Abstract
Lactic acid bacteria (LAB) and yeasts were enumerated and identified from naturally fermented buttermilk. Isolates were first subjected to chemical tests and then to molecular characterization. Molecular identification involved pure sequencing of 16s rRNA (LAB) and 18s rRNA (yeast) genes. Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (DGGE) was used for detection of microbiota composition. Eighty-five LAB and 26 yeast isolates obtained from 16 small-scale dairy farms were isolated and identified. The microbial composition was dominated by strains of Lactococcus lactis ssp. lactis, Lactobacillus plantarum and Saccharomyces cerevisiae. Molecular techniques enabled not only genetic confirmation but also detection of some uncultivated strains. The presence of diverse strains of LAB and yeasts in NFB indicated a potential for development of different starter cultures to make new dairy products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Buttermilk is a by-product of butter-making from sour milk or cream, and/or sweet cream. Naturally fermented buttermilk (NFB) is one of several fermented dairy products consumed in Northern Ethiopia. The processing and manufacturing methods of NFB have been previously described by Negussie et al. (2012). There are two types of NFB; Sour milk buttermilk (SMB) produced by individual farmers, and Sour cream buttermilk (SCB) made at individual private dairy farms and cooperative units where a separator is available. Cooperative dairy units are milk collection centres that receive milk from individual farmers. The pooled milk is then either sold to customers as it is, or processed and marketed after processing into other products such as butter and buttermilk. SMB is made by allowing whole milk to ferment in locally made equipment such as a cow dung pot for 2–3 days at ambient temperature (Gonfa et al. 2001). SCB involves cream separation followed by souring of the cream and churning (Negussie et al. 2012). One third of Ethiopia’s total milk production, about 1.5 million tons, is used for butter-making and buttermilk is a by-product of this process (CSA 2002). Buttermilk is consumed by all household members as it is, or as a side dish following processing into other products, such as, Ajibo, a variety of Ethiopian cottage cheese made from heated buttermilk and Hazo, spiced buttermilk (Negussie et al. 2012).
Like any other fermented product, fermentation is the principal process that takes place during souring of milk in NFB processing. The fermentation is spontaneous because lactic acid bacteria (LAB) and yeasts that gained access to the milk from the environment ferment it in an uncontrolled manner. NFB thus differs from commercially made cultured buttermilk, which is produced by the use of specific known strains of LAB added as a starter culture. Microorganisms involved in preparation of commercial buttermilk comprise Lc. lactis ssp. cremoris and lactis, Lc. lactis ssp. lactis biovar. diacetylactis and Leuconostoc spp (Doyle and Beuchat (2007).
Recently, interest has increased in LAB and yeasts found in naturally fermented milk products due to their biotechnological potential for contributing to better and novel processing methods and the nutritional status of fermented products (Holzapfel 2002). Although a considerable amount of research has been done on commercially utilized LAB and yeasts, there is a certain lack of modern knowledge about the identity of wild LAB and yeasts such as those involved in the fermentation of NFB. Mostly relevant studies have been based on phenotypic and morphological properties (Feresu and Muzondo 1990). Several authors isolated and identified LAB and yeasts from African naturally fermented milks using phenotypic and morphological techniques (Abdelgadir et al. 2001; Beukes et al. 2001; Lane 1991; Obodai and Dodd 2005; Saleh 2013). However, these conventional methods have some drawbacks, as they are time consuming and may be inaccurate. It is thus important also to use molecular methods to enable genetic identification of the microorganisms involved in fermentation. This study was therefore undertaken to quantify, isolate, identify and characterize LAB and yeast strains found in naturally fermented buttermilk through phenotypic characterization, pure strain sequencing and PCR-DGGE analyses.
Materials and methods
Collection of samples
Naturally Fermented Buttermilk producing farms in Mekelle area, Tigray region of Northern Ethiopia were categorized by the type of buttermilk produced. Sixteen dairy farms were included in the study: 10 individual dairy farms and 6 cooperative dairy processing units that produced SMB and SCB, respectively. Thirty-two naturally fermented buttermilk samples were collected from the 16 dairy farms in two consecutive years (2010 and 2011). Buttermilk samples were collected from products made at farm level following the manufacturing scheme outlined by Negussie et al. (2012). Collection took place in two seasons with different ambient temperature. In the colder months, a fermentation time of ca. 48 h was used, whereas in warmer months the fermentation was complete after 32 h. The pH of 48 h buttermilk products and microbial load were analysed immediately after churning of sour milk. Two samples were collected from each product, one for isolation of microorganisms at Mekelle University (MU), Ethiopia and the second for molecular analysis at the Norwegian University of Life Sciences (NMBU). The samples were collected in sterile bottles and kept refrigerated in an icebox for immediate transportation to the laboratory at MU.
Culture media for growth and isolation of pure strains
Enumeration of LAB was done on M17-lactose and MRS agars (Merck, Darmstadt, Germany) to estimate the numbers of presumptive coccus and bacillus, respectively. The inoculated agars were incubated aerobically at 30 °C for 48 h. Yeasts were enumerated by spread-plating dilutions of samples on Dichloran Rose-Bengal Chloramphenicol agar (Merck, Darmstadt, Germany) and incubated at 30 °C for 48–72 h. Tryptone Glucose Yeast Extract broth was used for overnight growth prior to DNA extraction from yeasts.
Isolation and preliminary identification of pure strains
Pure cultures of LAB and yeasts were isolated from M17 and MRS plates of buttermilk samples. Isolates were grown in broth, according to the original isolation medium, purified and stored at −20 °C. LAB and yeasts were further investigated by microscopic examination, Gram-staining and catalase reactions. Pure isolates of LAB and yeasts were inoculated into API 50 CHL and API 20 AUX kits respectively, to study carbohydrate metabolism profiles (Biomerieux, Marcy L’ Etoile, France).
Sequencing of pure LAB and yeast strains
DNA extraction
DNA was extracted from overnight broth cultures of pure LAB and yeast strains. About 2 ml was taken from overnight M17 and MRS broth cultures (for Lactococci and Lactobacilli respectively) and Tryptone Glucose Yeast Extract broth (for yeasts) and centrifuged at 17,500×g for 2 min and 4 °C. Extraction was then made following the manufacturers protocol for GenEluteTM bacterial genomic DNA kit (Sigma-Aldrich, Oslo, Norway) (Tables 1 and 2).
Amplification of PCR product
The extracted DNA was used to set up a conventional Polymerase Chain Reaction (PCR) to amplify target 16S rRNA gene of LAB and 18S rRNA gene of yeasts using a thermocycler (PTC-200, MJ research Inc., St. Bruno (Quebec), Canada). The PCR reaction (50 μl) was prepared with 3 μl of DNA and 47 μl of master mix which contained 5 μl 1x PCR buffer, 1 μl 10 mM deoxynucleotide triphosphate mixture (dNTP-mix), 0.5 μl of both primer sets (100 pmol), (Table 4) and 0.5 μl of Dynazyme (2u/ μl). The universal primer sets used to amplify the bacterial 16s rRNA gene were 1F (5′GAG TTT GAT CCT GGC TCA G 3′) [18] and 5R (5′GGT TAC CTT GTT ACG ACT T 3′) (Muyzer 1999). The ITS1-5.8S-ITS2 region of rRNA for yeasts was amplified using ITS1: ITS1 (5′TCC GTA GGT GAA CCT GCG G 3′) and ITS4 (5′TCC TCC GCT TAT TGA TAT GC 3′) [1, 8]. The PCR program involved initial denaturation at 94 °C for 3 min, 29 cycles of denaturation 94 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 3 min and a final extension step at 72 °C for 10 min. Similar PCR programs were used for LAB and yeast DNA except that the annealing temperature for yeasts was 53 °C.
PCR product purification and sequencing
PCR product purification was undertaken after the amount (ng/ μl) and quality of DNA were assessed using a spectrophotometer and 1 % agarose gel electrophoresis, respectively. The former measured DNA quantity in nano drop per μl while the later showed magnified gel marks in samples with high DNA concentrations. PCR purification followed QIAquick PCR purification kit protocol, first by adding 250 μl of buffer PB per 50 μl volumes of PCR product in 2 ml collection tube containing a spin column (QIAquick PCR purification kit protocol), and centrifuged at 15,700×g for 1 min. This mixture was then washed with 750 μl of buffer PE added with 5 volumes of ethanol, centrifuged at 15,700×g for 1 min. The supernatant was discarded and finally the column was transferred into a new 1.5 ml Eppendorf tube, added 30 μl elution buffer, held for 1 min at room temperature and centrifuged at 15,700×g for 2 min. Purified PCR products were sent to GATC (Biotech AG, Cologne, Germany) for sequencing.
Determination of microbial composition of buttermilk samples
Total genomic DNA extraction from buttermilk samples
Extraction of total genomic DNA was performed using GenEluteTM bacterial genomic DNA kit (Sigma-Aldrich) protocol. Buttermilk samples (1.5 ml) were mixed with 8.5 ml 0.9 % aqueous NaCl solution, homogenized by a Stomacher 400 (Seward Ltd., West Sussex, UK) at speed 4 for 2 min and centrifuged at 231×g. The supernatant was collected in a new tube and centrifuged at 3846×g for 15 min to obtain cell pellets. The cell pellets were suspended in 1 ml of 0.9 % NaCl in a new Eppendorf tube (1.5 ml), centrifuged at 10,000×g for 2 min, and the supernatant discarded.
PCR-DGGE of NFB
The DNA of LAB and yeast was amplified using a thermocycler (PTC-200). The PCR reaction mixture (50 μl) was prepared from 2 μl of DNA and 48 μl of master mix, which comprises 5 μl 10X Dream TaqTM Green Buffer (Fermentas, VWR International, Oslo, Norway), 8 μl deoxynucleotide triphosphate mixture, dNTP mix [1.25 mM], 1 μl of specific primer sets (5 mM), 0.25 μl of Dream TaqTM DNA Polymerase [5U/μl] (Fermentas) and 32.5 μl of milliQ3-water. The specific forward primers (Table 4) used to amplify 16 s rRNA gene of Lactococcus, Enterococcus, Streptococcus, Lactobacillus, Leuconostoc and Pediococcus were LAC3F (AGCAGTAGGGAATCTTCGG) and LAC1F (AGCAGTAGGGAATCTTCCA), respectively (Endo et al. 2009). The reverse primer used for both LAB species was LAC2R (ATTTCACCGCTACACATG) (Cocolin et al. 2000). The PCR program used for LAB had initial denaturation at 94 °C for 5 min, 35 cycles of actual denaturation 94 °C for 30 s, annealing temperature 61 °C for 30 s, extension 72 °C for 60 s and final extension 72 °C for 10 min. The PCR program for yeasts was the same as that used for LAB except that the annealing temperature was 53 °C and different primer sets were used. The forward primer NL1GC [5 mM μl−1] 5′ CGC CCG CCG CGC GCG GCG GGC GGG GCG GGG GCA TAT CAA TAA GCG GAG GAA AAG′3 and LS2 [5 pmol μl−1] 5′ATT CCC AAA CAA CTC GAC TC′ were used to amplify 26s rRNA gene of yeasts (Akabanda et al. 2013; Cocolin et al. 2000).
Preparation of the gel cassette and gel analysis
The urea-formamide denaturing gradients of gels used were 50 to 30 % for LAB and 55 to 40 % for yeasts. For polymerization, 5.2 μl (Tetramethylethylenediamine (TEMED and 52 μl 20 % amonium persulfate (APS) + 800 μl milliQ water) were used. The stacking gel was prepared from 2.25 ml 40 % bisacrylamide, 100 μl 50x TAE and milliQ water to 10 ml (7.65 ml) and 20 μl of PCR product loaded into the gel. The gel cassette was run at 120 V, Current 50 mA, Power 5 W, Time 16 h. When the gel run was finished the gel was stained in a solution of 250 ml of 1xTAE (Tris-acetate-EDTA) buffer + 25 μl ethidium bromide, placed on a shaker for approximately 30 min and visualization was made in the BioRad Gel Doc (Bio-Rad Laboratories, Hercules, CA, USA. DGGE bands of interest were excised with a sterile scalpel blade, transferred to a sterile Eppendorf tube, and DNA was eluted by 50 μl of 0.1xTAE buffer and kept overnight at 4 °C (Porcellato et al. 2013). A mixture of 5 μl purified PCR product and 5 μl primers were sent to GATC (Biotech AG, Cologne, Germany) for genome sequencing.
Data analysis
Averages and frequencies of observed numeric data were calculated using MS Excel and Minitab 16 (Minitab 2010). Carbohydrate profile results were analysed using apiTM web software version 5.1 (Biomerieux, Marcy I’Etoile, France). Bio edit software (Tom Hall, Ibis Biosciences, and Carlsbad, CA, USA) was used to edit and identify good and bad sequences. Good sequences were subjected to Nucleotide BLAST, Basic Local Alignment of Search Tool of NCBI, the National Center for Biotechnology Information, (http://www.ncbi.nlm.nih.gov/) to identify pure strains at species level (Altschul et al. 1990; Dolan et al. 2001).
Results and discussion
pH and microbial load
All but one sample of buttermilk had a pH below 4.6 after 48 h fermentation, indicating in general an adequate fermentation (Table 1). Counts on M17 agar and MRS agar ranged from log 6–9 and log 6–8.6 cfu ml−1, respectively. Samples with low counts of LAB (fewer than 7 cfu ml−1) had a pH value above 4.5. These results were comparable with other African naturally fermented milks. Mean counts on MRS and M17 agar from South African traditionally fermented milk were reported to be 7.7 × 10E8 and 7.05 × 10E8 cfu ml−1, respectively (Beukes et al. 2001). Ghanaian fermented buttermilk contained log 4–9 cfu ml−1 of LAB and log 1–7 cfu ml−1 of yeasts (Dolan et al. 2001).
All NFB samples contained yeasts, with numbers ranging from log 3.2 to log 7.7 cfu ml−1 after 48 h fermentation (Table 1). The average yeast load for the Ethiopian fermented milk products, Itutu and Moamatta, was reported as log 6.18 cfu ml−1 (Fekadu 1994) and for southern Ethiopian fermented milks log 5.8 cfu ml−1 (Mogessie 1990). Gadaga et al. (2001) showed that yeasts were present as log 6–8 cfu ml−1 in Zimbabwean fermented milks. The occurrence of such high numbers of yeasts in buttermilk indicates their ability to use milk components or metabolites produced by the other microorganisms in the product, and thereby possibly contribute towards the flavour of the products.
Biochemical isolation and identification of LAB and yeasts
Carbohydrate profile of LAB
All bacterial isolates from MRS and M17 that were Gram-positive and catalase negative were considered as presumptive LAB. The API profiles of these isolated LAB enabled identification of 85 strains of Lactococcus (Lc) lactis ssp. lactis (23 strains), Enterococcus (Ent.) faecalis (14 strains), En. faecium (11 strains), Lactobacillus (Lb.) pentosus (16 strains), Lb. plantarum (16 strains), Lb. brevis (3 strains) and Lb. paracasei (2 strains) (Table 2). API analysis results were based on the more frequently occurring forms of strains.
All LAB utilized galactose, maltose glucose, fructose, mannose and lactose, which are the main fermenters. More or less similar LAB species were identified phenotypically from some African naturally fermented products such as Itutu/Ergo (Gonfa et al. 2001) and Leban Zeer of Egypt (Saleh 2013).
Carbohydrate profile of isolated yeasts
Analysis of 26 yeast isolates using API 20 AUX identified four yeast species in NFB (Table 3). These were Saccharomyces (S.) cerevisiae (8 strains), Kluyveromyces (Kl.) marxianus (3 strains), Candida (Ca.) parapsilosis (9 strains) and Trichosporon (Tri.) asahii (2 strains). The first three yeast species were also isolated and identified from Zimbabwean fermented milk (Gadaga et al. 2001). All but not S. cerevisiae isolates were able to ferment lactose. All isolates of these four yeast species fermented glucose, galactose and sucrose. Saccharomyces cerevisiae had highest frequency of occurrence but utilized least number of carbohydrates only galactose, maltose, sucrose and rafinnose compared to other isolated yeasts.
Kluyveromyces (Kl.) marxianus was identified as Candida kefir in API analysis. This is because Kl. marxianus is the sexual form of Candida kefir. Capacity of this yeast to utilize lactose indicates its involvement in fermentation of SFB (Ramirez-Zavala et al. 2004). The yeast is utilized in the synthesis of commercial enzymes including: Inulinase production of fructose syrup (Rouwenhorst et al. 1990); β-galactosidase, reduction of lactose (Martins et al. 2002); protein phosphatase, modification of cheese making qualities of casein and carboxypeptidases (reduction of bitter taste) and aminopeptidases (direct processing and ageing of dairy and meat products (Ramirez-Zavala et al. 2004). This yeast has high metabolic diversity and is used for different industrial application (Lane and Morrissey 2010). Kl. marxianus is used for commercial lactase that hydrolyze lactose into glucose and galactose during milk processing (Seyis and Aksoz 2004). This yeast is utilized in ethanol production process by fuel ethanol producing industries (Hack and Marchant 1998). Some isolates of Candida and Trichosporon species were identified in buttermilk products. Post processing contamination of these two yeast species might occurred due to poor farm hygiene.
Molecular characterization
Sequencing of lactic acid bacteria
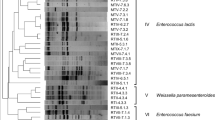
Conventional PCR amplification of pure strains gave PCR products that fulfill minimum requirements of spectrophotometry measurements and gel electrophoresis indicators for sequencing. All the 85 LAB strains isolated from 16 NFB samples were identified by sequencing their 16S rRNA genes (Table 5). These isolated LAB were identified using V3 region of 16S rRNA gene of LAB. The V3 region of 16S rRNA gene is a highly variable region which is able to differentiate LAB isolates at species level (Blaiotta et al. 2003). Thirty different strains of LAB were selected and their sequences with the accession numbers given in Fig. 1 were used for construction of the phylogenetic tree of LAB isolated from NFB. The remaining 55 strains were identical with the 30 representative strains and believed to have no effect in the Phylogenetic clustering. Accordingly, these thirty representative nucleotide sequences of LAB resulted in four main groups of lactic acid bacteria found in NFB (Fig. 1).
Phylogenetic analysis of 16s rRNA gene of 30 nucleotide sequences of LAB isolated from naturally fermented buttermilk based on Maximum Likelihood Method (Tamura et al. 2013). Estimation of similarity was done using maximum likelihood model (Tamura et al. 2013). Accession numbers of identified representative LAB strains 
Pair wise comparisons of 16s rRNA gene sequences of 30 representative LAB strains indicated the relative similarity between and within groups of LAB (Fig. 1). The overall sequence similarity within different strains of a species ranged between 92 and 98.5 %. The closest (99.5 %) similarity was obtained among strains of Lb. brevis followed by that of Lc. lactis strains where by 98.5 % of their genetic material was similar. These strains which are very close were isolated from different products. This phylogenetic grouping indicated that strains having similar sequences were clustered in the same group and presumably were considered as members of the same species.
Group I and II were composed of Lactobacillus species, mainly Lb. plantarum and Lb. brevis. The former was isolated from all buttermilk samples while the later was detected in buttermilk from only three farms. Thirty one isolates of Lb. plantarum (Table 5) were identified and had the most diversification of strains, 11 strains, compared with other identified LAB (Fig. 1).
Isolates of Enterococcus were identified as indicated in group III. The total number of isolates was 23 that were diversified in eight strains and mainly represented by En. faecalis (Fig. 1). Group IV consisted of Lc. lactis ssp lactis/ cremoris. A total of 23 isolates of Lc. lactis ssp. lactis varying in 6 different strains were identified using this method (Table 5).
Yeasts
PCR amplification of 18s rRNA gene, using ITS1 and ITS4 primers showed that the 26 strains belonged to four yeast species (Table 5). The 18s rRNA gene is a hyper-variable region which differentiate yeasts at species level (Saleh 2013). Phylogenetic tree made for 18s rRNA gene sequences of 16 isolates of representative yeasts clustered all identified yeasts into four clearly recognizable groups (Fig. 2). The sequence similarity of all yeasts ranged between 82 and 96 %. Sequence similarity was highest (96 %) within strains of Candida species while Trichosporon species had less (82 %) similarity than the other isolated species. Members of the species, Saccharomyces cerevisiae had a sequence similarity of 93 %.
A total of 9 candida strains were identified with 6 different strains all of which were assigned to Group I (Fig. 2). The presence of these organisms could be attributed to post production contaminations from the environment. Group II included a mixture of both Kluyveromysis and Trichosporon species with lower percentages of similarity (82–84 %). Group III consisted of 6 different strains of all the 12 isolated Saccharomyces cerevisiae species. The yeast population was dominated by S. cerevisiae and presence of this species indicated that yeasts contributed to flavour of buttermilk during souring of SFB.
PCR-DGGE analyses of LAB and yeast DNA in buttermilk samples
Isolates of Lactococcus, Enterococcus, Streptococcus and Lactobacillus were identified with PCR-DGGE in sample buttermilks. Amplified DNA, loaded onto standard gels, formed well-resolved amplicons of the 16s rRNA genes (Figs. 3 and 4). The numbers labeled for each band in a gel depict bands of organisms. Although many tiny bands were visually observed, only some were sequenced as the remaining contained too low DNA concentration. This indicates that buttermilk samples could contain even more diverse LAB and yeast species than the sequenced bands.
PCR-DGGE profile of isolates of Lactococcus. First lane: 100 bp ladder, lanes A and B pure strains of Lactococus lactis spp lactis/cremoris, Lanes 1–16: Buttermilk samples. Bands marked as 1–8 from top down direction across the gel represented identified strains given in Table 6
PCR-DGGE profile of Lactobacillus isolates. First lane: 100 bp ladder, lanes A and B pure strains of Lb. brevis and Lb plantarum, respectively isolated from naturally fermented buttermilk, identified in this study by 16s RNA sequencing, Lanes 1–16: Buttermilk samples. Bands marked as 1–8 from top down direction across the gel represented identified strains given in Table 6
Lactococcus, Streptococcus and Enterococcus
PCR-DGGE profiles of buttermilk samples indicated presence of 4 Lactococcus spp., one Streptococcus spp. and one Enterococcus spp. (Fig. 3). These species were determined by sequencing DNA extracted from gel amplicons. The reference strains used were pure strains of Lc. lactis ssp lactis strain 25 (A) and Lc. lactis ssp. cremoris (B) isolated in the present study and identified by 16s RNA sequencing. The first six bands (1–5) consisted of members of Lactococcus species as Lc. lactis ssp lactis dominated this group. The distribution of this species along different lanes could be related with mixed DNA extracted and indicated the diversity of this species in Buttermilk samples. Lc. lactis ssp lactis migrated slightly further than the Lc. lactis ssp cremoris (bands 5 and 6). This could be due to differences in the target region (V1 or V3) of the 16s rRNA amplified. These two species can be differentiated by amplifying the V1 region although they are similar in V3 region (Ward et al. 1998). Akabanda et al. (2013) investigated the microbial composition of a Ghanaian naturally fermented milk and identified Lactococcus lactis, Streptococcus thermophilus and Leuconostoc mesenteroides. Sour milk buttermilk products, 8, 9, 10, 11 and 13, had a higher number of bands (4 to 6) of Lactococcus strains than the other products. Sour cream buttermilk products, 3, 4, and 6 showed bands indicating 3–4 different strains (Fig. 3).
PCR-DGGE analysis of buttermilks showed the presence of two additional species compared to those identified using the pure strain sequencing method. Lc. raffinolactis (band 3) was present in almost half of the buttermilk samples (Table 5). This species is not used in commercial starter cultures but has been found in raw milk and also in traditional and industrial dairy products including sour milk, fresh and soft cheese, hard and semi-hard cheeses (Urbach 1995; Ward et al. 1998). St. thermophilus (band 8) was found in two buttermilk samples (SCB6 and SMB11).
Lactobacillus, Leuconostoc and Pediococcus
Eight amplicons were identified from the DGGE gel of the Lactobacillus, Leuconostoc and Pediococcus primers (Fig. 4). The reference strains used for this group were isolated pure strains of Lb. brevis (A) and Lb. plantarum (B). Sequenced bands obtained from Lactobacillus gel resulted in detection of 4 different species of Lactobacillus, 2 of Leuconostoc and 1 Pediococcus spp. in SFB (Table 6). Lb. plantarum was detected in all buttermilk samples (band 3) although the bands for some samples were faint (Fig. 4). As it can be seen from the figure, there were more amplicons that could not be identified because of inability to harvest adequate PCR products.
Yeasts
The primer set of NL1GC and LS2 amplified the 26S rRNA gene using conventional PCR and displayed six amplicons from buttermilk samples (Fig. 5). These bands represented four yeast species, namely S. cerevisiae, Yarrowia (Ya.) lipolytica, Ca. parapsilosis, Tri. lactis (Table 6). The diversity of identified yeast species was greater in PCR-DGGE than in pure strain sequencing. The presence of yeasts in NFB indicated the occurrence of yeast-lactic fermentation. This implies that this product could be source of starter cultures to make commercial fermented milks with yeast-lactic fermentation. Sc. cerevisiae was detected in eight samples when both pure sequencing and community analyses were combined. Ya. lipolytica is a lipolytic yeast that produces citric acid and also aroma compounds from sugar-containing materials (van Kranenburg et al. 2002; Zinjarde 2014).
PCR-DGGE profile of yeast species First lane: 100 bp ladder, lane A: pure strain of S. cerevisiae isolated from naturally fermented buttermilk, identified in this study by 18s RNA sequencing, Lanes 1–16: Buttermilk samples. Bands marked as 1–6 from top down direction across the gel represented identified strains given in Table 6
The identification of Lactococcus strains by API analysis was similar to the results obtained by pure strain sequencing. Slightly higher numbers of Lb. plantarum were detected using 16s sequencing than those determined by API analysis. Yet, members of these two species were detected in all of the analyzed products. The identified LAB are probably responsible for the main fermentation of the milk/cream and yeasts may also contribute to the aroma of NFB as reported from previous findings of (Urbach 1995). Van Kranenburg et al. (2002) described the functions of LAB in fermented milks as: catabolism of lactose to lactic acid, flavour formation and preservation. Yeasts, due to production of flavour compounds and metabolites, affect the characteristics of fermented milks (Roostita and Fleet 1996).
Conclusion and recommendation
Naturally fermented buttermilk was produced by several dairy farms and cooperative units in the study area. The product involved fermentation as a main processing tool via the action of LAB and yeasts. Carbohydrate profile of API analysis results of all LAB and some yeasts exhibited fermentation of lactose into lactic acid. Pure strain sequencing showed results more or less similar to identification methods used for Biochemical tests at species level. A total of 85 pure LAB and 26 yeast strains were isolated and identified with most abundant species of Lc. lactis ssp. lactis, Lb. plantarum and S. cerevisiae compared with other members of the isolated organisms. The presence of the identified LAB and yeasts, indicated that a yeast-lactic fermentation had probably taken place. The use of both culture-dependent and culture-independent molecular characterization enabled not only identification of more microbial diversity but also detection of DNA from additional organisms that were not found in the process of pure strain sequencing. It was suggested to more detailed research works to investigate mixed culture milk fermentations using the most frequently occurring isolates of Lc. lactis ssp. lactis, Lb. plantarum, Sc. cerevisiae, and Kl. marxianus in order to upgrading the current traditional processing methods into small scale processing technologies.
Abbreviations
- NUFU:
-
Norwegian Government Councils of Universities Committee for Development Research & Education
- IKBM:
-
Institutt for Kjemi Bioteknologi og Matvitenskap
- NFB:
-
Naturally fermented buttermilk
- SCB:
-
Sour cream buttermilk
- NMB:
-
Sour milk buttermilk
- LAB:
-
Lactic acid bacteria
- PCR:
-
Polymerase chain reaction
- DGGE:
-
Denaturing gradient gel electrophoresis
- rRNA:
-
Ribosomal ribonucleic acid
- DNA:
-
Deoxyribonucleic acid
- Bp:
-
Base pair
- LAB:
-
Lactic acid bacteria
- ssp.:
-
Sub species
- spp:
-
Species
- bp:
-
Base pairs
- cfu:
-
Colony-forming units
References
Abdelgadir WS, Hamad SH, Møller PL, Jakobsen M (2001) Characterisation of the dominant microbiota of Sudanese fermented milk Rob. Int Dairy J 11(1–2):63–70
Akabanda F, Owusu-Kwarteng J, Tano-Debrah K, Glover RLK, Nielsen DS, Jespersen L (2013) Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol 34(2):277–283
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Beukes EM, Bester BH, Mostert JF (2001) The microbiology of South African traditional fermented milks. Int J Food Microbiol 63(3):189–197
Blaiotta G, Pennacchia C, Ercolini D, Moschetti G, Villani F (2003) Combining denaturing gradient gel electrophoresis of 16S r DNA V3 Region and 16S–23S r DNA spacer region polymorphism analyses for the identification of staphylococci from Italian fermented sausages. Syst Appl Microbiol 26(3):423–443
Cocolin L, Bisson LF, Mills DA (2000) Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Immunol 189(1):81–87
CSA (2002) Federal Republic of Ethiopia. Agricultural sample enumeration. Report on size, characteristics and purpose of livestock and use of livestock products. Central Statistic Authority of Ethiopia, Addis Abeba
Dolan M, Babin P, Wollenzien P (2001) Construction and analysis of base-paired regions of the 16s Rrna in the 30s ribosomal subunit determined by constraint satisfaction molecular modelling. J Mol Graph Model 19:495–513
Doyle MP, Beuchat LR (2007) Food Microbiology. Fundamentals and frontiers. 3rd ed. Center for food safety, The University of Georgia, Griffin, GA 30223–1797. American Society for Microbiology, ASM press 1752 N Street, N.W. Washington, DC 20036–2804, USA
Endo A, Futagawa-Endo Y, Dicks L (2009) Lactobacillus and Bifidobacterium diversity in horse feces, revealed by PCR-DGGE. Curr Microbiol 59:651–655
Fekadu B (1994) Present situation and future aspects of milk production, milk handling and processing of dairy products in Southern Ethiopia. Ph.D. thesis, Agricultural University of Norway, Ås, Norway
Feresu SB, Muzondo MI (1990) Identification of some lactic acid bacteria from two Zimbabwean fermented milk products. World J Microbiol Biotechnol 6:178–186
Gadaga TH, Mutukumira AN, Narvhus JA (2001) The growth and interaction of yeasts and lactic acid bacteria isolated from Zimbabwean naturally fermented milk in UHT milk. Int J Food Microbiol 68(1–2):21–32
Gonfa A, Foster HA, Holzapfel WH (2001) Field survey and literature review on traditional fermented milk products of Ethiopia. Int J Food Microbiol 68(3):173–186
Hack C, Marchant R (1998) Characterization of a noble hermotolerant yeast, Kluyveromyces (Kl.) marxianus var marxianus: development of an ethanol fermentation process. Biotechnology research group, university of Ulster, Colorain, UK. J Ind Microbiol Biot 20:323–327
Holzapfel W (2002) Appropriate starter culture technologies for small-scale fermentation in developing countries. Int J Food Microbiol 75:197–212
Lane D (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. J Wiley & Sons, Chichester, pp 115C–175C
Lane MM, Morrissey JP (2010) Kluyveromyces marxianus: a yeast emerging from its sister’s shadow. Fungal Biol Rev 24(1–2):17–26
Martins DB, de Souza CG Jr, Simões DA, de Morais MA Jr (2002) The b-galactosidase activity in Kluyveromyces marxianus CBS6556 decreases by high concentrations of galactose. Curr Microbiol 44:379–382
Minitab (2010) Licensed to It-seksjionen UMB. 2010 Minitab Inc. Minitab
Mogessie A (1990) Effect of curd cooking temperatures on the microbiological quality of ayib, a traditional ethiopian cottage cheese. World J Microbiol Biotechnol 6:159–162
Muyzer G (1999) DGGE/TGGE a method for identifying genes from natural ecosystems. Curr Opin Microbiol 2:317–322
Negussie G, Abrahamsen RK, Fekadu B, Narvhus J (2012) A survey on spontaneously fermented buttermilk in Northern Ethiopia. AJFST 3(3):78–89
Obodai M, Dodd CER (2005) Characterization of dominant microbiota of a Ghanaian fermented milk product, nyarmie, by culture- and nonculture-based methods. Division of Food Sciences, School of Biosciences, University of Nottingham, Sutton Bonington Campus, Loughborough
Porcellato D, Østlie HM, Brede ME, Martinovic A, Skeie SB (2013) Dynamics of starter, adjunct non-starter lactic acid bacteria and propionic acid bacteria in low-fat and full-fat Dutch-type cheese. Int Dairy J 33(2):104–111
Ramirez-Zavala B, Mercado-Flores Y, Hernandez-Rodriguez C, Villa-Tanaca L (2004) Purification and characterization of a lysine aminopeptidase from Kluyveromyces marxianus. FEMS Microbiol Lett 235:369–375
Roostita R, Fleet GH (1996) The occurrence and growth of yeasts in Camembert and Blue-veined cheeses. Int J Food Microbiol 28(3):393–404
Rouwenhorst R, Hensing M, Verbakel J, Scheffers W, van Dijken J (1990) Structure and properties of the extracellular inulinase of Kluyveromyces marxianus CBS 6556. Appl Environ Microbiol 56:3337–3345
Saleh FA, (2013) Isolation and identification of microorganisms and antibacterial activity of Laban Zeer, an Egyptian traditional fermented milk product. Food and Nutrition Sciences Dep. College of Agriculture and Food Science, King Faisal University, Saudi Arabia 2:31–42
Seyis I, Aksoz N (2004) Production of lactase by Trichoderma spp. Food Technol Biotechnol 42(2):121–124
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Urbach G (1995) Contribution of lactic acid bacteria to flavour compound formation in dairy products. Int Dairy J 5:877–903
Vainio E, Hantula J (2000) Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Mycol Res 104:927–936
van Kranenburg R, Kleerebezem M, Van Hylckama Vlieg J, Ursing B, Boekhorst J, Smit B, Ayad E, Smit G, Siezen R (2002) Flavour formation from amino acids by lactic acid bacteria: predictions from genome sequence analysis. Int Dairy J 2:111–121
Ward L, Brown J, Davey G (1998) Two methods for the genetic differentiation of Lactococcus Lactis ssp. lactis and cremoris based on differences in the 16s Rrna gene sequence. FEMS Microbiol Lett 166:15–20
Zinjarde S (2014) Food-related applications of yarrowia lipolytica. Food Chem 152:1–10
Acknowledgments
Thanks are due to the Norwegian Government Councils of Universities Committee for Development, Research and Education (NUFU) which has funded this research project through the project NUFUGe 2008 10265 on “Women and Food Science: together towards national visibility” for funding this research work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gebreselassie, N., Abay, F. & Beyene, F. Biochemical and molecular identification and characterization of lactic acid bacteria and yeasts isolated from Ethiopian naturally fermented buttermilk. J Food Sci Technol 53, 184–196 (2016). https://doi.org/10.1007/s13197-015-2049-z
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13197-015-2049-z