Abstract
Protein–protein interaction plays a fundamental role in many biological processes and diseases. Characterizing protein interaction sites is crucial for the understanding of the mechanism of protein–protein interaction and their cellular functions. In this paper, we proposed a method based on integrated support vector machine (SVM) with a hybrid kernel to predict protein interaction sites. First, a number of features of the protein interaction sites were extracted. Secondly, the technique of sliding window was used to construct a protein feature space based on the influence of the adjacent residues. Thirdly, to avoid the impact of imbalance of the data set on prediction accuracy, we employed boost-strap to re-sample the data. Finally, we built a SVM classifier, whose hybrid kernel comprised a Gaussian kernel and a Polynomial kernel. In addition, an improved particle swarm optimization (PSO) algorithm was applied to optimize the SVM parameters. Experimental results show that the PSO-optimized SVM classifier outperforms existing methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Protein–protein interaction involves in numerous physiological and pathological processes [1]. Identifying protein seats involved in protein–protein interactions, which is called protein interaction site, can help to achieve a better understanding of the protein–protein interaction. To effectively predict the sites of protein interaction, computational intelligence methods such as support vector machine, bayesian, random forest and neural networks have been tested and achieve varying degree of success.
The prediction of protein interaction sites can be viewed as a binary classification problem which is equivalent to determine whether a given amino acid residue of a protein is an interaction site or not [2]. In the work of Yan et al. features were constructed based on a fragment of nine continuous amino acids. Support vector machine and bayesian network were used to predict interaction sites and their method achieved a result of 72 % accuracy and 0.3 Matthews’ correlation coefficient (MCC) relevance [3]. Chen et al. who predicted protein interaction sites based on random forest method had extracted features based on biochemical characteristics such as protein amino acid sequence, the residues’ distance matrix, conservatism of protein evolution and so on [4]. They obtained 0.28 MCC. Chen et al. used six features from the homology derived secondary structure of proteins database (HSSP) and the integrated radial basis function (RBF) neural network algorithm to predict protein interaction sites [5]. According to the related work, still there are several problems which limit the prediction accuracy. Because the theory of the biological properties responsible for protein–protein interaction are not clear yet, it is difficult to extract informative features from biological properties to represent protein. What more, because there are much fewer amount of data of interacting sites than that of non-interacting sites, which is called data imbalance, it is difficult to get a significant training set. In addition, the machine learning methods used to predict the interaction sites in the past was used without further modification according to the characteristics of this specific problem, efficient classification models were difficult to build there still has plenty of room to improve the performance of the prediction.
Inspired by recent works, we reported a novel method to predict protein interaction sites. After extracting relevant features of interaction sites from HSSP database, we used sliding window technique to construct a protein feature space based on the influence of the adjacent residues. Then, we used the boost-strap method to construct a balanced data set from the originally imbalanced data set. Furthermore, for constructing an efficient classification model, the hybrid kernel comprising a Gaussian kernel and a Polynomial kernel was applied in SVM, and an improved particle swarm optimization was used to optimize the parameters. To evaluate performance of the prediction, we applied our method to a data set from the related work [5]. The data set was downloaded from SPIN database.
2 Materials and methods
2.1 Data set
To be convenient for the comparison of our prediction method to previous work, the proposed method was tested with the data set used by Chen et al. [5]. As shown in Table 1, this data set contained 38 proteins (63 peptide chains). More details of the data set building process can be found in the Ref. [6].
2.2 Performance evaluation
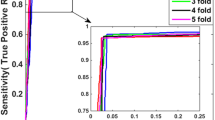
The performance of the prediction was evaluated using leave-one-out cross validation. In each experiment, we chose 37 proteins from the data set to build the train set and leave the rest one as the test set, and carried out the replication experiments 38 times. By choosing each individual protein from the data set as test set for each experiment, all of the proteins in the data set were traversed.
To analyze the effectiveness of the proposed method quantitatively, several widely used measures, including accuracy, Matthews’ correlation coefficient (MCC) and F-score were used. Accuracy is a simple but effective evaluation standard. The preliminary knowledge of the details of the experimental data is unnecessary, but if the data set was imbalance, accuracy muse be used with caution, as some researchers had shown that accuracy would be biased toward the majority one in such condition. MCC is an objective method to reflect the prediction performance. The F-score is determined by two parameters: recall and precision which are defined as (4) and (5) respectively, it is a useful measure to reflect the overall prediction performance. The precise definition of three measures above are as follows:
where
where, TP, TN, FP, FN present the number of interaction sites predicted correctly, the number of non-interaction sites predicted correctly, the number of non-interaction sites predicted as interaction sites, the number of interaction sites predicted as non-interaction sites, respectively.
2.3 Feature selection
In our data set, if an amino acid residue was an interaction site, it was labeled as ‘1’, otherwise it was labeled as ‘0’. Protein interaction site was defined according to Yohei Minakuchi et al. [7]. A surface residue is a residue of a single peptide whose ratio of its accessible surface area to its whole surface area was greater than 16 %, otherwise, it was called an internal residue. Interface residue was defined according to Chen et al. [5], if the difference between the accessible surface areas of a surface residue in a single chain (monomer accessible surface area, MASA) to the accessible surface areas when it was in a protein complex (complex accessible surface area, CASA) was more than 1(A 2), the residue would be defined as an interface residue, and it was also called as an interaction site. Otherwise it was defined as a non-interface residue [5], which was also called as a non-interaction site. All related terms we used in this paper had the same definition as above-mentioned.
In this paper, six features of the protein interaction site originally proposed by Chen et al. [5] were used to characterized the interaction site. They are sequence profiles, entropy, relative entropy, conservation weight, accessible surface area and sequence variability. These features could be downloaded from the biological database HSSP (ftp://ftp.ebi.ac.uk/pub/database/hssp/). More details of these six features can be learned from the Ref. [5]. The sliding window with a size of \( W \) was used to construct the feature vector space. \( \omega \) continuous residues flank each of a target residue are used to build the vector where \( \omega \) is defined in Eq. (7). The size of the sliding window \( W \) is in the range of between 3 and 11. For each residue in a protein. The dimensions of the feature vector are 25, including 20 for sequence profiles-related variables, and the other five variables for the rest five features. Thus, we defined a feature vector to represent the nth residue in a protein amino acid sequence as follows:
Where,
The data set used in this paper was unbalanced due to the number of non-interaction sites being much larger than the interaction sites’. Generally, the former was two-thirds more than the latter. We used boost-strap method to reduce the impact of the unbalanced problem to improve the performance of the prediction.
We proposed the following algorithm to transform the unbalanced data set to a balanced one.

2.4 The classifier
Support vector machine (SVM) initially proposed by Cortes and Vapnik in 1995 is based on the VC dimension theory and minimum structure risk principle. SVM is aimed at finding a balance between the complexity of the model and its learning ability [8]. In this paper, we adopted SVM to predict protein interaction sites. SVM is an effective method for problems with small number of samples, and predicting protein interaction sites is the case, so theoretically SVM can get the global optimal point for this problem. The kernel function of a SVM determines this learning ability [9], and there are some commonly used kernels as follows:
Linear kernel:
Polynomial kernel:
Radial basis function:
Sigmoid Tanh kernel:
Choosing an appropriate kernel can improve the prediction performance. Currently, SVM kernel function is determined based on the experience. In addition, a kernel can also be designed for a specific data set. It’s worth mentioning that the constructed kernel should meet the requirements of Mercer’s condition [10].
Some related works [11, 12] proved that the model with a mixture kernel could get better prediction performance than the ones with a simple kernel. In this paper, in order to improve the prediction accuracy, we constructed a model based on SVM with a hybrid kernel function which comprised a Gaussian kernel and a Polynomial kernel. The constructed hybrid kernel is defined as following:
Where, \( K_{POLY} \) is a Polynomial kernel as (9), which is a global kernel only considering the distant points. On the contrary, radial basis function \( K_{RBF} \) as (10), is a local kernel only considering the neighbors of the testing point. The global kernel is responsible for the universal search to avoid losing of the optimal solution, while the local kernel is responsible for speeding up the convergence. The proposed hybrid kernel here was thus designed to take advantages of the both of the two types of kernels above. And the influence of two kernels can be tuned by the parameter λ.
2.5 Parameter optimization
There are many advantages to apply particle swarm optimization (PSO) to optimize parameters of SVM with a hybrid kernel [13]. PSO has quick convergence, and possesses global search capability with a few parameters. In the PSO algorithm, particles were used to represent potential solutions of a optimization problem. In this paper, particles were used to represent λ which was defined as in (12), the weight of the basal classifiers was constructed by training the data subset as described in “Sect. 2.3”.
During PSO optimization, each particle flies in search space at a certain velocity. The velocity is usually adjusted in light of flight experiences. In this paper, in order to search the solution space more efficiently, a parameter \( K \) was introduced to adjust the velocity. If a particle’s ordinal number was odd, \( K \) was set to be ‘−1’, which meant this particle would fly in the direction opposite to the current direction. Otherwise, it was set to be ‘1’ which meant it would continue to fly in the current direction. Such treatment could make the particles flying in two directions at the same time, searching more widely. The iteration formula of the PSO method is as follows:
Where, \( d \) represents the dimension of the problem space respectively. \( V_{id} \) represents the velocity of the \( i^{th} \) particle in the swarm. \( pbest_{id} \) represents the best previous position of the \( i^{th} \) particle by which the best fitness value is achieved. \( pbest_{d} \) represents the best particle among all particles in the swarm. \( rand() \) represents random numbers in the range of 0–1. \( w \) and \( t \) represent the inertia and the iteration number, respectively. c 1 and c 2 are both positive constants. \( K \) is defined above. In this paper, the fitness was defined as the prediction accuracy.
2.6 Algorithm to predict protein interaction sites
We proposed an algorithm to predict protein interaction sites using six features mentioned in “Sect. 2.3” as input. The predicting algorithm is as follows:

3 Results
The effectiveness of the protein interaction site prediction method in this paper was evaluated by the leave-one-out cross validation method. By choosing one protein each time from the data set to construct a test set without repetition, we carried out the replication experiments 38 times, thus eventually all proteins were treated. Finally, we gave some performance evaluations according to the measures as introduced in “Sect. 2.2”.
3.1 Parameters setting
Some related parameters about PSO were set as follows, the size of the population \( N = 40 \), the max iteration \( DT = 100 \), the learning factor \( c_{1} = c_{2} = 1.4962 \), the max particle velocity \( V_{\hbox{max} } = (X_{\hbox{max} } - X_{\hbox{min} } ) \times 0.1 \), the max value of the target particle position \( X_{\hbox{max} } = 1 \) and the min value \( X_{\hbox{min} } = 0 \), the linear iterative inertia weight \( w_{0} = 0.9 \) and \( w_{1} = 0.4 \). Finally, particles in the swarm stopped iterating when the optimal prediction accuracy did not change in the continuous 20 times iteration.
3.2 Experimental results
Firstly, seven sliding windows were made, including 1, 3, 5, 7, 9, 11, to optimize the size of sliding windows which achieved the best accuracy. During this process, the basic classifier was based on hybrid kernel SVM, and the experiment data was the 38 proteins as listed in Table 1. The results were shown in Table 2.
In the table, each row stores the experiment result of different window size. The last number of the names of the first column is the size of the windows, for example, the WIN3 row stores the experiment result for window size equals to 3.
According to the results in Table 2, WIN9 which was represented the slipping window with the size of nine, obtained the best MCC and F-score than the others, this conclusion was the same as the result of previous related work [5, 14].
Secondly, we compared the performance of our method with an existing method, called integrated RBF neural network [5]. Here, we set the size of the slipping window to be nine. The result of the comparison was shown in Table 3.
According to Table 3, the MCC, F-score and accuracy were higher by 5.45, 4.08 and 2.84 % respectively in SVM experiment than RBF experiment. In conclusion, the proposed method on the basis of SVM with hybrid kernels achieved better performance in predicting the protein interaction sites.
The structures of proteins could be visualized at atomic scale by molecular graphic software such as Rasmol. In order to verify our method visually, we would show the details of prediction results of protein 1npo and 1 tmc as Chen et al. [5] had done to make the result comparison more convenient.
In the Fig. 1, for the protein 1npo, there are totally 95 yellow residues which are surface residues defined as “Sect. 2.3”, and each of these residues might be an interaction site or a non-interaction site. And our proposed method correctly predicted 79 residues of them as Fig. 2 showed. For the protein 1 tmc, as shown in Fig. 3, there are totally 175 yellow residues called surface residues, 151 of them were correctly predicted by using our method as Fig. 4 showed. So it is evident that we got much more correctly predicted protein residues than Chen et al. [5], showing the effectiveness of our method in another way.
4 Conclusion and discussions
With the advent of post-genome era, the researchers have put an increasing attention to study the proteomics, especially in the field of predicting protein interaction sites. In this paper, we proposed a method based on support vector machine with a hybrid kernel to predict protein interaction sites. Firstly, sliding window was applied to construct feature vector space for each amino acid residues on a peptide. Secondly, boost-strap was applied to transfer the imbalanced data set into a balanced one. Thirdly, a SVM with a hybrid kernel comprising a Gaussian kernel and a Polynomial kernel was constructed. In order to obtain the optimal parameters for the prediction performance, the parameters of hybrid kernel and the weight of basal classifiers were selected by PSO. Finally, for comparison convenience, prediction results were described in two different ways according to Chen et al. [5]. The comparison results showed the proposed method effectively improve the prediction performance.
Although the proposed method has improved the performance of prediction, it was mainly focused on classifier algorithm. In the future work, more research about feature extraction are expected to further improve the performance of prediction.
References
Alberts BD, Bray D, Lewis J et al (1989) Molecular biology of the cell. Garland, New York
Ni QS, Wang ZZ, Wang GY et al (2008) Prediction of protein–protein interactions based on local support vector machine. J Biomed Eng Res 02(9):1106–1109
Yan CH, Dobbs D, Honavar V et al (2004) A two stage classifier for identification of protein–protein interface residues. Bioinformatics 20(1):371–378
Chen XW, Jeong JC (2009) Sequence-based prediction of protein interaction sites with an integrative method. Bioinformatics 25(5):585–591
Chen YH, Xu JR, Bin Yang et al (2012) A novel method for prediction of protein interaction sites based on integrated RBF neural networks. Comput Biol Med 42:402–407
Meng W, Wang F, Peng X (2008) Prediction of protein–protein interaction sites using support vector machine. Appl Sci 26(4):403–408
Minakuehi Y, Satou K, Konagaya A (2002) Prediction of protein–protein interaction sites using support vector machines. Genome Inform 13:322–323
LiQin Jin (2007) Biological chemistry. Zhejiang University Press, Hangzhou (in Chinese)
Marangoni F, Barberis M, Botta M (2003) Large scale prediction of protein interactions by a SVM-based method. In: Neural Nets, vol 2859. Springer, Berlin Heidelberg, pp 296–301
Li Liu (2009) The research and validation of support vector machine (SVM) algorithm with different kernels. Jiangnan University, Wuxi, Jiangsu (in Chinese)
Cortes C, Vapnik V (1995) Support vector network. Mach, Learn
Chatterjee P, Basu S, Kundu M et al (2011) PPI_SVM: prediction of protein–protein interactions using machine learning domain–domain affinities and frequency tables. Cell Mol Biol Lett 16:264–278
Aimin Zhou, Bo-Yang Qub, Hui Li et al (2011) Multiobjective evolutionary algorithms: a survey of the state of the art. Swarm Evol Comput 1(1):32–49
Xing X, Chen Y, Yang B (2010) Dimensional reduction based on conservative adaptive K-nearest neighbor algorithm. Univ Jinan Sci Technol 2:159–162 (in Chinese)
Acknowledgments
This work was funded by the Natural Science Foundation of Fujian Province (2012J05114, 2013N5006), Special Project on the Integration of Industry, Fuzhou City Science Foundation (2012G106). We also want to thank for the help from Dr. Zhibin Fu.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Guo, H., Liu, B., Cai, D. et al. Predicting protein–protein interaction sites using modified support vector machine. Int. J. Mach. Learn. & Cyber. 9, 393–398 (2018). https://doi.org/10.1007/s13042-015-0450-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-015-0450-6