Abstract
The spontaneously fermented curdled milk product from Burkina Faso, lait caillé is prepared by traditional processing from raw unpasteurised milk. The fermentation lasts 1–3 days. This study aims to identify the predominant microbiota involved in lait caillé fermentation from cow milk. A survey on lait caillé end-products from local markets showed pH ranges of 3.5 to 4.2. Counts of total lactic acid bacteria (LAB) were 7.8 ± 0.06 to 10.0 ± 0.03 log CFU/g and yeast counts were 5.3 ± 0.06 to 8.7 ± 0.01 log CFU/g, together with considerate amounts of Enterobacteriaceae < 3.00 to 8.4 ± 0.14 log CFU/g. Sampling throughout the entire fermentation of lait caillé was performed at a traditional house-hold production site. A drop in pH from 6.7 ± 0.01 at 0 h to 4.3 ± 0.08 in the end-product (59 h) was found. Total LAB counts increased to 8.6 ± 0.02 log CFU/g in the end-product, while yeast and Enterobacteriaceae counts reached 6.4 ± 0.11 and 6.7 ± 0.00 log CFU/g, respectively. LAB and yeasts isolated during the fermentation were clustered by (GTG)5 repetitive-PCR fingerprinting followed by 16S and 26S rRNA gene sequencing, respectively. Microbial successions were observed with Leuconostoc mesenteroides being the predominant LAB followed by Pediococcus pentosaceus and Weissella paramesenteroides at the onset, while Lactococcus lactis and Enterococcus spp. where the predominant LAB after 7 h of fermentation. During the first 18 h Candida parapsilosis was the dominant yeast species, while from 35 h to the end-product, Saccharomyces cerevisiae predominated. The microbial safety risk pointed out in this study, showed the need for implementation of good manufacturing practices including pasteurisation and use of well-defined starter cultures.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spontaneously fermented curdled milk product from Burkina Faso, known as lait caillé, is consumed as a beverage or in combination with various cereal based products. Similar spontaneously fermented milk products exist in Africa with slight differences in the processing and consequently the final products. Specifically differences can be seen in microbial quality and sensorial characteristics. Lait caillé from Burkina Faso is prepared from raw cow milk or more seldom from raw goat milk by traditional processing using spontaneous fermentation. Typically the Fulani people of Burkina Faso prepare lait caillé by collecting the raw milk in calabashes, gourds, clay pots or plastic containers and leave it to ferment at ambient temperature until the preferred characteristics of lait caillé are obtained (Savadogo et al. 2004). Lait caillé processing is of social and socioeconomic value since it serves as income generation for both main processors and traders, in most cases involving vulnerable women groups (Duteurtre 2007; Gonfa et al. 2001).
Lactic acid bacteria (LAB) play a key role in the production of fermented milk products. In African fermented milk LAB counts ranging between 7.0 and 10.0 log CFU/mL were found in the “Masai” from Tanzania, kivuguto from Rwanda, leben from Tunisia, fènè from Mali as well as nyarmie and nunu from Ghana (Akabanda et al. 2013; Karenzi et al. 2012; Obodai and Dodd 2006; Samet-Bali et al. 2012; Wullschleger et al. 2013). The most commonly identified LAB in African fermented milk products are Lactococcus lactis, Lactobacillus spp. and Leuconostoc mesenteroides or Leuconostoc pseudomesenteroides. Other LAB, such as Enterococcus spp. and Streptococcus spp., especially Streptococcus infantarius, have also been reported with high frequencies in some traditional fermented milk products (Jans et al. 2012; Mathara et al. 2004; Obodai and Dodd 2006; Walsh et al. 2017; Wullschleger et al. 2013). Yeasts in African fermented milk products are less reported in the literature. However, their importance was highlighted in amasi, a fermented milk from Zimbabwe with counts up to 8.1 log CFU/g (Gadaga et al. 2000). Saccharomyces cerevisiae is most often the dominant yeast species in African fermented milk products, usually in association with other species, such as Candida spp. and Pichia kudriavzevii (Akabanda et al. 2013; Jespersen 2003; Obodai and Dodd 2006).
Characterisation of LAB and yeasts in African spontaneous fermented milk products has mostly been done on end-products, identifying the microorganisms by phenotypic techniques with low discriminatory power (Akabanda et al. 2010; Beukes et al. 2001; Gadaga et al. 2000; Mathara et al. 2004; Savadogo et al. 2004), while later studies used genotypic techniques for microbial identifications (Akabanda et al. 2013; Jans et al. 2017; Karenzi et al. 2012; Parker et al. 2018; Walsh et al. 2017; Wullschleger et al. 2013). Further, microbial successions during processing of African fermented milk products have only been studied to a limited extent (Akabanda et al. 2013; Wullschleger et al. 2013), and LAB and yeasts involved in lait caillé production in Burkina Faso have, to the best of our knowledge, not been investigated before. Understanding the microbial successions is important, since the overall quality of the fermented products highly depends on the microorganisms present during the fermentation. Moreover, microbial successions likely arise from changes in nutrient availability, pH, temperature, concentrations of organic acids and oxygen availability (Jespersen et al. 2005). Previous reports have additionally revealed low quality linked to the hygienic conditions and processing of lait caillé and similar fermented milks, limiting shelf life and income from the production (Broutin et al. 2007; Koussou et al. 2007; Walsh et al. 2017). The aim of this study was therefore to obtain a deeper understanding of lait caillé processing by identifying the predominant microbiota involved in the spontaneous fermentation of lait caillé in the South-West of Burkina Faso by combining phenotypic and genotypic techniques and to study microbial successions taking place during the fermentation.
Materials and methods
Lait caillé processing, sampling and pH measurements
A survey of ten lait caillé end-products was performed by sampling (S1–S10) from three local open markets in Bobo-Dioulasso (in the South-West of Burkina Faso), i.e. S1–S6 at Ouezzin-ville market, S7–S8 at Grand marché market and S9–S10 at Farakan market. The lait caillé was processed by the vendor at the market, in covered plastic containers at ambient temperature, using part of the unsold cow milk originating from farms in outlying or rural areas of Bobo-Dioulasso. The milk was fermented until optimal characteristics of the product were reached, which was determined by the vendor. After purchase, the sample (approximately 250 mL) was transported to the laboratory in an ice-box (0–4 °C) for analysis.
A detailed study on lait caillé fermentation was subsequently performed by sampling throughout the traditional processing of lait caillé at a house-hold scale production site in the village Tolotama outside Bobo-Dioulasso (in the South-West of Burkina Faso). Approximately 3.5 L of fresh cow milk was roughly filtered (pore size of approximately 1 mm) into the fermentation container (plastic bucket covered with a straw-woven lid). The fermentation container was subsequently placed inside a closed room and left for spontaneous fermentation. The fermentation took place at ambient temperature 22.5–29.0 °C for 59 h until the optimal characteristics of the fermented product were reached, which was determined by the producer. Sampling was performed at 9 specific time points (0 h, 7 h, 13 h, 18 h, 28 h, 35 h, 41 h, 53 h and 59 h) during the fermentation process. At each previous time point the fermenting milk was homogenised then 100–150 mL of sample was collected aseptically in duplicate in sterile screw-capped bottles and transported in an ice box (0–4 °C) to the laboratory for analyses. The pH of lait caillé was measured using a pH-meter (Hanna, USA).
Enumeration and isolation of microorganisms
Ten grams of homogenized sample were mixed with 90 g of diluent [0.1% peptone (Cultimed, Spain), 0.85% NaCl (Scharlau, Spain), pH 7.0] to yield a 1:10 dilution. Microbial enumerations were done from appropriate tenfold dilutions. According to Akabanda et al. (2010), total lactic acid bacteria (LAB) were enumerated on Man, Rogosa and Sharpe (MRS) agar (DIFCO, USA), incubated at 37 °C for 48 h in GasPak™ EZ containers with anaerobic container system sachets. Lactococcal counts were determined on M17 agar (Liofilchem, Italy), incubated aerobically at 37 °C for 48 h. Yeasts counts were determined on Sabouraud Chloramphenicol agar (Liofilchem, Italy) pH 5.6 ± 0.2, incubated aerobically at 30 °C for 72–96 h. Enterobacteriaceae counts were enumerated on Violet Red Bile Glucose (VRBG) agar (Liofilchem, Italy), incubated aerobically at 37 °C for 24 h. After enumeration, colonies were picked proportionally, based on their morphological differences (colour, shape, size, margin, surface) and purified by successive streaking on corresponding agar plates, aiming at 20 colonies per sampling time point. For long term storage, pure cultures of LAB were preserved at − 80 °C in corresponding broths (MRS, M17, respectively) with 19% (v/v) glycerol and yeasts in MYGP agar (3 g of yeast extract (Oxoid), 3 g of malt extract (Oxoid), 5 g of bactopeptone (Oxoid), 10 g of glucose (Sigma, Milan, Italy) and 20 g of agar (Oxoid) per liter of distilled water, pH 5.6) with 19% (v/v) glycerol.
Identification of LAB and yeasts by phenotypic- and biochemical characterisation
Micro- and macro morphological descriptions of the isolates were performed according to Akabanda et al. (2010). For bacteria and yeast colonies, colour, shape, size, surface and margin characteristics were examined from agar plates. All bacterial isolates were examined for Gram reaction (Gregersen 1978) and catalase activity (Taylor and Achanzar 1972). After genotypic identification, isolates of the Enterococcus genus were distinguished from isolates of the Leuconostoc genus by determination of CO2 production from glucose, acid production from mannitol and the ability to grow at 45 °C (Facklam and Elliott 1995). Acid production from L-arabinose, mannitol, amygdalin and galactose was examined in order to differentiate between the species Enterococcus durans/hirae/faecalis/lactis and faecium (Manero and Blanch 1999). Leuconostoc mesenteroides and Leuconostoc pseudomesenteroides were differentiated by growth in NaCl 6.5% (w/v) (Facklam and Elliott 1995) and acid production from L-arabinose, mannitol, amygdalin and galactose (Björkroth et al. 2014).
Genotypic characterisation
After the preliminary phenotypical characterisation, DNA was extracted using InstaGene (Bio-Rad Laboratories, Hercules, CA, USA) following the instructions of the manufacturer. The rep-PCR procedure was carried out as described by (Nielsen et al. 2007). In brief, the rep-PCR products were separated by 1.5% agarose gel electrophoresis (5 h, 120 V) in Tris–Borate–EDTA buffer (0.5× TBE) with a 1-kb DNA ladder as reference marker (Thermo Scientific, Lithuania). Afterwards, gels were stained with ethidium bromide and documented using a digital camera (Computer ® TV Zoom LENS, Japan). Cluster analysis of rep-PCR-profiles were performed using Bionumerics 7.1 (Applied Maths, Sint-Martens-Latem, Belgium) based on Dice’s Coefficient of similarity with the unweighted pair group method with arithmetic averages clustering algorithm (UPGMA). Based on the clusters, representatives of each cluster (approximately the square root of the number of isolates within each cluster) were selected for sequencing of the 16S rRNA or 26S rRNA gene for LAB and yeasts, respectively. For 16S rRNA gene sequencing, the primers 27f and 1540r (Jensen et al. 2009) were used for amplification under the following conditions: initial denaturation at 95 °C for 5 min, 35 cycles at 95 °C for 30 s, 60 °C for 30 s and 72 °C for 120 s, followed by a final extension at 72 °C for 10 min. For the D1/D2-region of 26S rRNA gene, primers NL-1 and NL-4 were used for amplification (Kurtzman and Robnett 1998) under the following conditions: initial denaturation at 95 °C for 5 min, 30 cycles at 95 °C for 90 s, 53 °C for 30 s, 72 °C for 90 s followed by a final extension step at 72 °C for 7 min. The PCR products were sent to a commercial sequencing facility (MacroGen, Germany). Subsequently, the LAB and yeasts sequences were manually corrected and assembled with CLC Genomics Workbench (version 8.5.1) software (CLC bio, Aarhus, Denmark). LAB sequences were aligned to 16S rRNA gene sequences in EzBioCloud (Yoon et al. 2017) and yeast sequences aligned to 26S rRNA gene sequences in NCBI GenBank database, using BLAST algorithm (Zhang et al. 2000). Nucleotide sequences obtained in this study have been assigned the GenBank Accession Nos. MH43170–MH431829 for LAB and MH447333–MH447352 for yeasts, as indicated in Tables 3 and 4. Isolates identified as Lactobacillus plantarum or Lactobacillus pentosus were differentiated by sequencing of multiplex PCR products targeting the recA gene according to Torriani et al. (2001) and identified by BLAST searches in NCBI GenBank database. Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis were differentiated by sequencing of internal transcribed spacer (ITS)-regions with the primers ITS1 and ITS4, according to Esteve-Zarzoso et al. (1999) and identified by BLAST searches in NCBI GenBank database.
Results
Microbial counts of lait caillé end-products
In Table 1, pH and microbial counts are listed for the ten investigated lait caillé end-products obtained from the three local markets in Bobo-Dioulasso. The pH of the ten end-product market samples varied from pH 3.5 to 4.2. LAB were predominating all ten investigated samples with total LAB counts ranging from 7.8 ± 0.06 to 10.0 ± 0.03 log CFU/g and similar lactococcal counts of 7.9 ± 0.17 to 10.1 ± 0.05 log CFU/g. The Enterobacteriaceae counts ranged from < 3.0 log CFU/g and up to 8.4 ± 0.14 log CFU/g, hence in five of the samples comprising a significant part of the microbiota with 6.2 ± 0.10 to 8.4 ± 0.14 log CFU/g (S1, S6, S7, S8 and S9). For the yeasts, counts between 5.3 ± 0.03 and 8.7 ± 0.01 log CFU/g were obtained.
Microbial counts during lait caillé fermentation
Table 2 lists the pH and microbial counts from the detailed study of traditional lait caillé fermentation from the house-scale production site in the village Tolotama outside Bobo-Dioulasso. During the lait caillé fermentation, the pH decreased from 6.7 ± 0.01 in the fresh milk to 4.3 ± 0.08 at the end of the fermentation (59 h). Meanwhile, the total LAB counts increased during the fermentation from 5.4 ± 0.04 in fresh milk to 8.6 ± 0.02 log CFU/g in the end-product, with similar lactococcal counts of 5.4 ± 0.01 to 8.5 ± 0.15 log CFU/g throughout the fermentation. Enterobacteriaceae counts increased from 2.7 ± 0.01 log CFU/g at the onset of fermentation to high amounts of 6.7 ± 0.00 log CFU/g at the end of the fermentation. The counts for both total LAB and lactococci peaked at 53 h of fermentation at 9.0 ± 0.04 and 9.1 ± 0.06 log CFU/g, respectively, followed by a slight decrease towards the end of the fermentation. Enterobacteriaceae counts peaked at 28 h with 8.3 ± 0.13 log CFU/g and from this points decreased until the end of the fermentation. For yeasts counts, an increase throughout the fermentation was observed from 4.6 ± 0.08 log CFU/g (7 h) to a final load of 6.4 ± 0.11 log CFU/g.
Identification of LAB during lait caillé fermentation
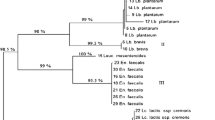
From the production site in Tolotama, a total of 251 Gram positive, catalase negative, rod or coccoid isolates, obtained throughout the lait caillé fermentation, were presumptively identified as LAB and genotypically characterized by (GTG)5-based rep-PCR fingerprinting followed by clustering. Table 3 shows the identities of representative LAB isolates from the eight clusters obtained, including GenBank Accession numbers. The representative LAB isolates were identified based on their 16S rRNA gene sequences and BLAST searches at EzBioCloud. Separate clusters for each identified LAB species, including all isolates from lait caillé, are presented in Online Resource 1 to 3.
Representative isolates of each LAB species are shown in Fig. 1. In total eight bacterial species were identified. The predominant part of the LAB isolates from lait caillé (39% of the total LAB isolates) were identified as Lactococcus lactis (cluster VIII) with similarities between 99.8 and 100% to EzBioCloud sequences. The most abundant genus in lait caillé was Enterococcus, clearly separated into three clusters (III, IV and VI) with 99.1–99.8% similarity to EzBioCloud sequences of several Enterococcus spp. Based on biochemical tests the three clusters were identified to species level. Isolates in cluster IV being the predominant Enterococcus spp. (21% of the total LAB isolates) were identified as Enterococcus lactis as the isolates produced acid from arabinose but not from amygdalin (Manero and Blanch 1999). Isolates in cluster III (16% of the total LAB isolates) were identified as Enterococcus hirae as the isolates were not able to produce acid from either L-arabinose or mannitol but possessed α-galactosidase activity (Manero and Blanch 1999). Isolates in cluster VI (6% of the total LAB isolates) were identified as Enterococcus faecium as the isolates were unable to produce CO2 from glucose but able to grow at 45 °C and to produce acid from arabinose, mannitol and galactose (Manero and Blanch 1999). Isolates in cluster II were identified as Leuconostoc mesenteroides (9% of the total isolated LAB) based on biochemical tests since the 16S rRNA gene sequences could not differentiate between L. mesenteroides and Leuconostoc pseudomesenteroides with 99.1–99.3% similarity to EzBioCloud sequences. Hence, isolates in cluster II had no growth at 45 °C but could grow in the presence of 6.5% NaCl (w/v). Further, no acid was formed from L-arabinose, mannitol and amygdalin, but acid was formed from galactose and CO2 was produced from glucose (Facklam and Elliott 1995). Isolates in cluster V were identified as Weissella paramesenteroides (4% of the total LAB isolates) and had 99.1–99.4% similarity to EzBioCloud sequences. Isolates in cluster VII were identified as Pediococcus pentosaceus (2% of the total LAB isolates) with 99.9% similarity to EzBioCloud sequences. Isolates in cluster I (1% of the total LAB isolates) were identified as Lactobacillus plantarum or Lactobacillus pentosus with 99.7–99.8% similarity to EzBioCloud sequences. By multiplex PCR of recA gene (Torriani et al. 2001) cluster I isolates were identified as L. plantarum with 100% similarity to EzBioCloud sequences (results not shown).
Identification of yeasts during lait caillé fermentation
From the production site in Tolotama, 169 isolates obtained during lait caillé fermentation were presumptively identified as yeasts following phenotypic characterisation. Table 4 shows the identities of representative yeast isolates from the four clusters obtained by analysis of (GTG)5-based rep-PCR fingerprinting, including GenBank Accession numbers. Separate clusters for each identified yeast species with all isolates from lait caillé are presented in Online Resource 4 and 5.
Representative isolates of each yeast species are shown in Fig. 2. In total four yeast species were identified in lait caillé. The predominant part of the yeast isolates (52% of the total isolated yeasts) were identified as Saccharomyces cerevisiae (cluster C) with 99.5–100% similarity to GenBank sequences. The second most abundant group of yeasts were found to belong to the Candida parapsilosis group (comprising Candida parapsilosis, C. orthopsilosis and C. metapsilosis) with 99.2–100% similarity to 26S rRNA gene sequences in GenBank, clearly separated into two clusters (A and D). Based on sequencing of ITS, isolates in cluster A (43% of the total isolated yeasts) were identified as C. parapsilosis with 100% similarity to GenBank sequences and isolated in cluster D (1% of the total isolated yeasts) were identified as C. orthopsilosis with 100 similarity to GenBank sequences (results not shown). Isolates in cluster B were identified as the genus Coniochaeta spp. (anamorph: Lecythophora) (3% of the total yeast isolates) with 99.4–100% similarity to 26S rRNA gene sequence in GenBank.
LAB and yeasts succession during lait caillé fermentation
Microbial successions of the identified LAB and yeasts were observed during lait caillé fermentation at the production site in Tolotama (Table 5). At the onset of the fermentation (0 h) the isolated LAB were dominated by P. pentosaceus comprising 50.0% of the isolated LAB, followed by W. paramesenteroides and L. mesenteroides accounting for 30.0% and 20.0% of the isolated LAB, respectively. After 7 h of fermentation the highest LAB species diversity was observed with seven identified species. At this time point (7 h), E. lactis, L. lactis and E. hirae were the most abundant species (21.3–31.9% of the isolated LAB) followed by E. faecium, W. paramesenteroides, L. mesenteroides and L. plantarum (2.5–10.6% of the isolated LAB). Throughout the rest of the fermentation, W. paramesenteroides and P. pentosaceus were not observed. From 13 h to the end of the fermentation (59 h), L. lactis was generally dominating ranging from 20.2 to 55.2% of the isolated LAB with an additional high abundance of E. lactis (8.6–64.9%) and E. hirae (7.2–47.2% of the isolated LAB). At the end of the fermentation lait caillé was predominated by L. lactis (31.4%) followed by E. hirae, L. mesenteroides and E. lactis (15.7–22.9% of the isolated LAB). Less frequently isolated LAB, were L. plantarum (2.5–5.2%) and E. faecium (3.4–16.6% of the isolated LAB) both occurring at the early stages and at the end of the lait caillé fermentation.
The yeasts in the lait caillé samples was dominated by C. parapsilosis during the first 18 h of fermentation ranging from 48.5 to 80.0%, followed by S. cerevisiae comprising 10.5–45.5% of the isolated yeasts. From 28 h and throughout the rest of the fermentation, yeast species diversity changed and S. cerevisiae was the predominating yeast, ranging between 47.4 and 100.0% of the isolated yeasts. From 53 h until the end of the fermentation (59 h) the only identified yeast species was S. cerevisiae. Other less frequently isolated yeasts during the fermentation were Coniochaeta spp. (3.0–10.5%) and C. orthopsilosis (3.0% to 5.9% of the isolated yeasts), occurring from 7 h to 35 h in the lait caillé fermentation.
Discussion
The samples from the local markets differed from those sampled at the end of fermentation in the house-hold production site in terms of fermentation conditions. In addition, the market samples were already in a state of stored end-products under ambient conditions, which may explain the differences in pH and e.g. Enterobacteriaceae counts. The pH of lait caillé from market samples (3.7 to 4.2) and from production site (4.3 ± 0.08) are comparable to the pH reported for other African fermented milk products, i.e. kivuguto (pH 4.5) from Rwanda (Karenzi et al. 2012) and nyarmie (pH 3.5 to 4.2) from Ghana (Obodai and Dodd 2006), with fermentation times of 24–48 h. Further, the results on LAB counts (7.8 ± 0.06 to 10.0 ± 0.03 log CFU/g) in the present study of both market samples of lait caillé end-products and from the production site in Tolotama showed predominance of LAB and are comparable to the findings for other African spontaneous fermented milk products where 8.1 to 9.0 log CFU/mL of LAB was reported in nyarmie (Obodai and Dodd 2006) and 8.0 to 8.7 log CFU/mL in nunu (Akabanda et al. 2013), both from Ghana. The high level of the presumptive LAB count (5.4 ± 0.04 log CFU/g), recorded at the beginning of the fermentation, may arise from the hygienic practices used at the farm. Previously, especially milk containers have been associated with relatively high total bacterial counts (Bonfoh et al. 2003), while also cow’s skins have been associated with noticeable lactobacilli counts (Monsallier et al. 2012). Similarly, the variability of microbial counts and pH observed among the samples from market could be explained by the variable, uncontrolled fermentation conditions; particularly the handling of the milk before and after the spontaneous fermentation.
Microbial successions are generally reported for spontaneous fermented food products (Akabanda et al. 2013; Greppi et al. 2013; Jespersen 2003; Nielsen et al. 2007; Wullschleger et al. 2013). In our study, eight different LAB species were identified in lait caillé from the production site in the village Tolotama, namely L. lactis, L. mesenteroides, E. lactis, E. hirae, E. faecium, P. pentosaceus, W. paramesenteroides and L. plantarum. At the end of the lait caillé fermentation L. lactis was the dominating LAB identified, which is similar to findings from end-products of e.g. kivuguto from Rwanda, the Masai fermented milk from Tanzania and Fulani fermented milk from Burkina Faso (Isono et al. 1994; Karenzi et al. 2012; Savadogo et al. 2004). In fact, L. lactis is typical of most of African fermented milk products, and commonly used as the acidifying starter culture for most industrialised fermented milk products. It has been reported as the most widely detected species in a survey covering up to 25 African fermented dairy products and raw milk (Jans et al. 2017). However, sometimes L. lactis is not the predominant LAB most probably due to differences in the environment and the processing. Thus, in nunu, a fermented milk from Ghana, Lactobacillus fermentum was reported as the dominant LAB (Akabanda et al. 2013) and in a recent study, Parker et al. (2018) reported Streptococcus and Lactobacillus as the dominant genera in another type of lait caillé (curdle milk based on pasteurised milk) from Northern Senegal. From the present study, L. lactis may be considered as the main acidifier of lait caillé. Furthermore, some strains of L. lactis have been reported to produce exopolysaccharides (Cerning 1995; Ruas-Madiedo et al. 2002) functioning as bio-thickening agents (Duboc and Mollet 2001). Hence, L. lactis might also improve the rheological properties of lait caillé (Kleerebezem et al. 1999). L. mesenteroides was isolated throughout the fermentation in the present study. This heterofermentative LAB has previously been reported among the dominant LAB of the Tunisian leben and the Rwandese kivuguto (Karenzi et al. 2012; Samet-Bali et al. 2012). Leuconostoc spp. are generally associated with production of aroma compounds in dairy products, by conversion of citrate into flavour compounds like diacetyl (Lore et al. 2005). Thus, L. mesenteroides may add to the development of the characteristic aroma of lait caillé. Furthermore, a noticeable amount of E. lactis (9–65%) and E. hirae (7–47% of the isolated LAB) were obtained throughout the lait caillé fermentation. Enterococcus spp. has been reported in other fermented milk products e.g. leben from Tunisia (Samet-Bali et al. 2012) and nunu from Ghana (Akabanda et al. 2013), as well as in many traditional cheeses from the Mediterranean countries (Moreno et al. 2006). Only few studies have dealt with the ability of enterococci as milk acidifiers, however, it has been reported that some species of E. faecium and E. faecalis grown in camel, ovine or caprine milk could produce relatively high amounts of lactic acid (El Hatmi et al. 2018; Freitas et al. 1999). Additionally, it has been reported that certain species of enterococci exhibit probiotic properties (Moreno et al. 2006, Quirós et al. 2007). On the contrary, the perceived safety of enterococci is affected by the fact that some are opportunistic pathogens, especially, E. faecalis, whereas E. faecium appears to pose a lower risk (Franz et al. 2003) and further antibiotic resistance and virulence factors have been reported for both enterococci species (Ogier and Serror 2008).
The yeast counts obtained in the present study, from the end-products of lait caillé at the production site in Tolotama (6.4 ± 0.11 log CFU/g), were for some samples higher than previously reported for nunu i.e. 5.0 to 5.8 log CFU/mL (Akabanda et al. 2013). Considering the level of yeasts in lait caillé, it could be assumed that the yeasts might play a role in the fermentation process, probably in the sensorial acceptability of the product by the consumer. The yeasts identified in lait caillé from Tolotama included S. cerevisiae, C. parapsilosis, C. orthopsilosis and Coniochaeta spp. Among these, S. cerevisiae was the only yeast detected at the end of the lait caillé fermentation. S. cerevisiae has often been isolated from African traditional fermented milk products (Akabanda et al. 2013; Gadaga et al. 2000; Obodai and Dodd 2006). Sudun et al. (2013) suggested that during the fermentation of airag, an alcoholic fermented milk product containing a co-culture of LAB and yeasts, the galactose resulting from degradation of lactose by the LAB promoted the growth of yeasts able to utilise galactose, such as S. cerevisiae (Sudun et al. 2013). A higher production of aroma compounds, such as ethanol, acetaldehyde and malty compounds has been reported to occur for some co-cultures of LAB and yeasts during fermentation of milk (Gadaga et al. 2001). Consequently, the authors suggested a possible interaction between LAB and yeasts during the fermentation process. Such interactions may also exist in lait caillé fermentation. Coniochaeta spp. has been isolated from wood or bark from different trees as well as from dung of various mammals (Damm et al. 2010), hence the Coniochaeta spp. identified in the lait caillé samples in the present study, were most likely introduced into the fermentation from the traditional straw-woven lids used to cover the fermentation containers or during the milking. It is though clear that the Coniochaeta spp. does not play a major role in the lait caillé fermentation.
Identifications of microorganisms based on 16S or 26S rRNA gene sequences pose, in some cases, difficulties due to high similarities between sequences of closely related species (Schleifer 2009). In the present study, it was not possible to differentiate between the isolated Enterococcus spp. based on the 16S rRNA sequences due to high similarity between sequences for these species (Švec and Franz 2014). Contrary, the rep-PCR fingerprinting profiles clearly separated the different Enterococcus spp. contributing to the identification of the isolated microorganisms from lait caillé. These findings are supported by previous studies, where rep-PCR was used to differentiate Enterococcus spp. (Pangallo et al. 2008; Švec et al. 2005). For C. parapsilosis and C. orthopsilosis it has previously been reported that only few nucleotides differ and that multi-gene analysis or ITS sequencing could be applied as techniques for identifying to species level (Tavanti et al. 2005). When studying the rep-PCR fingerprints obtained in the current study, clearly separated profiles were obtained for the two species, indicating that rep-PCR likewise could aid in the separation of C. parapsilosis and C. orthopsilosis.
In the present study it became clear that high amounts of Enterobacteriaceae occurred both in the ten lait caillé end-products sold at markets in Bobo-Dioulasso (up to 8.4 ± 0.14 log CFU/g) and at the production site in Tolotama (6.7 ± 0.00 log CFU/g), making these bacteria a considerable part of the microbiota in some of the lait caillé samples. Similar levels of Enterobacteriaceae have been reported in a previous study on hygienic quality of raw and sour milk products from Burkina Faso, sampled at different markets, with total coliform counts of 5.6 ± 0.59 log CFU/mL (Tankoano et al. 2016). Other traditional fermented milk products from Namibia and South Africa, sampled at household level, reported mean counts of coliforms to 6.5 log CFU/mL (Beukes et al. 2001). In a study on the microbiota in nunu, high levels of Enterobacteriales including Escherichia coli (7–13% relative abundance) and Klebsiella pneumoniae (3–71% relative abundance) were detected by shotgun amplicon sequencing (Walsh et al. 2017). The high level of Enterobacteriaceae in lait caillé can be explained by the poor hygienic conditions surrounding the milking step and milk storage, as observed during sampling. In addition, the manufacturing practices did not include pasteurisation or back-slopping and relied on spontaneous fermentation, which are major risk factors that enable various microorganisms, including potential pathogenic microorganisms, to grow during the fermentation (Broutin et al. 2007; Fondén et al. 2006). Moreover, C. parapsilosis was the dominant yeast until 28 h of fermentation. C. parapsilosis is a common spoilage yeast in fermented dairy products due to production of lipolytic and proteolytic enzymes (Fröhlich-Wyder 2003) and it is moreover recognised as an opportunistic pathogen (Silva et al. 2012). The high abundance of C. parapsilosis could indicate poor hygiene and ineffective cleaning procedures. Importantly, this yeast species decreased during the lait caillé fermentation in our study. This could be due to inhibitory compounds produced during the fermentation including possible antagonism from other yeasts present during the fermentation, e.g. through killer toxin production (Viljoen 2006).
Conclusion
Our results showed that lait caillé end-products sold at markets were partly dominated by LAB and yeasts along with considerable amounts of Enterobacteriaceae in some of the samples. These results were confirmed in the detailed study of the lait caillé fermentation. Further, microbial successions occurred during lait caillé fermentation with L. mesenteroides, P. pentosaceus and W. paramesenteroides being present at the onset of the fermentation. After 7 h the LAB diversity changed and L. lactis, E. lactis and E. hirae became the predominant LAB for the remaining of the fermentation. For yeasts C. parapsilosis was dominating the first 18 h of lait caillé fermentation. After 35 h S. cerevisiae became the most abundant yeast species and after 53 h the only yeast species identified. The deeper understanding of the microbiota involved in the lait caillé fermentation obtained in this study and the fact that high levels of especially Enterobacteriaceae were detected, point out the need to upgrade the lait caillé process by introducing pasteurisation combined with the use of acidification cultures, specifically made for lait caillé, to enhance the quality and food safety.
References
Akabanda F, Owusu-Kwarteng J, Glover RLK, Tano-Debrah K (2010) Microbiological characteristics of Ghanaian traditional fermented milk product, nunu. Nat Sci 8:178–187
Akabanda F, Owusu-Kwarteng J, Tano-Debrah K, Glover RLK, Nielsen DS, Jespersen L (2013) Taxonomic and molecular characterization of lactic acid bacteria and yeasts in nunu, a Ghanaian fermented milk product. Food Microbiol 34:277–283
Beukes EM, Bester BH, Mostert JF (2001) The microbiology of South African traditional fermented milks. Int J Food Microbiol 63:189–197
Björkroth J, Dicks LMT, Endo A, Holzapfel WH (2014) The genus Leuconostoc. In: Holzapfel WH, Wood BJB (eds) Lactic acid bacteria. Wiley, Chichester, pp 391–404. https://doi.org/10.1002/9781118655252.ch23
Bonfoh B, Wasem A, Traoré AN, Fané A, Spillmann H, Simbé CF, Alfaroukh IO, Nicolet J, Farah Z, Zinsstag J (2003) Microbiological quality of cow’s milk taken at different intervals from the udder to the selling point in Bamako (Mali). Food Control 14:495–500
Broutin C, François M, Niculescu NLN (2007) Gestion de la qualité dans la transformation laitière: expérimentation d’une démarche d’élaboration concertée de guides de bonnes pratiques d’hygiène au Sénégal et au Burkina Faso. Revue Elev Méd Vét Pays Trop 60:163
Cerning J (1995) Production of expolysaccharides by lactic acid bacteria and dairy propionibacteria. Lait 75:463–472
Damm U, Fourie PH, Crous PW (2010) Coniochaeta (Lecythophora), Collophora gen. nov. and phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 24:60–80
Duboc P, Mollet B (2001) Applications of exopolysaccharides in the dairy industry. Int Dairy J 11:759–768
Duteurtre G (2007) Commerce et développement de l’ élevage laitier en Afrique de l’ Ouest: une synthèse. Revue Elév Méd Vét Pays Trop 60:209–223
El Hatmi H, Jrad Z, Oussaief O, Nasri W, Sbissi I, Khorchani T et al (2018) Fermentation of dromedary camel (Camelus dromedarius) milk by Enterococcus faecium, Streptococcus macedonicus as a potential alternative of fermented cow milk. LWT Food Sci Technol 90:373–380
Esteve-Zarzoso B, Belloch C, Uruburu F, Querol A (1999) Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int J Syst Bacteriol 49:329–337
Facklam R, Elliott JA (1995) Identification, classification, and clinical relevance of catalase-negative, gram-positive cocci, excluding the streptococci and enterococci. Clin Microbiol Rev 8:479–495
Fondén R, Leporanta K, Svensson U (2006) Nordic/scandinavian fermented milk products. In: Tamime A (ed) Fermented milks. Blackwell Publishing Ltd, Oxford, pp 156–173. https://doi.org/10.1002/9780470995501.ch7
Franz CMAP, Stiles ME, Schleifer KH, Holzapfel WH (2003) Enterococci in foods—a conundrum for food safety. Int J Food Microbiol 88:105–122
Freitas AC, Pintado AE, Pintado ME, Malcata FX (1999) Organic acids produced by lactobacilli, enterococci and yeasts isolated from Picante cheese. Eur Food Res Technol 209:434–438
Fröhlich-Wyder MT (2003) Yeasts in dairy products. In: Boekhout T, Robert V (eds) Yeasts in food. Elsevier, Hamburg, pp 209–237. https://doi.org/10.1533/9781845698485.209
Gadaga TH, Mutukumira AN, Narvhus JA (2000) Enumeration and identification of yeasts isolated from Zimbabwean traditional fermented milk. Int Dairy J 10:459–466
Gadaga TH, Mutukumira AN, Narvhus JA (2001) The growth and interaction of yeasts and lactic acid bacteria isolated from Zimbabwean naturally fermented milk in UHT milk. Int J Food Microbiol 68:21–32
Gonfa A, Foster HA, Holzapfel WH (2001) Field survey and literature review on traditional fermented milk products of Ethiopia. Int J Food Microbiol 68:173–186
Gregersen T (1978) Rapid method for distinction of gram-negative from gram-positive bacteria. Eur J Appl Microbiol 5:123–127
Greppi A, Rantisou K, Padonou W, Hounhouigan J, Jespersen L, Jakobsen M et al (2013) Yeast dynamics during spontaneous fermentation of mawè and tchoukoutou, two traditional products from Benin. Int J Food Microbiol 165:200–207
Isono Y, Shingu I, Shimizu S (1994) Identification and characteristics of lactic acid bacteria isolated from Masai fermented milk in northern Tanzania. Biosci Biotechnol Biochem 58:660–664
Jans C, Bugnard J, Njage PMK, Lacroix C, Meile L (2012) Lactic acid bacteria diversity of African raw and fermented camel milk products reveals a highly competitive, potentially health-threatening predominant microflora. LWT Food Sci Technol 47:371–379
Jans C, Meile L, Kaindi DWM, Kogi-Makau W, Lamuka P, Renault P et al (2017) African fermented dairy products—overview of predominant technologically important microorganisms focusing on African Streptococcus infantarius variants and potential future applications for enhanced food safety and security. Int J Food Microbiol 250:27–36
Jensen MP, Ardö Y, Vogensen FK (2009) Isolation of cultivable thermophilic lactic acid bacteria from cheeses made with mesophilic starter and molecular comparison with dairy-related Lactobacillus helveticus strains. Lett Appl Microbiol 49:396–402
Jespersen L (2003) Occurrence and taxonomic characteristics of strains of Saccharomyces cerevisiae predominant in African indigenous fermented foods and beverages. FEMS Yeast Res 3:191–200
Jespersen L, Nielsen D, Honholt S, Jakobsen M (2005) Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS Yeast Res 5:441–453
Karenzi E, Dauphin RD, Mashaku A, Majad L, Munyanganizi B, Thonart P (2012) Fermentation of kivuguto, a Rwandese traditional milk: selection of microbes for a starter culture. Sci Technol C Biotechnol 36:9–17
Kleerebezem M, van Kranenburg R, Tuinier R, Boels IC, Zoon P, Looijesteijn E et al (1999) Exopolysaccharides produced by Lactococcus lactis: from genetic engineering to improved rheological properties? Antonie Van Leeuwenhoek 76:357–365
Koussou MO, Grimaud P, Mopaté LY (2007) Evaluation de la qualité physico-chimique et hygiénique du lait de brousse et des produits laitiers locaux commercialisés dans les bars laitiers de N’Djamena au Tchad. Revue Elev Méd Vét Pays Trop 60:45
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Lore TA, Mbugua SK, Wangoh J (2005) Enumeration and identification of microflora in suusac, a Kenyan traditional fermented camel milk product. LWT Food Sci Technol 38:125–130
Manero A, Blanch AR (1999) Identification of Enterococcus spp. with a biochemical key. Appl Environ Microbiol 65:4425–4430
Mathara JM, Schillinger U, Kutima PM, Mbugua SK, Holzapfel WH (2004) Isolation, identification and characterisation of the dominant microorganisms of kule naoto: the Maasai traditional fermented milk in Kenya. Int J Food Microbiol 94:269–278
Monsallier F, Verdier-Metx I, Agabriel C, Martin B, Montel MC (2012) Variability of microbial teat skin flora in relation to farming practices and individual dairy cow characteristics. Dairy Sci Technol 92:265–278
Moreno MR, Sarantinopoulos P, Tsakalidou E, De Vuyst L (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106:1–24
Nielsen DS, Teniola OD, Ban-Koffi L, Owusu M, Andersson TS, Holzapfel WH (2007) The microbiology of Ghanaian cocoa fermentations analysed using culture-dependent and culture-independent methods. Int J Food Microbiol 114:168–186
Obodai M, Dodd CER (2006) Characterization of dominant microbiota of a Ghanaian fermented milk product, nyarmie, by culture- and nonculture-based methods. J Appl Microbiol 100:1355–1363
Ogier J-C, Serror P (2008) Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol 126:291–301
Pangallo D, Drahovska H, Harichova J, Karelova E, Chovanova K, Aradska J et al (2008) Evaluation of different PCR-based approaches for the identification and typing of environmental enterococci. Antonie Van Leeuwenhoek 93:193–203
Parker M, Zobrist S, Donahue C, Edick C, Mansen K, Hassan ZNM et al (2018) Naturally fermented milk from northern Senegal: bacterial community composition and probiotic enrichment with Lactobacillus rhamnosus. Front Microbiol 9:2218. https://doi.org/10.3389/fmicb.2018.02218
Quirós A, Ramos M, Muguerza B, Delgado MA, Miguel M, Aleixandre A et al (2007) Identification of novel antihypertensive peptides in milk fermented with Enterococcus faecalis. Int Dairy J 17:33–41
Ruas-Madiedo P, Tuinier R, Kanning M, Zoon P (2002) Role of exopolysaccharides produced by Lactococcus lactis subsp. cremoris on the viscosity of fermented milks. Int Dairy J 12:689–695
Samet-Bali O, Ennouri M, Dhouib A, Attia H (2012) Characterisation of typical Tunisian fermented milk: Leben. Afr J Microbiol Res 6:2169–2175
Savadogo A, Ouattara CAT, Savadogo PW, Ouattara AS, Barro N, Traore AS (2004) Microorganisms involved in Fulani traditional fermented milk in Burkina Faso. Pak J Nutr 3:134–139
Schleifer KH (2009) Classification of Bacteria and Archaea: past, present and future. Syst Appl Microbiol 32:533–542
Silva S, Negri M, Henriques M, Oliveira R, Williams DW, Azeredo J (2012) Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol Rev 36:288–305
Sudun, Wulijideligen, Arakawa K, Miyamoto M, Miyamoto T (2013) Interaction between lactic acid bacteria and yeasts in airag, an alcoholic fermented milk. Anim Sci J 84:66–74
Švec P, Franz CMAP (2014) The genus Enterococcus. In: Holzapfel WH, Wood BJB (eds) Lactic acid bacteria. Wiley, Chichester, pp 175–211. https://doi.org/10.1002/9781118655252.ch15
Švec P, Vancanneyt M, Seman M, Snauwaert C, Lefebvre K, Sedláček I et al (2005) Evaluation of (GTG)5-PCR for identification of Enterococcus spp. FEMS Microbiol Lett 247:59–63
Tankoano A, Kabore D, Savadogo A, Soma A, Fanou Fogny N, Compaore-Sereme D et al (2016) Evaluation of microbiological quality of raw milk, sour milk and artisanal yoghurt from Ouagadougou, Burkina Faso. Afr J Microbiol Res 10:535–541
Tavanti A, Davidson AD, Gow NAR, Maiden MCJ, Odds FC (2005) Candida parapsilosis Groups II and III. J Clin Microbiol 43:284–292
Taylor WI, Achanzar D (1972) Catalase test as an aid to the identification of Enterobacteriaceae. Appl Microbiol 24:58–61
Torriani S, Clementi F, Vancanneyt M, Hoste B, Dellaglio F, Kersters K (2001) Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum species by RAPD-PCR and AFLP. Syst Appl Microbiol 24:554–560
Viljoen CB (2006) Yeast ecological interactions. Yeast–yeast, yeast–bacteria, yeast–fungi interactions and yeasts as biocontrol agents. In: Querol A, Fleet GH (eds) Yeast in food and beverages. Springer, Berlin, pp 83–110. https://doi.org/10.1007/978-3-540-28398-0_4
Walsh AM, Crispie F, Daari K, O’Sullivan O, Martin JC, Arthur CT et al (2017) Strain-level metagenomic analysis of the fermented dairy beverage nunu highlights potential food safety risks. Appl Environ Microbiol. https://doi.org/10.1128/aem.01144-17
Wullschleger S, Lacroix C, Bonfoh B, Sissoko-Thiam A, Hugenschmidt S, Romanens E et al (2013) Analysis of lactic acid bacteria communities and their seasonal variations in a spontaneously fermented dairy product (Malian fènè) by applying a cultivation/genotype-based binary model. Int Dairy J 29:28–35
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgements
The authors are grateful to the local producers and retailers who contributed to the study. The authors would like to acknowledge Ministry of Foreign Affairs of Denmark (Danida) for funding through the project Preserving African food microorganisms for Green Growth (Project Number DFC No. 13-04KU).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11274_2019_2672_MOESM1_ESM.pptx
Supplementary material 1 (PPTX 5773 kb) Online Resource 1: Dendrogram of rep-PCR cluster analysis of all Lactococcus lactis isolates from lait caillé during the fermentation, based on Dice’s coefficient of similarity with the unweighted pair group method with arithmetic average clustering algorithm (UPGMA) with sub-clusters indicated
11274_2019_2672_MOESM2_ESM.pptx
Supplementary material 2 (PPTX 6335 kb) Online Resource 2: Dendrogram of rep-PCR cluster analysis of all Enterococcus spp. isolates from lait caillé during the fermentation, based on Dice’s coefficient of similarity with the unweighted pair group method with arithmetic average clustering algorithm (UPGMA) with sub-clusters indicated
11274_2019_2672_MOESM3_ESM.pptx
Supplementary material 3 (PPTX 5538 kb) Online Resource 3: Dendrograms of rep-PCR cluster analysis of all less frequent LAB isolates from lait caillé during the fermentation, based on Dice’s coefficient of similarity with the unweighted pair group method with arithmetic average clustering algorithm (UPGMA) with sub-clusters indicated, a) Leuconostoc mesenteroides, b) Lactobacillus plantarum, c) Pediococcus pentosaceus, d) Weissella paramesenteroides
11274_2019_2672_MOESM4_ESM.pptx
Supplementary material 4 (PPTX 6856 kb) Online Resource 4: Dendrogram of rep-PCR cluster analysis of all Saccharomyces cerevisiae isolates from lait caillé fermentation, based on Dice’s coefficient of similarity with the unweighted pair group method with arithmetic average clustering algorithm (UPGMA) with sub-clusters indicated
11274_2019_2672_MOESM5_ESM.pptx
Supplementary material 5 (PPTX 6414 kb) Online Resource 5: Dendrogram of rep-PCR cluster analysis of all a) Candida parapsilosis, b) Candida orthopsilosis and c) Coniochaeta spp. isolates from lait caillé fermentation, based on Dice’s coefficient of similarity with the unweighted pair group method with arithmetic average clustering algorithm (UPGMA) with sub-clusters indicated
Rights and permissions
About this article
Cite this article
Bayili, G.R., Johansen, P., Nielsen, D.S. et al. Identification of the predominant microbiota during production of lait caillé, a spontaneously fermented milk product made in Burkina Faso. World J Microbiol Biotechnol 35, 100 (2019). https://doi.org/10.1007/s11274-019-2672-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-019-2672-3