Abstract
Breast cancer (BC) is the leading cause of cancer mortality in women and a major risk to world health. Therefore, effective strategies are required for prompt diagnosis and treatment. Nowadays, non-coding RNAs (ncRNAs), particularly long ncRNAs (lncRNAs), have assumed a significant role in the prognosis and diagnosis of diseases, including cancer. In the present study, surveying the bioinformatic tools, including the lncRNADisease v2.0, OncoDB, InteractiVenn, GEPIA, RAID, COXPRESdb, DAVID v6.8, GEO2R, and LncSEA, we proposed the Maternally Expressed Gene (MEG3) as a potential biomarker in BC. This lncRNA significantly downregulates in BC and is associated with tumor size, metastasis, and pathological stage. MEG3 expression is downregulated in several types of primary human cancers and tumor cell lines, which raises the possibility that it could act as a tumor suppressor. The results suggest that MEG3 may play a crucial role in fundamental pathways, including apoptosis, and interact with essential genes and proteins such as P53. It may also be associated with the prognosis, proliferation, migration, invasion, and metastasis of BC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With an anticipated 2.26 million cases reported in 2020, breast cancer (BC) is the most often diagnosed disease in the world. BC is the main cause of cancer mortality in women, considered a serious threat to global health [1]. Genetic factors, hormones, lifestyle, and proliferative breast lesions with atypia are all risk factors for developing BC [2]. It is divided into three main subtypes, including triple-negative, ERBB2-positive, and hormone receptor–positive/ERBB2-negative, depending on the presence or absence of molecular markers for the human epidermal growth factor 2 (ERBB2; formerly HER2), progesterone receptor (PR), and estrogen receptor (ER) [3].
The benefit of transcriptomics has evolved recently thanks to the expansion of high-throughput sequencing methods. It has illustrated the function of non-coding RNAs (ncRNAs), especially long ncRNAs (lncRNAs), in human illness and cellular function [4]. LncRNAs are a class of recently discovered non-coding transcripts with a length of more than 200 nucleotides [5]. Recent studies have defined distinct roles for either particular lncRNAs or the process of lncRNA synthesis, despite the fact that at least some lncRNA production may represent transcriptional “noise” [6]. It has been suggested that lncRNAs play a role in tumorigenesis and tumor metastasis by controlling gene expression at the transcriptional and translational levels [7]. The tumor microenvironment (TME), autophagy, apoptosis, cell cycle, epithelial-mesenchymal transition (EMT), DNA repair, epigenetic alteration, and drug efflux are merely some of the recent studies that have demonstrated the essential and various functions of lncRNAs in BC chemoresistance [8]. Progress in analytical methods gradually made the early detection of BC possible through biomarker scanning in nipple aspirate fluid, sweat, tears, blood, breath, and urine [9]. The cellular damage produced by lncRNAs may be alleviated by targeting them [10]. RNA sequencing (RNA-seq) identified numerous up- and downregulated genes involved in many biological pathways, such as EMT and paths related to mammary gland development [11]. Although RNA-seq presents an unequaled platform for identifying appropriate targets for BC precision medicine, current molecular-based prognostic markers are not the best fit for RNA-seq data, necessitating comprehensive genomic testing [12]. Two major regulators of gene expression, lncRNAs and microRNAs (miRNAs), functionally intercede the mechanisms of pathogenesis. The lncRNA-miRNA axis might have oncogenic or tumor suppressor effects depending on the unique lncRNA/miRNA interaction, making it crucial to define the lncRNA-miRNA axis for evaluating targetability [13]. The identified pathways, genes, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, and gene ontology (GO) terms may serve as a foundation for further research in BC and the potential creation of new therapies, diagnostics, and prognostic biomarkers [14].
In order to study potential markers associated with disease phenotype and prognosis, we can develop networks of interactions between genes and proteins by analyzing differentially expressed RNAs, such as lncRNAs, miRNAs, and mRNAs. In this study, following a thorough examination of MEG3 lncRNA and its interactions with RNAs, proteins, and medicines, we determined that MEG3 is a crucial gene involved in the invasion, prognosis, and other properties of BC.
Methods
Identifying lncRNA MEG3
The LncRNA and Disease Database version 2.0 (LncRNADisease v2.0) (https://www.cuilab.cn/lncrnadisease) was used to choose lncRNAs associated with BC. Based on patient information from OncoDB (https://oncodb.org/), differentially expressed genes (DEG) enrolling in tumor size, metastasis, and clinical stage of BC were separated. Common lncRNA between all four mentioned criteria was chosen by drawing a Venn diagram using InteractiVenn web-based tool (http://www.interactivenn.net/index.html).
Expression of lncRNA MEG3 in the Cancer Genome Atlas (TCGA) and GTEx Tissues
The GEPIA version 2.0 (http://gepia2.cancer-pku.cn) was used to compare the MEG3 lncRNA expression levels in BC and non-tumor tissues. A web-based tool, Kaplan–Meier plotter (https://kmplot.com/analysis/) and GEPIA were utilized to analyze the expression levels of MEG3 in relation to the prognosis of BC, which includes overall survival, disease-free survival, relapse-free survival, and progression-free survival.
Gene–Gene and Gene-Protein Network of the lncRNA MEG3
RAID database version 2.0 (https://www.rna-society.org/raid2/) was used to analyze MEG3-Gene and MEG3-Protein interactions, and the networks were created using Cytoscape version 3.9.1.
KEGG Pathway and GO Term Analysis
The COXPRESdb database version 8.1 (https://coxpresdb.jp/) was utilized to determine the genes associated with MEG3. Afterwards, Database for Annotation, Visualization and Integrated Discovery (DAVID) version 6.8 (https://david.ncifcrf.gov/) was used to carry out KEGG pathway and GO analyses of MEG3-associated genes.
Validation of MEG3 Expression Profiling in the GSE Dataset
Gene expression omnibus (GEO) dataset GSE2219 was utilized to confirm differential expression of the MEG3 in BC cell lines and mammary epithelial cells. Using the GEO2R web-tool (https://www.ncbi.nlm.nih.gov/geo/geo2r/), volcano plot of the DEGs included in the GSE1299 dataset was displayed.
Enrichment Analysis of MEG3
Correlation between MEG3 and cancer hallmarks and interactions with drugs were investigated using LncSEA version 2.0 (https://bio.liclab.net/LncSEA/index.php). Using OncoDB, the correlation between MEG3 and clinical parameters was investigated.
Results
Identifying lncRNA MEG3
Surveying the LncRNADisease v2.0, 200 distinct lncRNAs were discovered associated with BC. On the other hand, the OncoDB revealed 1026, 1673, and 386 DEGs in BC patients related to different tumor sizes, metastasis stages, and pathological stages, respectively. Based on the Venn diagram (Fig. 1a), MEG3 was chosen as the common lncRNA between these four criteria.
Selecting the common lncRNA related to BC clinicopathological traits and expression levels of MEG3 lncRNA. a MEG3 was identified as the common gene, using the InteractiVenn web-based tool. Each digit in the figure is related to the number of common genes between different sections. b Expression levels of MEG3 in BC (1085) and in normal breast (291) tissues indicate a downregulated level of its expression in BC. c Expression of MEG3 in different clinical stages of BC represents a significant difference. Red: BC tissues. Blue: normal breast tissues. Black dots: individual cases
Expression and Survival Analysis of lncRNA MEG3
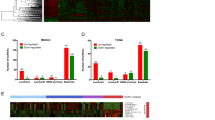
MEG3 expression in BC tissues was lower than in normal breast tissues, according to the results of gene expression in TCGA and GTEx samples (Fig. 1b). There was a significant difference between the clinical stages of BC and the levels of MEG3 expression (Fig. 1c; P = 0.03). Lower MEG3 expression levels were not shown to be substantially related to a worse prognosis for BC patients’ survival (Fig. 2a–f).
Kaplan–Meier survival curves based on MEG3 lncRNA expression level. a There is no significant relation between the overall survival rate of patients with BC and lncRNA MEG3 expression levels. b MEG3 expression level has no noticeable effect on the disease-free survival rate of patients with BC. c–f There is no significant relationship between MEG3 lncRNA expression level and c overall survival rate of patients with BC, d relapse-free survival rate of patients with BC, e disease-specific survival rate of patients with BC, and f progression-free survival rate of patients with BC
Gene–Gene and Gene-Protein Network of the MEG3 lncRNA
Based on the RAID database, 73 miRNAs, 3 mRNAs, 2 lncRNAs, and 20 proteins had interactions with MEG3 (Fig. 3a and b). The most significant interactions of MEG3 were with TP53 mRNA, TUG1 and CRNDE lncRNAs, has-miR-140-5p miRNA, and TP53 protein, based on the combined score given to each interaction on the RAID database.
RAID analysis presented by Cytoscape and GO and KEGG analysis. a Using Cytoscape 3.9.1, a gene–gene interaction network and b a gene-protein interaction network for the MEG3 was created. Protein-coding genes are represented by blue rectangles, lncRNAs by orange ovals, and miRNAs by green diamonds. Symbolizing proteins are blue circles. Highest combined score is represented by edges with thicker and deeper hue (yellow to red). c GO term and d KEGG pathways enrichment plots of the MEG3 lncRNA and its associated genes enriched the cell adhesion, extracellular region, extracellular matrix structural constituent, and PI3K-Akt signaling pathway as biological process, cellular component, molecular function, and KEGG pathway, respectively. Deeper color (red to blue) bars represent higher log10 (P value)
KEGG Pathway and GO Term Analysis
Using the COXPRESdb database, the top 200 genes associated with MEG3 were determined. Eight biological processes (BP), 6 cellular components (CC), and 8 molecular functions (MF) were significantly enriched, according to the GO enrichment results (Fig. 3c). Three KEGG pathways were considerably enriched, according to KEGG enrichment analyses (Fig. 3d). The most significant BP, CC, MF and KEGG pathway were cell adhesion (GO:0007155, P = 1.9E − 9), extracellular region (GO:0005576, P = 1.2E − 14), extracellular matrix structural constituent (GO:0005201, P = 7.6E − 22), and PI3K-Akt signaling pathway (hsa04151, P = 6.2E − 6), separately.
Enrichment Analysis of MEG3
In order to analyze drug interactions and cancer hallmarks, the LncSEA database was employed. The results revealed strong interactions between cisplatin and temozolomide with MEG3 as well as the major contribution of MEG3 with cancer hallmarks (Table 1). Based on OncoDB database, MEG3 is significantly downregulated in BC patients and significantly affected the tumor size, pathological stages, and metastasis stages (Fig. 4a).
Correlation between the expression of lncRNA MEG3 and different metastasis, tumor size, and pathological stages of patients with BC and volcano plot of DEGs in GSE1299. a MEG3 expression decreased nearly 2.5-fold in BC samples than normals, and this reduction significantly affected the tumor sizes, pathological stages, and metastasis stages of BC. b DEGs between mammary epithelial cells and BC cell lines have been identified. There were 2396 DEGs in GSE1299. Among them, 530 and 600 genes were significantly (|log2FC|≥ 1) up- and downregulated. MEG3 is one of the significantly downregulated genes in this dataset and was represented with the yellow star. The red dots represent upregulated genes, and the blue ones represent downregulated genes. Black dots represent the insignificant data
Validation of MEG3 Expression Profiling in the GSE Dataset
The volcano plot of the DEGs found in the GSE1299 dataset was displayed in Fig. 4b. Based on these findings, the expression of MEG3 in BC cell lines was lower compared to mammary epithelial cells (adj. P-value = 0.01, log2(fold change) = − 0.955).
Discussion
BC is the most frequent cancer diagnosed globally, and its incidence has increased over the past few decades [15]. Developing precise diagnostic and prognostic biomarkers may increase the survival rates of BC patients. LncRNAs can act alone or in association with miRNAs and other molecules as part of multiple pathways, to stimulate or repress gene expression [16]. There is expanded evidence that lncRNAs play crucial roles in numerous biological processes, including the initiation and progress of cancer [17].
In this study, the LncRNADisease v2.0 was used to choose lncRNAs associated with BC. Using patients’ data from OncoDB, DEGs in terms of tumor size, metastasis, and pathological stage were isolated. Using the InteractiVenn web-based application, the MEG3 was selected as the common lncRNA between the four criteria.
MEG3 is a lncRNA located on chromosome 14q32.3 [18] and modifies the expression of target genes through transcription, translation, post-translational alterations, and epigenetic regulation [19]. Several primary human tumors exhibit downregulated MEG3 expression, which suggests that it could serve as a tumor suppressor [20]. According to the research by Wang et al., MEG3 is downregulated in colorectal cancer [21]. Overexpression of MEG3 lncRNA suppresses cell proliferation and invasion and induces apoptosis in ovarian cancer by sponging miR-205-5p [22] and its abnormal expression prognosticates low survival in ovarian cancer [23]. This lncRNA also prevents the development of non-small cell lung cancer by inhibiting telomere function, cell proliferation, telomerase activity, cell migration, and invasion through control of the expression of the DKC1 protein [24]. Moreover, the downregulation of MEG3 plays a significant role in the development of hepatocellular carcinoma [25], prostate [26], and ovarian cancer [27, 28] and findings point to the possibility of MEG3 expression as a prognostic marker and potential immunotherapeutic target for gliomas [29] bladder cancer [30] and cervical cancer [31]. Besides, multiple studies indicate the downregulation of MEG3 in BC which is consistent with our findings. For BC patients, the lower MEG3 levels is a diagnostic and unfavorable prognostic feature [32, 33].
Methylation of the MEG3 promoter by DNMT1 activity downregulates the MEG3 expression and promotes the malignant behavior of BC cells due to the freedom of miR-494-3p from the MEG3 trap [34]. Zhang et al. proposed that MEG3 inhibits the growth of BC and triggers apoptosis through the activation of the ER stress, NF-κB, and p53 pathways where NF-κB signaling is necessary for MEG3-induced p53 activation in BC cells [35]. As we proposed, the MEG3 level may affect the tumor sizes, metastasis, and pathological stages of BC. According to Zhang et al. [36], the expression rate of this lncRNA is associated with lymph node metastasis, various grades, and the TNM stage of BC. Although our results indicate no noticeable prognostic value for MEG3 in BC, it is potentially related to poor overall survival, progression-free survival, and 5-year survival rate, based on the research [36, 37]. MEG3 level may also differentiate between the triple-negative status of BC—negatively correlated with MEG3 expression level—and estrogen or progesterone receptor–positive status—which are positively related [37]. Though, our results show no significant differences between various grades of BC in the MEG3 rate.
In mining the RAID database, we achieved the most significant interactions of MEG3 with TP53 mRNA, TUG1 and CRNDE lncRNAs, has-miR-140-5p miRNA, and TP53 protein. TP53, an effective tumor suppressor gene, frequently targeted by tumorigenesis processes, and MEG3, like some other lncRNAs, interferes in regulating its expression in BC [38]. This lncRNA affects the transcriptional level of MDM2 and stabilizes TP53 to prevent the progression of BC [39]. As in BC [35], MEG3 induces apoptosis in hepatocellular carcinoma by promoting the NF‑κB-related ER stress and p53 pathway [40]. MEG3 is also correlated with TP53 by regulating the FOXP3 expression and upregulating the miR-149-3p [41]. Sequestration of miR-140-5p with MEG3 has a confirmed role in the osteogenic differentiation of human adipose tissue–derived stem cells (hASCs) [42] which authenticates the competing endogenous task of MEG3 about the miR-140-5p, as our study proposed. This correlation also protects the number of circulating endothelial progenitor cells (EPCs) in metabolic syndrome [43]. TUG1 expression indicates a coordinated rhythm with MEG3 expression in cisplatin-resistant tissues [23] and diabetic patients [44, 45]. It may present a close relationship between these two lncRNAs.
Gene Ontology analysis predicted that MEG3 participates in cell adhesion, and most of its target genes are constituents of the extracellular matrix. MEG3 is one of the genes that regulate epithelial-mesenchymal transition (EMT), so its downregulation induces EMT in C2C12 myoblasts and inhibits myogenesis [46]. This lncRNA represses the intracellular cell adhesion molecule-1 (ICAM-1) expression by sponging the miR-147 to reduce cell viability and migration [47]. miR-147 has a target site in the promoter of the ICAM-1 gene and positively regulates its expression [47].
Due to the importance of PI3K/Akt signaling in controlling the main biological processes like cell growth, apoptosis, and migration [48], this pathway is exploited by multiple genes, including MEG3, to interfere with tumorigenesis [48]. Zhu et al. indicated that MEG3 acts as a tumor inhibitor by reversing miR-21-mediated activation of PI3K/Akt pathway in BC cells [48]. The inhibitory interference of MEG3 in the PI3K/AKT signaling is demonstrated in hepatocellular carcinoma [49], inflammatory disorders [50], and diabetic retinopathy (DR) [51]. Consistent with them, we have also predicted the PI3K/AKT signaling as the crucial pathway exploited by MEG3 to prevent the BC progression based on the KEGG analysis.
LncSEA data guided our study toward a significant interaction between MEG3 and cisplatin. This interaction is not strange because, in triple-negative breast cancer (TNBC), cisplatin (DDP) utilizes MEG3 expression to exert its anti-tumor effect [52]. DDP-related upregulation of MEG3 promotes the NLRP3/caspase-1/GSDMD pyroptosis pathway and sensitizes TNBC to cisplatin [52]. The low expression level of MEG3 in cisplatin-resistant cells of non-small cell lung cancer (NSCLC) is another proof demonstrating the effect of MEG3 on the tumor cell response to cisplatin [53].
Some obstacles should be mentioned despite the scientific relevance of the findings. First, the results are obtained using organic bioinformatics techniques and are entirely reliant on the TCGA database. Even if certain conclusions had been confirmed by the GEPIA database, the discoveries needed to be evaluated. Second, a typical follow-up period should be used to examine the prognostic signature of MEG3 (three to 5 years).
Conclusion
According to the findings, MEG3 may have a significant role in critical pathways such as apoptosis and interactions with crucial genes and proteins, as well as in the prognosis, proliferation, migration, invasion, and metastasis of BC. More clinical data is necessary to determine the precise role of MEG3.
Data Availability
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
References
Wilkinson L, Gathani T (2022) Understanding breast cancer as a global health concern. Br J Radiol 95(1130):20211033. https://doi.org/10.1259/bjr.20211033
Grogan Fleege NM, Cobain EF (2022) Breast cancer management in 2021: a primer for the obstetrics and gynecology. Best Pract Res Clin Obstet Gynaecol 82:30–45. https://doi.org/10.1016/j.bpobgyn.2022.02.004
Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L (2019) lncRNA OSTN-AS1 may represent a novel immune-related prognostic marker for triple-negative breast cancer based on integrated analysis of a ceRNA network. Front Genet 10:850. https://doi.org/10.3389/fgene.2019.00850
Sideris N, Dama P, Bayraktar S, Stiff T, Castellano L (2022) LncRNAs in breast cancer: a link to future approaches. Cancer Gene Ther 29(12):1866–1877. https://doi.org/10.1038/s41417-022-00487-w
Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T (2018) Emerging roles of long non-coding RNA in cancer. Cancer Sci 109(7):2093–2100. https://doi.org/10.1111/cas.13642
Nojima T, Proudfoot NJ (2022) Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat Rev Mol Cell Biol 23(6):389–406. https://doi.org/10.1038/s41580-021-00447-6
Wu P, Mo Y, Peng M, Tang T, Zhong Y, Deng X et al (2020) Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer 19(1):22. https://doi.org/10.1186/s12943-020-1147-3
Ye P, Feng L, Shi S,Dong C (2022) The mechanisms of lncRNA-mediated multidrug resistance and the clinical application prospects of lncRNAs in breast cancer. Cancers (Basel) 14(9). https://doi.org/10.3390/cancers14092101
Li J, Guan X, Fan Z, Ching LM, Li Y, Wang X et al (2020) Non-invasive biomarkers for early detection of breast cancer. Cancers (Basel) 12(10). https://doi.org/10.3390/cancers12102767
Jiang N, Zhang X, Gu X, Li X, Shang L (2021) Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discovery 7(1):30. https://doi.org/10.1038/s41420-021-00407-1
Shafaroudi AM, Sharifi-Zarchi A, Rahmani S, Nafissi N, Mowla SJ, Lauria A et al (2021) Expression and function of C1orf132 long-noncoding RNA in breast cancer cell lines and tissues. Int J Mol Sci 22(13). https://doi.org/10.3390/ijms22136768
(2020) A novel prognostic signature of mRNA-lncRNA in breast cancer. DNA and Cell Biology 39(4), 671–82. https://doi.org/10.1089/dna.2019.5223
Venkatesh J, Wasson M-CD, Brown JM, Fernando W, Marcato P (2021) LncRNA-miRNA axes in breast cancer: novel points of interaction for strategic attack. Cancer Lett 509:81–88. https://doi.org/10.1016/j.canlet.2021.04.002
Ashekyan O, Abdallah S, Shoukari AA, Chamandi G, Choubassy H, Itani ARS et al (2022) Spotlight on exosomal non-coding RNAs in breast cancer: an in silico analysis to identify potential lncRNA/circRNA-miRNA-target axis. Int J Mol Sci 23(15). https://doi.org/10.3390/ijms23158351
Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M et al (2022) Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast 66:15–23. https://doi.org/10.1016/j.breast.2022.08.010
Volovat SR, Volovat C, Hordila I, Hordila DA, Mirestean CC, Miron OT et al (2020) MiRNA and LncRNA as potential biomarkers in triple-negative breast cancer: a review. Front Oncol 10:526850. https://doi.org/10.3389/fonc.2020.526850
Butova R, Vychytilova-Faltejskova P, Souckova A, Sevcikova S,Hajek R (2019) Long non-coding RNAs in multiple myeloma. Noncoding RNA 5(1). https://doi.org/10.3390/ncrna5010013
Ghafouri-Fard S, Taheri M (2019) Maternally expressed gene 3 (MEG3): a tumor suppressor long non coding RNA. Biomed Pharmacother 118:109129. https://doi.org/10.1016/j.biopha.2019.109129
Zhang L, Zhao F, Li W, Song G, Kasim V,Wu S (2022) The biological roles and molecular mechanisms of long non-coding RNA MEG3 in the hallmarks of cancer. Cancers (Basel) 14(24). https://doi.org/10.3390/cancers14246032
Sun H, Peng G, Wu H, Liu M, Mao G, Ning X et al (2020) Long non-coding RNA MEG3 is involved in osteogenic differentiation and bone diseases (Review). Biomed Rep 13(1):15–21. https://doi.org/10.3892/br.2020.1305
Wang W, Xie Y, Chen F, Liu X, Zhong LL, Wang HQ et al (2019) LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol 25(29):3972–3984. https://doi.org/10.3748/wjg.v25.i29.3972
Tao P, Yang B, Zhang H, Sun L, Wang Y, Zheng W (2020) The overexpression of lncRNA MEG3 inhibits cell viability and invasion and promotes apoptosis in ovarian cancer by sponging miR-205-5p. Int J Clin Exp Pathol 13(5):869–879
El-Khazragy N, Mohammed HF, Yassin M, Elghoneimy K, Bayoumy W, Hewety A et al (2020) Tissue-based long non-coding RNAs “PVT1, TUG1 and MEG3” signature predicts cisplatin resistance in ovarian cancer. Genomics 112(6):4640–4646. https://doi.org/10.1016/j.ygeno.2020.08.005
Yang Z, Wang Z, Duan Y (2020) LncRNA MEG3 inhibits non-small cell lung cancer via interaction with DKC1 protein. Oncol Lett 20(3):2183–2190. https://doi.org/10.3892/ol.2020.11770
Dong H, Zhang Y, Xu Y, Ma R, Liu L, Luo C et al (2019) Downregulation of long non-coding RNA MEG3 promotes proliferation, migration, and invasion of human hepatocellular carcinoma cells by upregulating TGF-β1. Acta Biochim Biophys Sin (Shanghai) 51(6):645–652. https://doi.org/10.1093/abbs/gmz046
Zhou Y, Yang H, Xia W, Cui L, Xu R, Lu H et al (2020) LncRNA MEG3 inhibits the progression of prostate cancer by facilitating H3K27 trimethylation of EN2 through binding to EZH2. J Biochem 167(3):295–301. https://doi.org/10.1093/jb/mvz097
Wang L, Yu M, Zhao S (2019) lncRNA MEG3 modified epithelial-mesenchymal transition of ovarian cancer cells by sponging miR-219a-5p and regulating EGFR. J Cell Biochem 120(10):17709–17722. https://doi.org/10.1002/jcb.29037
Buttarelli M, De Donato M, Raspaglio G, Babini G, Ciucci A, Martinelli E et al (2020) Clinical value of lncRNA MEG3 in high-grade serous ovarian cancer. Cancers 12(4):966. https://doi.org/10.3390/cancers12040966
Xu X, Zhong Z, Shao Y, Yi Y (2021) Prognostic value of MEG3 and its correlation with immune infiltrates in gliomas. Front Genet 12:679097. https://doi.org/10.3389/fgene.2021.679097
Huang C, Liao X, Jin H, Xie F, Zheng F, Li J et al (2019) MEG3, as a competing endogenous RNA, binds with miR-27a to promote PHLPP2 protein translation and impairs bladder cancer invasion. Mol Ther Nucleic Acids 16:51–62. https://doi.org/10.1016/j.omtn.2019.01.014
Wan S,Zhao H (2020) Analysis of diagnostic and prognostic value of lncRNA MEG3 in cervical cancer. Oncology Letters 20(5), 1-. https://doi.org/10.3892/ol.2020.12044
Ali MA, Shaker OG, Alazrak M, AbdelHafez MN, Khalefa AA, Hemeda NF et al (2020) Association analyses of a genetic variant in long non-coding RNA MEG3 with breast cancer susceptibility and serum MEG3 expression level in the Egyptian population. Cancer Biomark 28(1):49–63. https://doi.org/10.3233/CBM-191072
Shaker O, Ayeldeen G, Abdelhamid A (2021) The impact of single nucleotide polymorphism in the long non-coding MEG3 gene on microRNA-182 and microRNA-29 expression levels in the development of breast cancer in Egyptian women. Front Genet 12:683809. https://doi.org/10.3389/fgene.2021.683809
Zhu X, Lv L, Wang M, Fan C, Lu X, Jin M et al (2022) DNMT1 facilitates growth of breast cancer by inducing MEG3 hyper-methylation. Cancer Cell Int 22(1):56. https://doi.org/10.1186/s12935-022-02463-8
Zhang Y, Wu J, Jing H, Huang G, Sun Z, Xu S (2019) Long noncoding RNA MEG3 inhibits breast cancer growth via upregulating endoplasmic reticulum stress and activating NF-κB and p53. J Cell Biochem 120(4):6789–6797. https://doi.org/10.1002/jcb.27982
Zhang JJ, Guo SH, Jia BQ (2016) Down-regulation of long non-coding RNA MEG3 serves as an unfavorable risk factor for survival of patients with breast cancer. Eur Rev Med Pharmacol Sci 20(24):5143–5147
Cui X, Yi Q, Jing X, Huang Y, Tian J, Long C et al (2018) Mining prognostic significance of MEG3 in human breast cancer using bioinformatics analysis. Cell Physiol Biochem 50(1):41–51. https://doi.org/10.1159/000493956
Ghafouri-Fard S, Sohrabi B, Hussen BM, Mehravaran E, Jamali E, Arsang-Jang S et al (2022) Down-regulation of MEG3, PANDA and CASC2 as p53-related lncRNAs in breast cancer. Breast Dis 41(1):137–143. https://doi.org/10.3233/bd-210069
Sun L, Li Y, Yang B (2016) Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53’s transcriptional activity. Biochem Biophys Res Commun 478(1):323–329. https://doi.org/10.1016/j.bbrc.2016.05.031
Chen RP, Huang ZL, Liu LX, Xiang MQ, Li GP, Feng JL et al (2016) Involvement of endoplasmic reticulum stress and p53 in lncRNA MEG3-induced human hepatoma HepG2 cell apoptosis. Oncol Rep 36(3):1649–1657. https://doi.org/10.3892/or.2016.4919
Xu QR, Tang J, Liao HY, Yu BT, He XY, Zheng YZ et al (2021) Long non-coding RNA MEG3 mediates the miR-149-3p/FOXP3 axis by reducing p53 ubiquitination to exert a suppressive effect on regulatory T cell differentiation and immune escape in esophageal cancer. J Transl Med 19(1):264. https://doi.org/10.1186/s12967-021-02907-1
Li Z, Jin C, Chen S, Zheng Y, Huang Y, Jia L et al (2017) Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem 433(1–2):51–60. https://doi.org/10.1007/s11010-017-3015-z
Liu HZ, Wang QY, Zhang Y, Qi DT, Li MW, Guo WQ et al (2016) Pioglitazone up-regulates long non-coding RNA MEG3 to protect endothelial progenitor cells via increasing HDAC7 expression in metabolic syndrome. Biomed Pharmacother 78:101–109. https://doi.org/10.1016/j.biopha.2016.01.001
Heydari N, Sharifi R, Nourbakhsh M, Golpour P, Nourbakhsh M (2023) Long non-coding RNAs TUG1 and MEG3 in patients with type 2 diabetes and their association with endoplasmic reticulum stress markers. J Endocrinol Invest 46(7):1441–1448. https://doi.org/10.1007/s40618-023-02007-5
Su M, Yu T, Yu Y, Cheng Q, Zheng Y, Liao R et al (2022) hsa-miR-607, lncRNA TUG1 and hsa_circ_0071106 can be combined as biomarkers in type 2 diabetes mellitus. Exp Biol Med (Maywood) 247(18):1609–1618. https://doi.org/10.1177/15353702221110648
Dill TL, Carroll A, Pinheiro A, Gao J,Naya FJ (2021) The long noncoding RNA Meg3 regulates myoblast plasticity and muscle regeneration through epithelial-mesenchymal transition. Development 148(2). https://doi.org/10.1242/dev.194027
Xu D, Liu T, He L, Han D, Ma Y, Du J (2020) LncRNA MEG3 inhibits HMEC-1 cells growth, migration and tube formation via sponging miR-147. Biol Chem 401(5):601–615. https://doi.org/10.1515/hsz-2019-0230
Zhu M, Wang X, Gu Y, Wang F, Li L, Qiu X (2019) MEG3 overexpression inhibits the tumorigenesis of breast cancer by downregulating miR-21 through the PI3K/Akt pathway. Arch Biochem Biophys 661:22–30. https://doi.org/10.1016/j.abb.2018.10.021
Sun Y, Cao FL, Qu LL, Wang ZM,Liu XY (2019) MEG3 promotes liver cancer by activating PI3K/AKT pathway through regulating AP1G1. Eur Rev Med Pharmacol Sci 23(4), 1459–67. https://doi.org/10.26355/eurrev_201902_17103
Tang ZL, Zhang K, Lv SC, Xu GW, Zhang JF, Jia HY (2021) LncRNA MEG3 suppresses PI3K/AKT/mTOR signalling pathway to enhance autophagy and inhibit inflammation in TNF-α-treated keratinocytes and psoriatic mice. Cytokine 148:155657. https://doi.org/10.1016/j.cyto.2021.155657
He Y, Dan Y, Gao X, Huang L, Lv H, Chen J (2021) DNMT1-mediated lncRNA MEG3 methylation accelerates endothelial-mesenchymal transition in diabetic retinopathy through the PI3K/Akt/mTOR signaling pathway. Am J Physiol Endocrinol Metab 320(3):E598-e608. https://doi.org/10.1152/ajpendo.00089.2020
Yan H, Luo B, Wu X, Guan F, Yu X, Zhao L et al (2021) Cisplatin induces pyroptosis via activation of MEG3/NLRP3/caspase-1/GSDMD pathway in triple-negative breast cancer. Int J Biol Sci 17(10):2606–2621. https://doi.org/10.7150/ijbs.60292
Wang P, Chen D, Ma H, Li Y (2017) LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21-5p/SOX7 axis. Onco Targets Ther 10:5137–5149. https://doi.org/10.2147/ott.S146423
Acknowledgements
This work was supported by the research deputy of University of Tabriz.
Author information
Authors and Affiliations
Contributions
AA and MK designed the study. AA, AR, MKK, and MK performed the statistical analysis. AA and AR were the major contributor in the data visualization. AR and MKK drafted the manuscript. MKK and MK critically revised the whole work. All authors read and commented on the final draft of the manuscript.
Corresponding author
Ethics declarations
Consent for Publication
All authors have given their consent for publication in this journal.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahmadi, A., Rezaei, A., Khalaj-Kondori, M. et al. A Comprehensive Bioinformatic Analysis Identifies a Tumor Suppressor Landscape of the MEG3 lncRNA in Breast Cancer. Indian J Surg Oncol (2024). https://doi.org/10.1007/s13193-024-01992-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13193-024-01992-0