Abstract
Breast cancer, the most common cancer in women, is characterized by high morbidity and mortality worldwide. Recent evidence has shown that long non-coding RNAs (lncRNAs) play a crucial role in the development and progression of breast cancer. However, despite increasing data and evidence indicating the implication of lncRNAs in breast cancer, no web resource or database exists primarily for lncRNAs associated with only breast cancer. Therefore, we developed a manually curated, comprehensive database, “BCLncRDB,” for lncRNAs associated with breast cancer. For this, we collected, processed, and analyzed available data on breast cancer-associated lncRNAs from different sources, including previously published research articles, the Gene Expression Omnibus (GEO) Database of the National Centre for Biotechnology Information (NCBI), The Cancer Genome Atlas (TCGA), and the Ensembl database; subsequently, these data were hosted at BCLncRDB for public access. Currently, the database contains 5324 unique breast cancer-lncRNA associations and has the following features: (i) a user-friendly, easy-to-use web interface for searching and browsing about lncRNAs of the user’s interest, (ii) differentially expressed and methylated lncRNAs, (iii) stage- and subtype-specific lncRNAs, and (iv) drugs, subcellular localization, sequence, and chromosome information of these lncRNAs. Thus, the BCLncRDB provides a one-stop dedicated platform for exploring breast cancer-related lncRNAs to advance and support the ongoing research on this disease. The BCLncRDB is publicly available for use at http://sls.uohyd.ac.in/new/bclncrdb_v1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
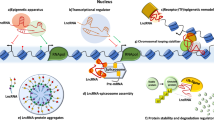
Long non-coding RNAs (lncRNAs) are a class of non-coding RNA (ncRNA) molecules with a length that ranges from 200 base pairs (bp) to 100 kilobases (KB) (Kung et al. 2013; Li et al. 2013). Initially, this class of ncRNAs is thought to be noisy without any biological functions. However, recent studies have indicated that lncRNAs are involved in various cellular physiological regulations such as polypeptide encoding, epigenetic regulation, transcriptional regulation, post-transcriptional regulation, and signal transducer (Jin et al. 2021). For example, lncRNA MEG3 suppresses the regulatory activity of the MDM2 gene, a negative regulator of the P53 gene, via phosphorylation resulting in tumor suppression by P53. Similarly, lncRNA LOC572558 acts as a negative regulator of MDM2, leading to tumor suppression by P53 (Zhu et al. 2016). Moreover, experimental evidence suggests the prominent roles of lncRNAs in the development and tumorigenesis of various cancers (Kopp and Mendell 2018; Pang et al. 2019; Sun et al. 2015; Chen et al. 2019).
Breast cancer is a highly heterogeneous disease with multiple subtypes and a major cause of concern for females at risk around the globe (Sung et al. 2021). Currently, available treatment regimes are insufficient to tackle the problem that arises due to metastasized tumors, disease recurrence, limited subtype-specific treatment options, and resistance development. Thus, there is an urgent need for an in-depth understanding and exploration of other regulatory molecules, such as lncRNAs, known for their roles and potentials associated with breast cancer. This, in turn, may lead to the identification and development of novel, more effective, and subtype-specific therapeutic and diagnostic candidates.
Many databases on cancer-related lncRNAs have been developed to store a wide range of information available for cancer-related lncRNAs. For example, the Lnc2cancer database has information on lncRNAs and human cancer associations. All the lncRNAs-cancer association information stored at the Lnc2cancer is manually curated from the previously published literature (Ning et al. 2016). Another database named LncRNADisease provides information on lncRNA-disease associations across various diseases, including cancers. These relationships have mainly been identified through in vitro experiments or computational predictions (Chen et al. 2012). Next, the LnCaNet provides a resource for a comprehensive co-expression network between lncRNAs and non-neighboring cancer genes (Liu and Zhao 2016). Furthermore, Lnc2catlas is a database that provides quantitative associations between lncRNAs and various cancers (Ren et al. 2018). Although these databases are useful in studying and researching cancer-related lncRNAs, there is a lack of dedicated databases or web resources on lncRNAs associated with only breast cancer.
To this end, we developed a manually curated comprehensive database of breast cancer-associated lncRNAs, named BCLncRDB. For creating the database, firstly, we collected various information on lncRNAs associated with breast cancer, including expression and methylation patterns, targets, effects of drugs on lncRNA expression, subcellular localization of lncRNAs, sequence, and chromosomal location from previously published literature on lncRNAs in breast cancer, The Cancer Genome Atlas (TCGA) (Weinstein et al. 2013), Gene Expression Omnibus (GEO) (Barrett et al. 2012), and various other sources including Ensembl database (Hubbard et al. 2002). Secondly, this information on lncRNAs was stored with the help of MySQL, while the front end of the database was constructed with the use of HyperText Markup Language (HTML) and Hypertext Pre-processor (PHP). We envision that BCLncRDB will be a productive resource designed explicitly for lncRNAs associated with breast cancer to accelerate future research in this area.
Materials and methods
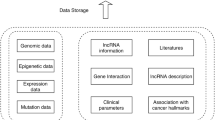
Various information on lncRNAs associated with human breast cancer was collected to develop the database. This information was collected not only from published literature but also from publicly available data (like RNA-seq, microarray, methylation, etc.) on lncRNAs available at various databases such as GEO of NCBI and TCGA. Figure 1 illustrates the schematic schema of database construction.
Data collection from published literature
To collect valuable information on lncRNAs associated with breast cancer from various published literature, all literature (published till 21st April 2023) was extracted from the PubMed database using keywords such as “long non-coding RNA,” “lncRNA,” and “long non-coding” along with “breast.” Secondly, all selected literature was curated manually and filtered based on criteria such as (i) research articles containing experiments on breast cancer patient samples; (ii) samples were not treated with any chemicals or drugs; and (iii) experimental findings have been validated using expression data of breast cancer patients, cell-line data, or animal models induced with breast cancer. All review articles and articles on experiments with samples treated with any chemicals or drugs were excluded from collecting information on lncRNAs associated with breast cancer.
Data collection from GEO database
The GEO database hosts array and sequence-based data on gene expression profiles and DNA methylation data. To collect useful GEO datasets, the same keywords were used to search GEO as used to search literature in PubMed. The GEO datasets were retrieved based on the following criteria: (i) samples should be from Homo sapiens, (ii) expression profiling should be by array or high-throughput sequencing, (iii) tissues should not be treated with any drugs/chemicals, and (iv) samples should include tumor and paired normal tissues. Those datasets that did not match these criteria were discarded. Differentially expressed lncRNAs were identified by comparing expression levels of lncRNAs between tumor and normal tissues using the limma R package (Ritchie et al. 2015).
Data collection from TCGA database
TCGA is a large, public repository of various cancer-related genomic data, including raw sequence reads, gene expression profiles, lncRNA and miRNA expression profiles, DNA methylation data, simple nucleotide variations, copy number variations, and clinical information of tumor and adjacent normal tissues. The differential expression, differential methylation, subtype-specific, and stage-specific information of lncRNAs associated with breast cancer were extracted by analyzing various TCGA data. Differentially expressed lncRNAs were identified by comparing expression levels of lncRNAs between tumor and normal tissues using the DESeq2 R package (Love et al. 2014). The differentially methylated lncRNAs were identified by analyzing TCGA methylation data of breast cancer using the R/Bioconductor package ELMER (Silva et al. 2019).
Other information on lncRNAs
Other information like Ensembl ID, sequences, and chromosome information of all lncRNAs associated with breast cancer were identified from various sources, including TCGA and Ensembl database. Published literature from PubMed was used to retrieve information on the influence of drugs on lncRNA expression associated with breast cancer. The lncLocator database (Lin et al. 2021) and published literature were used to find information on the subcellular localization of lncRNAs.
Database construction
All collected data on lncRNAs were stored and managed into a database named “BCLncRDB” using MySQL, a relational database structured query language; the user interface, or the front end of the database, was built using HTML and PHP for browsing and searching the data contained therein.
Results
Database content
More than 3500 published research articles on lncRNAs associated with breast cancer were retrieved from the PubMed database. These articles were systematically reviewed and curated, as discussed in the “Materials and methods” section. Only 1370 articles were further considered for extracting information on 696 unique lncRNAs associated with breast cancer. After completing this process, data retrieved from these articles included the lncRNA name, Ensembl ID, chromosome information, expression patterns (upregulated or downregulated), experimental techniques (e.g., qRT-PCR), experimentally validated targets of lncRNAs, experimental samples (tissues, cell lines, etc.), subtype information of breast cancer (e.g., Her-2, Basal/Triple-negative breast cancer (TNBC), Luminal A (LumA), Luminal B (LumB), and Normal-like (NormL)), drug information on lncRNA expression, subcellular localization of lncRNAs, and associated literature (PubMed ID, year of publication, and the title of the paper). Further, based on the availability of target information of all breast cancer-associated lncRNAs, functional annotation of these lncRNAs was also carried out to understand their roles in terms of Gene Ontology (GO)-based biological processes and molecular functions.
Moreover, publicly available microarray and RNAseq-based expression profile data of lncRNAs were also analyzed to identify differentially expressed lncRNAs between breast tumor and normal tissues. Four microarray datasets (GSE60689, GSE64790, GSE113851, and GSE119233) from the GEO database were found useful after filtering based on criteria as discussed in the “Materials and methods” section. After analyzing these data using the limma package, significantly differentially expressed lncRNAs were identified based on the criteria of adjusted P-value (adjP-value) < 0.05 and |log2FoldChange| ≥ 2 as used in previous studies (Xue et al. 2020; Hozhabri et al. 2022). Thus, we got 2944 lncRNA-breast cancer associations from these GEO datasets, including 2905 unique lncRNAs. The detailed information of all four GEO datasets with microarray platform information and distribution of differentially expressed lncRNAs across them is described in Table 1.
From the RNAseq-based lncRNA expression data of TCGA, we identified 734 lncRNAs associated with breast cancer, out of which 720 lncRNAs were unique. In the case of the expression data (GEO and TCGA), the association of lncRNAs with breast cancer has been determined based on the differential expression of these lncRNAs in tumor tissues compared to adjacent normal tissues of the breast.
For stage-specific and subtype-specific lncRNAs, we collected the samples (tumor and normal tissues) from TCGA as discussed in the “Materials and methods” section. Stages I, II, III, and IV contained 179, 607, 241, and 19 tumor samples, respectively, while subtypes viz. LumA, Lum B, Her-2, Basal, and NormL contained 559, 201, 81, 187, and 40 tumor samples, respectively. Further, 112 adjacent normal tissue samples were used to identify the stage- and subtype-specific lncRNAs (Table 2). The DESeq2 package was used to determine the differentially expressed stage-specific and subtype-specific lncRNAs. Based on previous studies (Xue et al. 2020; Hozhabri et al. 2022), the adjP-value < 0.05 and |log2FoldChange| ≥ 2 were used as the criteria to identify upregulated and downregulated stage- and subtype-specific lncRNAs. Thus, breast cancer stages viz. I, II, III, and IV had 193, 215, 213, and 252 differentially expressed lncRNAs, respectively, whereas subtypes viz. Lum A, Lum B, Her-2, Basal, and NormL had 228, 406, 403, 387, and 61 differentially expressed lncRNAs, respectively (Table 2).
From the above data, we got 12 lncRNAs which were present in all subtypes (Lum A, Lum B, Her-2, Basal, and NormL) with their expression patterns as upregulated; on the other hand, 27 lncRNAs were present in all subtypes (Lum A, Lum B, Her-2, Basal, and NormL) with their expression patterns as downregulated. Further, 42 lncRNAs were present in all stages (I, II, III, and IV) with expression patterns upregulated, whereas 101 lncRNAs were present in all stages (I, II, III, and IV) with expression patterns downregulated.
There were 9, 8, 14, and 63 lncRNAs specific to stages I, II, III, and IV, respectively, while 10, 66, 79, and 158 lncRNAs specific to subtype Lum A, Lum B, Her-2, and Basal, respectively. Detailed information on lncRNAs for stage-specific and subtype-specific is given in Table 3.
No lncRNA was common among all three sources, i.e., literature, TCGA datasets, and GEO datasets. Among all downregulated lncRNAs, 187 lncRNAs from the literature, 401 lncRNAs from the TCGA dataset, and 1292 lncRNAs from the GEO datasets. Similarly, among all upregulated lncRNAs, there were 487 lncRNAs from the literature, 306 lncRNAs from the TCGA dataset, and 1650 lncRNAs from the GEO datasets.
When we compared the content of our database, i.e., BCLncRDB, with other publicly available databases on lncRNAs associated with diseases including breast cancer such as LncRNADisease, Lnc2Cancer, lnCaNet, and Lnc2catlas, it was found that the BCLncRDB hosts a wide variety of information on lncRNAs associated with breast cancer and this information are not available in any of these prior-available databases or web resources. For example, information such as drug, methylation, subtypes, and stages of breast cancer and associated differential lncRNAs are unique features of our database. The total number of lncRNAs associated with breast cancer, as reported in our database as a single public platform, is also comparatively higher than the number of lncRNAs associated with breast cancer as available in any other databases or web resources. A detailed comparison of all available information on the cancer-associated lncRNAs across different databases is available in Table 4.
Database interface
The BCLncRDB, freely available at http://sls.uohyd.ac.in/new/bclncrdb_v1, provides several search functionalities on the navigation bar for end users to retrieve useful information on lncRNAs available therein (Fig. 2a). By clicking on any search option, users can enter the lncRNA name or Ensembl ID of interest in the search box to retrieve various information about that particular lncRNA (Figs. 2 and 3). For a specific lncRNA name or Ensembl ID, the following details in the general search option will be displayed: lncRNA name, Ensembl ID, chromosome information, expression patterns of lncRNAs, experimental techniques, experimental samples, stage, subtype information of breast cancer, PubMed ID, year of publication, and title of the paper (Fig. 2b). The lncRNA-target search option will display the following details: lncRNA name, Ensembl ID, target, regulatory direction, experimental method for lncRNA target, expression patterns, experimental method for lncRNA expression, lncRNA position, the title of the paper, and PubMed ID (Fig. 2c). Next, the methylation search option will display the following information: lncRNA name, Ensembl ID, methylation pattern, and source (Fig. 3a). Further, the drug-target search option will provide the following information: lncRNA name, Ensembl ID, expression pattern, method, target gene, pathway, drug ID, drug name, drug method, drug response, experimental samples, the title of the paper, and paper’s PubMed ID (Fig. 3b). We also have provided the search option for lncRNA-target functions based on GO terms and this displays the following information: lncRNA name, Ensembl ID, target, target Ensembl ID, GO ID, and GO terms description (Fig. 3c).
Users can also retrieve the complete data provided in the database from the Download page available at the database interface. On the Download page, users can download the data in XLS format. Further, a help page provides detailed information for any user to ensure easy access to the data as a first-time visitor to the database.
Data statistics
All lncRNAs associated with breast cancer were collected from various sources, viz., GEO, lncLocator, published literature, and TCGA. Among the total lncRNAs, 5324 lncRNAs were identified as unique lncRNAs associated with breast cancer. Figure 4 shows the detailed distribution of these lncRNAs across different sources as a histogram. Further, the detailed distributions of lncRNAs across different subtypes and stages of breast cancer are depicted as a histogram in Figs. 5 and 6, respectively. Across different subtypes and stages of breast cancer, subtype LumB (407) and stage IV (252) had the highest number of lncRNAs, whereas subtype Normal-like or NormL (61) and stage I (193) had the lowest number of lncRNAs (Figs. 5 and 6).
Further, 25 lncRNAs were found differentially methylated in the case of breast cancer from TCGA data (Fig. 7). Of these 25 lncRNAs, 15 were hypo-methylated, while ten were hyper-methylated. Moreover, target information of 153 lncRNAs associated with breast cancer was retrieved from published research articles. Also, 20 unique lncRNAs related to breast cancer were found to have drug information based on lncRNA expression as obtained from published literature (Fig. 7). A total of 1653 unique lncRNAs have information on subcellular localization as collected from published literature and the lncLocator database (Fig. 7). The BCLncRDB also hosts chromosome and sequence information of 1419 and 795 unique lncRNAs, respectively.
Discussion
LncRNAs play several critical roles in the etiology and progression of breast cancer. For example, the well-known lncRNA HOTAIR located on chromosomal location 12q13.13 has been found differentially upregulated in breast cancer tissues (Su et al. 2014) and promotes proliferation and metastasis via targeting miR-130a-3p and Suv39H1 (He et al. 2022). LncRNA PVT1 located on the chromosomal location 8q24.21 has also been reported to be upregulated and promote the invasion and cell migration via regulation of miR-148a-3p and hence ROCK1, a target of miR-148a-3p (Li et al. 2022). Similarly, lncRNA MALAT1, located on the chromosomal location 11q13.1 and upregulated in breast cancer, facilitates proliferation and invasion by targeting miR-129-5p in the case of triple-negative breast cancer (Zuo et al. 2017). Further, it has been shown that lncRNA MEG3, located on the chromosomal location 14q32.2, is downregulated in breast cancer, and its overexpression regulates malignant behaviors of breast cancer cells such as proliferation, migration, and invasion by activating p53’s transcriptional activity (Sun et al. 2016). Moreover, lncRNA GAS5, located on the chromosomal location 1q25.1 and reported to be downregulated in breast cancer, confers resistance to Trastuzumab (Li et al. 2016). These studies on lncRNAs indicate that lncRNAs may be key biomarkers of breast cancer diagnosis and treatment with great potential. Currently, several databases are available with information on lncRNAs associated with various diseases, including cancers, e.g., Lnc2cancer (Ning et al. 2016), LncRNADisease (Chen et al. 2012), LnCaNet (Liu and Zhao 2016), and Lnc2catlas (Ren et al. 2018), that provide useful information. Nevertheless, these existing databases have limited information on breast cancer-associated lncRNAs. In this regard, we developed a comprehensive and manually curated database on breast cancer-associated lncRNAs called BCLncRDB.
The BCLncRDB (http://sls.uohyd.ac.in/new/bclncrdb_v1/) is the first dedicated database on lncRNAs associated with breast cancer that stores a large number of information such as lncRNA name, Ensembl ID, breast cancer subtype, breast cancer stage, expression pattern, methylation pattern, chromosomal location, targets, pathway, drug information, sequence, subcellular localization, PubMed ID, and experimental techniques. Our breast cancer-associated lncRNA database can be useful for researchers working on breast cancer in various ways: (i) researchers can access breast cancer-associated lncRNAs at stage and subtype levels, along with expression and methylation patterns that can be utilized for exploring stage- and subtype-specific biomarkers and therapeutic candidates; (ii) they can seek drugs, resistance, and targets information to infer more efficient and new drug targets; and (iii) users can download the lncRNAs data such as targets, methylation, drugs, sequence, and subcellular localization for other related studies of their interest. Further, as the high-throughput sequencing costs have decreased and technologies have advanced, more tumor tissues paired with adjacent normal or normal tissue samples will be sequenced in the near future. We will continue to make enhancements to the database content, such as the addition of extra information on lncRNAs associated with breast cancer, including newly reported lncRNAs in breast cancer, simple nucleotide variations, and copy number variations, along with missing information on existing lncRNA entries of the database.
Conclusion
We developed a unique and comprehensive database of lncRNAs associated with breast cancer named “BCLncRDB.” It not only provides the experimentally validated data but also provides the information from the TCGA dataset, GEO datasets, and Ensembl database. The development of BCLncRDB aims to provide a more orientated, curated database of lncRNAs associated with breast cancer that hosts information on subtypes, stages, gene expression, methylation, sequences, chromosomal location, targets, drugs, and many others. Several studies suggest that the deregulation of lncRNAs has a vital role in the development and progression of breast cancer. Thus, we believe that this comprehensive and expandable resource of lncRNAs associated with breast cancer will facilitate scientists with an extended platform to accelerate breast cancer research to identify more effective and specific therapeutics for the disease.
Data availability
The BCLncRDB is publicly available at http://sls.uohyd.ac.in/new/bclncrdb_v1. The downloadable information of this database is available at http://sls.uohyd.ac.in/new/bclncrdb_v1/Downloads.php.
Change history
14 May 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s10142-024-01372-5
References
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A (2012) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41(D1):D991–D995. https://doi.org/10.1093/nar/gks1193
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen X, Zhang Q, Yan G, Cui Q (2012) LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41(D1):D983–D986. https://doi.org/10.1093/nar/gks1099
Chen H, Xu Z, Liu D (2019) Small non-coding RNA and colorectal cancer. J Cell Mol Med 23(5):3050–3057. https://doi.org/10.1111/jcmm.14209
He W, Li D, Zhang X (2022) LncRNA HOTAIR promotes the proliferation and invasion/metastasis of breast cancer cells by targeting the miR-130a-3p/Suv39H1 axis. Biochem Biophys Rep 30:101279. https://doi.org/10.1016/j.bbrep.2022.101279
Hozhabri H, Ghasemi Dehkohneh RS, Razavi SM, Razavi SM, Salarian F, Rasouli A, Azami J, Ghasemi Shiran M, Kardan Z, Farrokhzad N, Mikaeili Namini A (2022) Comparative analysis of protein-protein interaction networks in metastatic breast cancer. PLoS One 17(1):e0260584. https://doi.org/10.1371/journal.pone.0260584
Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R (2002) The Ensembl genome database project. Nucleic Acids Res 30(1):38–41. https://doi.org/10.1093/nar/30.1.38
Jin H, Du W, Huang W, Yan J, Tang Q, Chen Y, Zou Z (2021) lncRNA and breast cancer: progress from identifying mechanisms to challenges and opportunities of clinical treatment. Mol Ther Nucleic Acids 25:613–637. https://doi.org/10.1016/j.omtn.2021.08.005
Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long non-coding RNAs. Cell 172(3):393–407. https://doi.org/10.1016/j.cell.2018.01.011
Kung JT, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193(3):651–669. https://doi.org/10.1093/genetics/193.3.NP
Li HL, Wang CP, Zhang Y, Du JX, Han LH, Yang HY (2022) Long non-coding RNA PVT1 facilitates cell migration and invasion by regulating miR-148a-3p and ROCK1 in breast cancer. Clin Transl Oncol 24(5):882–891. https://doi.org/10.1007/s12094-021-02736-0
Li J, Xuan Z, Liu C (2013) Long non-coding RNAs and complex human diseases. Int J Mol Sci 14(9):18790–18808. https://doi.org/10.3390/ijms140918790
Li W, Zhai L, Wang H, Liu C, Zhang J, Chen W, Wei Q (2016) Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 7(19):27778–27786. https://doi.org/10.18632/oncotarget.8413
Lin Y, Pan X, Shen HB (2021) lncLocator 2.0: a cell-line-specific subcellular localization predictor for long non-coding RNAs with interpretable deep learning. Bioinformatics 37(16):2308–2316. https://doi.org/10.1093/bioinformatics/btab127
Liu Y, Zhao M (2016) lnCaNet: pan-cancer co-expression network for human lncRNA and cancer genes. Bioinformatics 32(10):1595–1597. https://doi.org/10.1093/bioinformatics/btw017
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):1–21
Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, Gao Y, Guo M, Yue M, Wang L, Li X (2016) Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res 44(D1):D980–D985. https://doi.org/10.1093/nar/gkv1094
Pang B, Wang Q, Ning S, Wu J, Zhang X, Chen Y, Xu S (2019) Landscape of tumor suppressor long non-coding RNAs in breast cancer. J Exp Clin Cancer Res 38:1–18. https://doi.org/10.1186/s13046-019-1096-0
Ren C, An G, Zhao C, Ouyang Z, Bo X, Shu W (2018) Lnc2Catlas: an atlas of long non-coding RNAs associated with risk of cancers. Sci Rep 8(1):1–8. https://doi.org/10.1038/s41598-018-20232-4
Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47–e47. https://doi.org/10.1093/nar/gkv007
Silva TC, Coetzee SG, Gull N, Yao L, Hazelett DJ, Noushmehr H, Lin DC, Berman BP (2019) ELMER v. 2: an R/Bioconductor package to reconstruct gene regulatory networks from DNA methylation and transcriptome profiles. Bioinformatics 35(11):1974–1977. https://doi.org/10.1093/bioinformatics/bty902
Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, Esteva FJ (2014) Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget 5(20):9864–9876. https://doi.org/10.18632/oncotarget.2454
Sun L, Li Y, Yang B (2016) Downregulated long non-coding RNA MEG3 in breast cancer regulates proliferation, migration and invasion by depending on p53’s transcriptional activity. Biochem Biophys Res Commun 478(1):323–329. https://doi.org/10.1016/j.bbrc.2016.05.031
Sun M, Gadad SS, Kim DS, Kraus WL (2015) Discovery, annotation, and functional analysis of long non-coding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell 59(4):698–711. https://doi.org/10.1016/j.molcel.2015.06.023
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander SJM (2013) The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 45(10):1113–1120. https://doi.org/10.1038/ng.2764
Xue JM, Liu Y, Wan LH, Zhu YX (2020) Comprehensive analysis of differential gene expression to identify common gene signatures in multiple cancers. Med Sci Monit 26:e919953–1. https://doi.org/10.12659/MSM.919953
Zhu Y, Dai B, Zhang H, Shi G, Shen Y, Ye D (2016) Long non-coding RNA LOC572558 inhibits bladder cancer cell proliferation and tumor growth by regulating the AKT–MDM2–p53 signaling axis. Cancer Lett 380(2):369–374. https://doi.org/10.1016/j.canlet.2016.04.030
Zuo Y, Li Y, Zhou Z, Ma M, Fu K (2017) Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed Pharmacother 95:922–928. https://doi.org/10.1016/j.biopha.2017.09.005
Acknowledgements
Vaibhav Vindal would like to acknowledge the Indian Council of Medical Research (ICMR), GoI (ISRM/12(72)/2020, ID: 2020-2951), Institution of Eminence (IoE), University of Hyderabad (No. UoH/IoE/RC3-21-052), and Department of Biotechnology, GoI (No. BUILDER-DBT-BT/INF/22/SP41176/2020) for their financial support. Swapnil Kumar would like to acknowledge the ICMR for providing financial assistance as the Senior Research Fellowship (Grant No. 3/2/2/113/2019/NCD-III, ID: 2019-6723). The authors would like to thank Mr. Sayed Saleem Javed for his technical assistance in database hosting on the server.
Funding
This study was supported by the extramural research grant from the Indian Council of Medical Research (ICMR) New Delhi (ISRM/12(72)/2020, ID: 2020-2951).
Author information
Authors and Affiliations
Contributions
AA and SK analyzed the data. AA curated the literature and developed the database. SK wrote the original draft. SK and VV conceived this study, proofread, and edited the manuscript. VV supervised the study. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Additional information
A preprint of this article is available at the link (https://www.biorxiv.org/content/10.1101/2022.12.05.519223).
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s10142-024-01372-5"
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, S., Agrawal, A. & Vindal, V. RETRACTED ARTICLE: BCLncRDB: a comprehensive database of LncRNAs associated with breast cancer. Funct Integr Genomics 23, 178 (2023). https://doi.org/10.1007/s10142-023-01112-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-023-01112-1