g for 10 min at 4 °C. The supernatant obtained was used in the experimental trials as follows: (1) the cell free supernatant (CFS) was used directly; (2) the CFS was neutralized to pH 6.5 using 6 N NaOH to exclude the antimicrobial effect of organic acids (NCFS); (3) the NCFS was treated with catalase (200 unit/ml) to eliminate the inhibitory effect of hydrogen peroxide (H2O2) and incubated at 37 °C for 1 h (NCFSC); and (4) proteolytic enzymes were added to NCFSC, including α-chymotrypsin, trypsin, proteinase K, or pronase E (1 mg/ml), and incubated at 37 °C for 3 h (NCFSC-proteolytic enzymes). All the samples were filter-sterilized (0.22 µm) and heated for 5 min at 90 °C before testing. Antibacterial activity spectrum was determined using an agar well diffusion assay. Soft agar media (1% agar, 45 °C) were first seeded with all the indicator strains (106 CFU/ml) and were poured into sterile petri dishes. Aliquots (50 µl) of each treatment were dispensed into each well and incubated at 37 °C for 24 h. Fifty microliters of MRS broth was applied as the control treatment, and the result reported is the diameter of inhibition zone, measured using a Vernier caliper.
Scanning electron microscopy (SEM)
Bacterial cells for SEM were prepared as described by Khan and Kang (2016) with minor modifications. Briefly, L. monocytogenes DMST 17303, as the test indicator, was cultured in TSB broth at 37 °C for 7 h to approximately 106 CFU/ml. Thereafter, the bacterial cells were treated with and without NCFS at equal proportions for 2 h. Then, the cells were centrifuged at 5000×g for 15 min at 4 °C and washed three times using 0.1 M phosphate buffer saline (PBS, pH 7.2). A thin smear was prepared on a glass slide and fixed in 2.5% glutaraldehyde for 1 h. Then the fixed samples were washed twice with 0.1 M PBS (pH 7.2) followed by washing twice with sterile distilled water for 15 min each time. In the next step, dehydration was performed using an ethanol series from 50 to 100%, with further drying using CO2 at 4 °C. The dried cells were sputter coated with gold and observed under SEM (Quanta 400, Czech Republic).
Tolerance to simulated human gastrointestinal tract (GI)
Survival of SKI19 under simulated GI tract conditions was assessed as described by Yu et al. (2013), with slight modifications. Freshly prepared culture was centrifuged at 5000×g for 5 min at 4 °C, and washed twice with 0.1 M PBS (pH 7.0). To simulate the hydrolysis of bacterium in the human oral cavity, the obtained cell pellets were resuspended in a sterile electrolyte solution (SES) containing 100 µg/ml lysozyme, and incubated at 37 °C for 10 min. To test for gastric juice tolerance, the bacterial suspension was centrifuged and washed twice with 0.1 M PBS (pH 7.0). Then, it was re-suspended in 0.1 M PBS (pH 2.5) containing 0.3% pepsin and incubated at 37 °C in a water bath for 4 h. Subsequently, the tolerance to the simulated small intestine was assessed. The bacterial suspension was centrifuged and washed twice with 0.1 M PBS (pH 7.0), before re-suspension in 0.1 M PBS (pH 8.0) containing 0.1% porcine pancreatin (Sigma, Germany) and 0.3% oxgall bile salt (Merck, Germany), and it was then incubated at 37 °C for 4 h. For cell counting, the sample was diluted and spread on MRS agar plates containing 0.01% bromocresol purple (Ajax Finechem, Australia). Survival counts for gastric tolerance and for intestinal tolerance were determined at the initial time (0 h) and at 1, 2, 3 and 4 h of exposure. The values are expressed as log CFU/ml.
Bacterial adhesion to hydrocarbons (BATH)
Bacterial adhesion to hydrocarbons was determined as described by Pieniz et al. (2015). Briefly, SKI19 was harvested by centrifugation at 5000×g for 5 min at 4 °C. The cells were washed twice and suspended in 10 mM PBS (pH 7.2). Initial OD600 nm (A0) was adjusted to 0.5 for optimizing the bacterial cells to 107 CFU/ml. Then, an equal volume of either xylene or chloroform was added to the bacterial cells and vortexed well for 5 min. The two phases were separated after incubation at room temperature for 1 h. The aqueous phase was carefully removed and measured at OD600 nm. The percentage of BATH was calculated according to the formula below:
A0 is the absorbance before mixing with xylene or chloroform, A is the absorbance after mixing with xylene or chloroform.
Competitiveness of SKI19 in co-cultivation with pathogenic bacteria
Co-cultivation of SKI19 with E. coli DMST 4212, S. aureus DMST 8840, and L. monocytogenes DMST 17303, was assessed as described by Hongpattarakere et al. (2012). The co-cultivations were evaluated in a minimal medium supplemented with 1% glucose (pH 6.5). For each experiment, the strains were inoculated in equal proportions of 105 CFU/ml for the co-cultures and for the monocultures that were used as controls. Each sample was incubated at 37 °C for 48 h under anaerobic conditions. After 0, 6, 12, 18, 24, 36, and 48 h of incubation, samples were collected to analyze for pH, SKI19, and remaining pathogenic bacteria. Ten-fold serial dilutions were spread-plated onto selective agars for enumeration of each individual strain. Growth of SKI19 was enumerated on MRS agar, while the enumerations of the pathogenic bacteria (E. coli DMST 4212, S. aureus DMST 8840, and L. monocytogenes DMST 17303) were determined on violet red bile agar, Bird-Parker agar, and PALCAM agar (HiMedia, India), respectively. The results are expressed as logCFU/ml.
Haemolytic activity
Fresh bacterial culture was streaked on TSA agar containing 5% human blood (Songklanakarin hospital, Thailand), and incubated at 37 °C for 48 h. The haemolytic reaction was labeled by observation of: clear zones around the colonies (β-haemolysis); green-hued zones around colonies (α-haemolysis); or no clear zone around the colonies (γ-haemolysis). The γ–haemolysis was considered as a negative case with missing haemolytic activity (Pieniz et al. 2015).
Susceptibility to antibiotics
Antibiotic susceptibility of SKI19 was determined according to the technical guidelines of the European Food Safety Authority (EFSA 2012), using the broth microdilution method. Briefly, SKI19 colonies were suspended in 10 ml sterile saline solution to a turbidity of 1.0 on McFarland scale, and diluted 500-fold in LSM medium (Iso-sensitestbroth:MRS, 9:1). Fifty-microliter aliquots of the diluted bacterial suspension were added to each well (containing the various antibiotic test concentrations in each 50 µl volume of LMS broth per well). The antibiotics tested in the concentration range 0.125–256 µg/ml were ampicillin, chloramphenicol, clindamycin, erythromycin, gentamicin, kanamycin, penicillin G, polymycin B, rifampicin, and tetracycline. SKI19 inoculated in MRS broth was used as a positive control, and bacteria-free case was used as a negative control. After 24 h incubation at 37 °C, the minimum inhibitory concentration (MIC) was defined as the lowest antibiotic concentration that inhibited visual bacterial growth. The growth inhibition of SKI19 was measured by OD600 nm using a microplate reader (Biotex, USA). The breakpoints (cutoff values) were used to distinguish susceptible and resistant strains. The strain with higher MIC than the respective cutoff value was considered resistant to an antibiotic.
Detection of virulence and biogenic amine genes
The presence of virulence factors was checked by PCR. The virulence genes detected were ace (encodes the adhesion collagen protein), asa1 (encodes the aggregation substance), efaAfs (encodes the cell wall adhesion), hyl (encodes the hyaluronidase), cylLS (encodes the cytolysin structural subunit), and gelE (encodes the protease GelE with gelatinase activity). The presence of biogenic amine genes encoding the decarboxylase enzymes involved in biogenic amine production was also investigated. The genes analyzed were hdc, encoding for histidine decarboxylase; tdc, encoding for tyrosine decardoxylase; odc, encoding for ornithine decarboxylase; and ldc, encoding for lysine decarboxylase. The amplified products were resolved by electrophoresis through a 1% agarose gel. The primer sequences and the PCR conditions are given in Table 1.
Statistical analysis
Statistical analysis was performed using SPSS statistical software (version 15.0; SPSS, Inc., Chicago, IL). One-way analysis of variance (ANOVA) was considered statistically significant at P < 0.05. All data are presented as mean ± standard deviation.
Results and discussion
Isolation and screening of potential bacteriocin-producing LAB
A total of 2257 LAB strains were directly isolated from 10 sai krok e-san mu samples. However, in the primary screening by the agar spot assay only 46 colonies produced inhibition zones against L. sakei subsp. sakei JCM 1157, with the radius ranging from 9 to 17 mm. Further investigation by the drop plate method revealed 16 strains of LAB with the radius of inhibition zone larger than 7 mm (data not shown). Of these isolates, SKI19 exhibited the largest 12 mm inhibition zone. Therefore, the strain SKI19 was selected for further experiments.
Identification of selected LAB strain SKI19
SKI19 is a rod-shaped, catalase negative, non-spore-forming, and Gram-positive bacterium (data not shown). Based on the 16S rRNA gene sequence, the strain was identified as L. plantarum, due to 99% sequence similarity in the GenBank database (the GenBank accession number is KY762263). However, the 16S rRNA gene sequence may not be sufficient to distinguish among the species L. plantarum, L. pentosus, and L. paraplantarum, since these species are closely related and share highly homologous phenotypes (Torriani et al. 2001) (Fig. 1a). The recA gene could be used to distinguish among them, and it had sequence similarity to L. plantarum subsp. plantarum (the GenBank accession number is KY762265) (Fig. 1b). The phylogenetic tree based on the dnaK gene obviously confirmed the distinction of SKI19 from L. plantarum subsp. argentoratensis, and indicated 99% homology to L. plantarum subsp. plantarum (the GenBank accession number is KY762264) (Fig. 1c). Thus, our data verified that the sequences of recA and dnaK genes provided resolution at a high discrimination level, particularly the dnaK gene. This suggests the dnaK gene sequence could be applied as a phylogenetic marker for accurate and rapid classification of L. plantarum strains at the subspecies level. This result is in agreement with Huang et al. (2010), where the dnaK gene sequence clearly distinguished between the phenotypically closely related species L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis, L. paraplantarum, L. pentosus, and L. fabifermentans. Therefore, based on 16S rRNA gene sequence analysis combined with recA and dnaK genes’ amplification, SKI19 was potentially identified as L. plantarum subsp. plantarum SKI19.
Phylogenetic trees were constructed by the neighbor-joining method, kimura’s two-parameter model. SKI19 had 99% nucleotide homology with L. plantarum strains and L. plantarum subsp. plantarum strains. E. coli strain BL21, P. acidilactici, and L. brevis KB290 were chosen as an out group for 16S rRNA, recA, and dnaK genes, respectively
Characterization of antibacterial compounds and inhibitory spectrum
The inhibitory spectrum of antibacterial compounds produced by SKI19 was assayed by the agar well diffusion method summarized in Table 2. The CFS in direct use exhibited inhibitory activity against the closely related bacteria Enterococcus spp. and L. sakei subsp. sakei JCM 1157. Also, it inhibited food spoilage and foodborne pathogenic bacteria, including Gram-positive bacteria (B. cereus DMST 5040, C. perfringens DMST 1663, L. monocytogenes DMST 17303, and S. aureus DMST 8840) and Gram-negative bacteria (E. coli DMST 4212, S. Typhimurium DMST 15674, and S. Enteritidis DMST 15676). The inhibition is probably due to the production of antibacterial compounds, like lactic and acetic acids, diacetyl, hydrogen peroxide (H2O2), bacteriocin, or their combination. After excluding the effects of organic acids (NCFS) and H2O2 (NCFSC), SKI19 showed unchanged activity in inhibiting the closely related bacteria. This indicates that SKI19 excreted a bacteriocin-like inhibitory substance (BLIS) (Aslim et al. 2005). Moreover, the BLIS was also effective against the foodborne bacterium L. monocytogenes DMST 17303, since the class IIa bacteriocins include bacteriocin-like Listeria-active peptides with a conserved N-terminal sequence Tyr–Gly–Asn–Gly–Val, and two cysteines forming a disulfide bridge in the N-terminal half of the peptide (Cleveland et al. 2001). Thus, our results indicate that the anti-Listeria organism in the inhibition region is predominantly this BLIS. In addition, the BLIS did not inhibit the tested probiotic strains such as L. plantarum TISTR 875 and P. pentosaceus TISTR 413, suggesting an interesting technological application in fermented foods for control of Listeria without affecting probiotic cultures (Barbosa et al. 2014). However, no inhibition by BLIS was observed for Gram-negative bacteria, due to their supplementary layers that make the outer membrane act as a permeability barrier hindering bacteriocin action (Gong et al. 2010). A similar observation has been reported regarding plantaricin NC8 (Maldonado et al. 2003) produced by L. plantarum NC8, which appeared to inhibit only Gram-positive bacteria. In contrast, plantaricin MG (Gong et al. 2010) was active against both Gram-positive and Gram-negative bacteria. This difference may depend on the bacteriocin-producing strain (Powell et al. 2007). Complete inactivation of the BLIS by treatments with proteolytic enzymes, including α-chymotrypsin, trypsin, proteinase K, and pronase E, provides preliminary affirmation that SKI19 is a bactericocin-producing strain.
Observation of changes in target bacterial cell morphology viewed by SEM
The SEM study allowed observation of the irregular cell morphology of anti-Listeria bacteriocin SKI19 treated L. monocytogenes DMST 17303, with untreated L. monocytogenes DMST 17303 serving as a control. The cell surfaces of the control were smooth and intact, with regular appearance (Fig. 2a), while membrane of the indictor strain appeared shrunken, corrugated, and changed in shape, after treatment at MIC (100 µl) of anti-Listeria bacteriocin SKI19 for 2 h, indicating that the cell wall probably was destroyed. Pores appeared in the cells, which led to cytoplasm leakage resulting in cell death (white arrows) (Fig. 2b). This provides evidence that the bactericidal activity exerted by anti-Listeria bacteriocin SKI19 was associated with damage to the cell membranes, as it destroyed the integrity of the membrane and caused subsequent cell lysis. This matches prior observations of other bacteriocins (Khan and Kang 2016).
Survival in simulated human gastrointestinal tract
Tolerance of SKI19 to the gastrointestinal tract was assessed by simulating exposure to gastric acid and passage through the small intestine by incubation at 37 °C for 4 h. In the presence of simulated gastric juice at pH 2.5, it was observed that SKI19 initially showed no obvious difference (P < 0.05) in cell count to the control. After incubation for 2, 3, and 4 h, the cell viability had declined to 8.13, 7.12 and 6.84 log CFU/ml, respectively. In the presence of simulated small intestinal juice at pH 8.0, the viable cell counts were approximately 3 log units below the control (6.60 log CFU/ml), and remained stable over the observation time span. Gobbetti et al. (2010) theorized that a potentially probiotic microorganism with benefits to the consumer’s health must reach the intestine with a large number of viable cells, between 105 and 106 CFU/ml. The pH in the stomach before a meal is typically between 2.5 and 3.5, and after a meal about 4.5 (Huang and Adams 2004). Our results were satisfactory in that SKI19 survived both simulated gastric and intestinal juices, and therefore might be appropriate for future development as a probiotic.
Hydrophobicity
The hydrophobicity of SKI19 was evaluated using two solvents. Xylene was used as an apolar (bacterial adhesion), while chloroform was considered a monopolar acid, an electron acceptor to the bacteria. SKI19 showed high adhesion to xylene at 90.14% whereas the cell wall adhered to acidic chloroform at 89.85%. The hydrophobic components of bacteria are generally present in their outer membrane and act hydrophobically due to net negative surface charge. These hydrophobic interactions are essential in the adhesion of bacteria to epithelial cells. The hydrophobicity of the cell surface could be affected by the growth medium, the age of the bacteria, the bacterial surface structures, and the composition changes of proteins from environmental stresses (Kaushik et al. 2009). Our results reveal that SKI19 has potential to adhere to and colonize on gut epithelial cells in the human intestine.
Inhibitory activity of SKI19 in co-cultivation with pathogenic bacteria
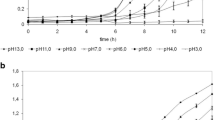
Probiotic strains inhibit pathogens by consuming nutrients that the pathogens need. This inhibition depends on the rate of nutrient absorption, the inherent metabolic velocity, the growth rate, and the excretion of specific inhibitors (Balcázar et al. 2006). SKI19 showed strong inhibitory effects against E. coli DMST 4212 and S. aureus DMST 8840, as the pathogen cell counts fell rapidly by approximately 3 log units at 24 h, and further to below detection limit after 36 h of incubation, and the pH level dropped to approximately 3.6 in these co-culture studies (Fig. 3a, b). The inhibition of those pathogens likely coincided with the decreased pH level. This observation correlates with the characterization of antibacterial compounds, as in Table 2, where organic acids are the major agents contributing to the antibacterial effect. Kongnum and Hongpattarakere (2012) reported that L. plantarum MR03.12 completely inactivated Vibrio harveyi to no detection around 18–24 h of co-culture. Also, L. plantarum or L. brevis totally inactivated Salmonella spp. in mixed cultures after 24 h or 48 h, respectively (Szala et al. 2012), indicating that organic acids produced by LAB are the main factors inhibiting the growth of Gram-negative bacteria, as observed also in our study. Moreover, SKI19 exhibited highly significant inhibitory effects against L. monocytogenes DMST 17303. The cell concentration sharply decreased by about 3 log units at 18 h to complete inhibition after 24 h of incubation in the co-culture (Fig. 3c). This reveals that the inhibition of SKI19 towards L. monocytogenes DMST 17303 is probably caused by a combination of antibacterial compounds, as mentioned in Table 2. SKI19 growth in the co-culture remained steady until the end of fermentation, while SKI19 and pathogen cell concentrations in monoculture remained at constant levels (106–108 CFU/ml) throughout the experiment. Our results thus clearly highlight that SKI19 has a very high competitive ability against foodborne pathogenic bacteria, which is pertinent to its potential application in food products.
Growth of E. coli DMST 4212 (a), S. aureus DMST 8840 (b), and L. monocytogenes DMST 17303 (c) in co-culture system.
 Pathogen alone;
Pathogen alone;
 SKI19 alone;
SKI19 alone;
 Pathogen in co-culture;
Pathogen in co-culture;
 SKI19 in co-culture;
SKI19 in co-culture;
 pH in co-cultivation. The results (Mean ± S.D) of three experiments with lowercase letters (a–f) indicate a significant difference (P <0.05)
pH in co-cultivation. The results (Mean ± S.D) of three experiments with lowercase letters (a–f) indicate a significant difference (P <0.05)
Haemolytic activity
The absence of haemolytic activity is required for safety of probiotic strains (FAO/WHO 2002). In the present study, SKI19 showed no haemolysis of blood cells (γ-hemolysis, Fig. 2c). Our result is consistent with the study of Oh and Jung (2015), in which L. plantarum and Pediococcus strains exhibited no haemolytic activity.
Susceptibility to antibiotics
Antibiotic resistance is assessed relative to the MIC breakpoints as given by EFSA (2012). SKI19 was susceptible to ampicillin and penicillin that inhibit cell wall synthesis. However, resistance to polymixin B was observed in this study. Susceptibility to protein synthesis inhibitors was observed for chloramphenicol, clindamycin, erythromycin, gentamicin, and tetracycline, but not for kanamycin. Also, SKI19 was sensitive to rifampicin that inhibits nucleic acid synthesis. In a similar study by Yu et al. (2013), antibiotic resistance to cell wall (polymyxin B) and protein synthesis inhibitors (kanamycin) was observed in L. plantarum strains. Most lactobacilli carry natural or intrinsic resistance to certain antibiotics. The various mechanisms involved in intrinsic resistance to aminoglycosides lead to absence of cytochrome-mediated electron transport, thus preventing uptake of antibiotics. However, the probiotics with intrinsic antibiotic resistance, like resistance to glycopeptides and aminoglycosides that are antibiotics regularly administered to humans, could be beneficial to maintain the gastrointestinal tract balance and avoid antibiotic-induced diarrheas (Fraqueza 2015). It seems reasonable to conclude that consumption of SKI19 does not present a health risk to humans due to its antibiotic resistance.
Detecting the presence of virulence and biogenic amine genes
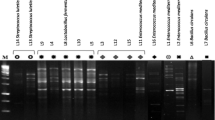
Although LAB are food-grade organisms with a long history of safe use, potential virulence factors or capacity to produce toxic compounds (such as biogenic amines) must be considered and assessed. To guarantee the safe use of SKI19 as either probiotic or starter culture, the absence of virulence and biogenic amine genes must be proven. SKI19 did not harbor any of the virulence genes (ace, asa1, cylLS, efaAfs, hyl, and gelE), whereas the expected PCR products were observed for E. faecalis CM6CR07 and E. faecalis CF1GI14 used as positive controls (Fig. 4a, b). This agrees with the results of Landeta et al. (2013) who detected none of these virulence genes in L. plantarum strains. The presence of virulence factors causes various risks of infections. The ace gene affects adhesion to collagen and is expressed conditionally after growth in serum. The efaAfs gene is expressed for cell wall adhesions in serum (H-Kittikun et al. 2015). The asa1 gene is located on pheromone-responsive plasmids and contributes to the severity of endocarditis in animal models. Cytolysin coding gene (cylLS) is a bacterial toxin expressed by haemolysis and contributes to the severity of enterococcal disease in humans. The gelE gene has been shown to exacerbate endocarditis in an animal model. The hyl gene is associated with invasive disease (Vankerckhoven et al. 2008).
The presence of biogenic amines in foods is considered potentially unsafe to human health. SKI19 was PCR-negative for the hdc, tdc, odc, and ldc genes tested, whereas the expected PCR products were observed for E. faecium CE5-1 (odc and tdc genes) and S. aureus DMST 8840 (ldc gene) used as positive controls (Fig. 4c). An analysis without the positive control showed absence of hdc gene. This result agrees with what was reported by Landeta et al. (2013), namely the absence of biogenic amine genes in L. plantarum strains. However, some L. plantarum strains (CK06, LM11, and ZS11) were found to decarboxylate tyrosine into tyramine. Histamine and tyramine cause toxic effects by their vasoactive and psychoactive properties and are the most common biogenic amines present in fermented foods and starter cultures, in which the presence, activity, and specificity of decarboxylases are strain-specific (Belicova et al. 2013). Putrescine and cadaverine cause toxic effects due to their relevant biogenic amines by histamine and tyramine (Perin et al. 2014). Our results prove that SKI19 is safe for consumption.
Conclusion
Lactobacillus plantarum subsp. plantarum SKI19 was isolated from sai krok e-san mu, a traditional Thai fermented pork sausage, and was found to have BLIS acting against closely related bacteria as well as the foodborne pathogenic bacterium L. monocytogenes DMST 17303. An evaluation of potential probiotic properties and safety characteristics demonstrated that SKI19 is a candidate probiotic for application in certain food products. However, further studies are necessary to characterize its BLIS, with a view to the biopreservation of meat products and related foods.
References
Aslim B, Yuksekda ZN, Sarikaya E, Beyatli Y (2005) Determination of the bacteriocin-like substances produced by some lactic acid bacteria isolated from Turkish dairy products. LWT Food Sci Technol 38:691–694
Balcázar JL, Decamp O, Vendrell D, Blas ID, Ruiz-Zarzuela I (2006) Health and nutritional properties of probiotics in fish and shellfish. Microb Ecol Health Dis 18:65–70
Barbosa MS, Todorov SD, Belguesmia Y, Choiset Y, Rabesona H, Ivanova IV, Chobert JM, Haertlé T, Franco BDGM (2014) Purification and characterization of the bacteriocin produced by Lactobacillus sakei MBSa1 isolated from Brazilian salami. J Appl Microbiol 116:1195–1208
Belicova A, Mikulasova M, Dusinsky M (2013) Probiotic potential and safety properties of Lactobacillus plantarum from Slovak bryndza cheese. Biomed Res Int. https://doi.org/10.1155/2013/760298
Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: safe, natural antimicrobials for food preservation. Int J Food Microbiol 71:1–20
de las Rivas B, Marcobal A, Carrascosa AV, Muñoz R (2006) PCR detection of foodborne bacteria producing the biogenic amines histamine, tyramine, putrescine, and cadaverine. J Food Prot 69:2509–2514
Eaton TJ, Gasson MJ (2001) Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl Environ Microbiol 67:1628–1635
EFSA (2012) Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740
FAO/WHO (2002) Guidelines for the evaluation of probiotics in food. London, Ontario, Canada, April 30 and May 1. http://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf. Accessed 16 July 2017
Fraqueza MJ (2015) Antibiotic resistance of lactic acid bacteria isolated from dry-fermented sausages. Int J Food Microbiol 212:76–88
Gobbetti M, Di Cagno M, De Angelis M (2010) Functional microorganisms for functional food quality. Crit Rev Food Sci Nutr 50:716–727
Gong HS, Meng XC, Wang H (2010) Plantaricin MG active against Gram-negative bacteria produced by Lactobacillus plantarum KLDS1.0391 isolated from “Jiaoke”, a traditional fermented cream from China. Food Control 21:89–96
H-Kittikun A, Biscola V, El-Ghaish S, Jaffrès E, Dousset X, Pillot G, Haertlé T, Chobert JM, Hwanhlem N (2015) Bacteriocin-producing Enterococcus faecalis KT2W2G isolated from mangrove forests in southern Thailand: purification, characterization and safety evaluation. Food Control 54:126–134
Hongpattarakere T, Cherntong N, Wichienchot S, Kolida S, Rastall RA (2012) In vitro prebiotic evaluation of exopolysaccharides produced by marine isolated lactic acid bacteria. Carbohydr Polym 87:846–852
Huang Y, Adams MC (2004) In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int J Food Microbiol 91:253–260
Huang CH, Lee FL, Liou JS (2010) Rapid discrimination and classification of the Lactobacillus plantarum group based on a partial dnaK sequence and DNA fingerprinting techniques. Antonie Van Leeuwenhoek 97:289–296
Hwanhlem N, Ivanova T, Biscola V, Choiset Y, Haertlé T (2017) Bacteriocin producing Enterococcus faecalis isolated from chicken gastrointestinal tract originating from Phitsanulok, Thailand: isolation, screening, safety evaluation and probiotic properties. Food Control 78:187–195
Kaushik JK, Kumar A, Duary RK, Mohanty AK, Grover S, Batish VK (2009) Functional and probiotic attributes of an indigenous isolate of Lactobacillus plantarum. PLoS ONE. https://doi.org/10.1371/journal.pone.0008099
Khan I, Kang SC (2016) Probiotic potential of nutritionally improved Lactobacillus plantarum DGK-17 isolated from Kimchi—a traditional Korean fermented food. Food Control 60:88–94
Kongnum K, Hongpattarakere T (2012) Effect of Lactobacillus plantarum isolated from digestive tract of wild shrimp on growth and survival of white shrimp (Litopenaeus vannamei) challenged with Vibrio harveyi. Fish Shellfish Immunol 32:170–177
Landeta G, Curiel JA, Carrascosa AV, Munõz R, de las Rivas B (2013) Technological and safety properties of lactic acid bacteria isolated from Spanish dry-cured sausages. Meat Sci 95:272–280
Maldonado A, Ruiz-Barba JL, Jiménez-Díaz R (2003) Purification and genetic characterization of plantaricin NC8, a novel co-culture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl Environ Microbiol 69:383–389
Martín-Platero AM, Valdivia E, Maqueda M, Martínez-Bueno M (2009) Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int J Food Microbiol 132:24–32
Oh YJ, Jung DS (2015) Evaluation of probiotic properties of Lactobacillus and Pediococcus strains isolated from Omegisool, a traditionally fermented millet alcoholic beverage in Korea. LWT Food Sci Technol 63:437–444
Paukatong KV, Kunawasen S (2001) The Hazard Analysis and Critical Control Points (HACCP) generic model for the production of Thai fermented pork sausage (Nham). Berl Munch Tierarztl Wochenschr 114:327–330
Perin LM, Miranda RO, Todorov SD, de Melo Franco BDG, Nero LA (2014) Virulence, antibiotic resistance and biogenic amines of bacteriocinogenic lactococci and enterococci isolated from goat milk. Int J Food Microbiol 185:121–126
Pieniz S, de Moura TM, Cassenego APV, Andreazza R, Frazzon APG, de Oliveira Camargo FA, Brandelli A (2015) Evaluation of resistance genes and virulence factors in a food isolated Enterococcus durans with potential probiotic effect. Food Control 51:49–54
Powell JE, Witthuhn RC, Todorov SD, Dicks LMT (2007) Characterization of bacteriocin ST8KF produced by a kefir isolate Lactobacillus plantarum ST8KF. Int Dairy J 17:190–198
Semedo T, Santos MA, Martins P, Lopes MFS, Marques JJF, Tenreiro R, Crespo MTB (2003) Comparative study using type strains and clinical and food isolates to examine hemolytic activity and occurrence of the cyl operon in enterococci. J Clin Microbiol 41:2569–2576
Swetwiwathana A, Visessanguan W (2015) Potential of bacteriocin-producing lactic acid bacteria for safety improvements of traditional Thai fermented meat and human health. Meat Sci 109:101–105
Szala B, Paluszak Z, Motyl I (2012) Antagonistic effect of Lactic acid bacteria on Salmonella Senftenberg in mixed cultures. Pol J Environ Stud 21:1399–1403
Torriani S, Felis GE, Dellaglio F (2001) Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl Environ Microbiol 67:3450–3454
Vankerckhoven V, Huys G, Vancanneyt M, Vael C, Klare I, Romond MB, Entenza JM, Moreillon P, Wind RD, Knol J, Wiertz E, Pot B, Vaughan EE, Kahlmeter G, Goossens H (2008) Biosafety assessment of probiotics used for human consumption: recommendations from the EU-PROSAFE project. Trends Food Sci Technol 19:102–114
Visessanguan W, Plengvidhya V, Chokesajjawatee N, Bakar JA (2015) Lactic meat fermentation. In: Owens JD (ed) Indigenous fermented foods of Southeast Asia. CRC Press, New York, pp 313–358
Vuyst LD, Falony G, Leroy F (2008) Probiotics in fermented sausages. Meat Sci 80:75–78
Yu Z, Zhang X, Li S, Li C, Li D, Yang Z (2013) Evaluation of probiotic properties of Lactobacillus plantarum strains isolated from Chinese sauerkraut. World J Microbiol Biotechnol 29:489–498
Acknowledgements
This work was supported by the Higher Education Research Promotion and the Thailand’s Education Hub for Southern Region of ASEAN Countries Project Office of the Higher Education Commission. Also, the research was funded by the Graduate School, Prince of Songkla University, the Thailand Research Fund and the Commission on Higher Education for Project No. MRG5080138, and the Research and Development Office, Prince of Songkla University. The authors would like to thank Associate Professor Seppo J. Karrila, Ph.D. (Chem Eng) for the professional English proofreading service.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Botthoulath, V., Upaichit, A. & Thumarat, U. Identification and in vitro assessment of potential probiotic characteristics and antibacterial effects of Lactobacillus plantarum subsp. plantarum SKI19, a bacteriocinogenic strain isolated from Thai fermented pork sausage. J Food Sci Technol 55, 2774–2785 (2018). https://doi.org/10.1007/s13197-018-3201-3
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13197-018-3201-3