Abstract
Background
This work aims to explore circ_ROBO1’s function in nasopharyngeal carcinoma (NPC).
Methods
circ_ROBO1 expression in NPC tissues and cell lines was measured. The regulation of circ_ROBO1 and/or miR-324-3p on the proliferation, migration, invasion, and apoptosis of NPC cells was investigated by functional experiments. The interplay between circ_ROBO1, miR-324-3p, and NME1 was explored. Tumor growth and metastasis were studied in mice.
Results
circ_ROBO1 was overexpressed in NPC. Knockdown of circ_ROBO1 repressed proliferation, migration, and invasion and induced apoptosis of NPC cells. Loss of circ_ROBO1 reduced tumor growth and metastasis in mice. circ_ROBO1 competed with miR-324-3p to upregulate NME1. Lowering miR-324-3p expression impaired the effect of knockdown of circ_ROBO1on NPC cells.
Conclusion
Overexpressed circ_ROBO1 promotes NPC development by modifying the miR-324-3p/NME1 axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Nasopharyngeal cancer (NPC) is a local malignant tumor with high incidence in middle age and males in China [1]. In 2020, the number of newly discovered cases and deaths of NPC worldwide are 133,000 and 80,000 respectively [2]. The risk factors associated with NPC pathogenesis include genetic susceptibility, EBV infection [3], and environmental factors [4]. Due to the hidden location of NPC tumors, the early symptoms are not obvious, causing misdiagnosis and missed diagnosis. About 1/3 of NPC cases are found in the advanced stage of distant metastasis, and the cancer progresses rapidly leading to death [5]. For advanced NPC, the 5-year survival rate can be as high as 90% after effective radiotherapy or radiotherapy [6]. Therefore, it is essential to develop better diagnostics and treatments based on understanding the molecular mechanisms of NPC progression.
Circular RNA (circRNA) is a class of ncRNAs that are commonly found in eukaryotic cells and are made by reverse splicing linear mRNA precursors into continuous covalent rings [7]. circRNAs are involved in tumorigenesis and cancer progression [8,9,10], such as hsa_circ_0004872 in gastric cancer [11], circLIFR in bladder cancer [12], and circDLC1 in hepatocellular carcinoma (HCC) [13]. Circ_ROBO1 remains a mystery as to whether it plays a role in NPC and tumor progression.
Many mechanisms have been proposed for how circRNAs perform biological functions, such as competitively binding miRNAs or interacting with RNA-binding proteins [14,15,16]. This work mainly explored the interplay between circ_ROBO1, miR-324-3p, and nucleoside diphosphokinase 1 (NME1) in NPC development.
2 Materials and methods
2.1 Patient specimens
NPC tissues and normal tissues (35 pairs) were obtained from patients undergoing biopsy in Xiangtan Central Hospital in 2021–2022. All specimens were pathologically confirmed. No radiation or chemotherapy was given to the patients before biopsy. Those with infection, autoimmune disease, or other tumor were excluded. Table 1 lists patients’ clinicopathological data. All subjects signed written informed consent. This study was approved by the Ethics Committee of Xiangtan Central Hospital (No. 202010HN93).
2.2 Cell culture
RPMI-1640 medium with 10% FBS (Gibco, USA) and 1% penicillin–streptomycin (Life Technologies, USA) was taken as a culture medium for NPC cell lines (CNE2, HK1, HONE1, HNE2) and immortalized NPE cell line (NP69). Cell culture was done with 5% CO2 at 37 °C.
2.3 RT-qPCR
TRIzol reagents (Invitrogen, USA) were taken to collect total RNA. The collected RNA was subjected to reverse-transcription using PrimeScript™ RT Kit (Takara, China). Then, RT-qPCR was conducted using TaqMan Universal PCR Master Mix (Applied Biosystems, USA) in AB7300 (Applied Biosystems). In the case of circ_ROBO1 and NME1, GAPDH was used as an endogenous control, which for miR-324-3p was U6. The 2−ΔΔCt method was utilized to calculate gene expression. Primers (Table 2) were obtained from GenePharma (Shanghai, China). The subcellular localization of circ_ROBO1 was determined by RT-qPCR using PARISTM Kit (Ambion, Life Technologies, USA), with U6 and GAPDH as controls in the nucleus and cytoplasm, respectively.
2.4 Treatment of actinomycin D and RNAse R
Actinomycin D treatment (2 mg/ml, Sigma-Aldrich, USA) in cells was performed to block transcription. RNAse R (3 U/μg, Epicenter Technologies, USA) was incubated with 5 μg total RNA at 37 °C for 15 min. After treatment, circ_ROBO1 and ROBO1 levels were detected by RT-qPCR.
2.5 Cell transfection
circ_ROBO1 small interfering RNA (si-circ_ROBO1#1/#2), circ_ROBO1 overexpressed plasmid, NME1 siRNA (si-NME1) or siRNA negative control (si-NC) at 100 nM, miR-324-3p mimics/inhibitors, or corresponding negative controls at 50 nM (GenePharma) were transfected into HONE1 and HK1 cells using Lipofectamine 3000 (Invitrogen). RT-qPCR was performed 36 h later to verify the transfection efficiency.
2.6 CCK-8 assay
Cells were plated in the 96-well plate (1 × 103 cells/well) and incubated with 10 μL CCK-8 reagent (Dojindo, Japan) at 0, 24, 48, 72, and 96 h, respectively for additional 2 h. Optical density values at 450 nm were read on a microplate reader.
2.7 Bromodeoxyuridine (BrdU) assay
Cells were incubated with 10 μM BrdU solution (BD Pharmingen, USA) for 12 h and fixed with 4% paraformaldehyde for 30 min. Then, anti-BrdU (1:1000, Sigma-Aldrich) or IgG was detected for 1 h. After that, a secondary antibody (1:500, Beyotime, China) was added for another 1 h. Cells were then stained with DAPI (Beyotime) and cleared with PBS, and BrdU-positive cells in 3 fields of view were microscopically counted.
2.8 Transwell analysis
Transwell chambers (BD Biosciences, USA) were coated with matrigel (BD Biosciences). Matrigel was only used in the invasion detection. HONE1 and HK1 cells were suspended in FBS-free DMEM to reach 5 × 106 cells/ml concentration. Next, 200 μl cell suspension was added in the upper compartment, and 500 μl DMEM + 20% FBS was supplemented in the lower compartment. After 24 h, after the removal of non-migratory and non-invasive cells, the remaining cells were fixed with 4% paraformaldehyde and stained with 0.5% crystal violet. Cells were counted in 5 fields of view under an inverted microscope (Olympus, Japan).
2.9 Flow cytometry
FITC Annexin V Apoptosis Detection Kit (BD Biosciences) was taken to assess cellular apoptosis. HONE1 and HK1 cells were re-suspended in 500 μl 1 × binding buffer and stained with 5 μl Annexin V-FITC and 5 μl PI solutions, respectively. After 15 min, apoptotic cells were measured on the FACScan flow cytometer (BD Biosciences).
2.10 Luciferase reporter gene assay
Binding sites between miR-324-3p 3'UTR fragments and circ_ROBO1 were predicted by starBase 3.0. The target of miR-324-3p was searched by TargetScan 3.0 7.1 and starBase. Wild type or mutant circ_ROBO1 and NME13ʹUTR sequences containing miR-324-3p binding sites were inserted into the pmirGLO vector and transfected into cells with miR-324-3p mimics or control vector based on Lipofectamine® 2000. After 48 h, cells were lysed with a lysis buffer (100 μL/well) in the dual luciferase reporter assay kit (Beyotime) and centrifuged at 10,000–15,000 g for 3–5 min for the detection of luciferase activities.
2.11 RIP assay
RIP was implemented based on the EZMagna RIP kit (Millipore, USA). HONE1 and HK1 cells were lysed in the RIP lysis buffer, added to RIP buffer containing anti-AgO2 or IgG (Millipore), and incubated with magnetic beads. Then, cells were treated with protease K and DNase (Beyotime), and RNA was purified for RT-qPCR detection.
2.12 RNA pull-down
RNA pull-down test was performed using Pierce Magnetic RNA–protein Pull-Down kit (Thermo Fisher Scientific, USA). Bio-miR-324-3p-WT and Bio-miR-324-3p-MUT were generated by biotinizing miR-324-3p WT/MUT sequences were biotinized to obtain Bio-miR-324-3p-WT and Bio-miR-324-3p-MUT. HONE1 and HK1 cells were transfected with Bio-miR-324-3p-WT and Bio-miR-324-3p-MUT and lysed with a lysis buffer (Sigma) for 10 min. After centrifugation, the sample was detected with streptavidin magnetic beads overnight. After elution, RNA was extracted and detected by RT-qPCR.
2.13 Western blot analysis
Cells or tissues were added with a lysis buffer (Beyotime) and placed on ice for 20 min. After protein concentration detection based on Bradford assay (Bio-Rad, USA), proteins were separated using 15% SDS-PAGE, loaded onto PVDF membranes, and sealed with 5% skim milk powder for 1 h. Then, E-cadherin (Abcam, ab76319, 1:2000), Vimentin (Abcam, ab92547, 1:2000), or NME1 (Sigma-Aldrich, HPA041113, 1:1000) was incubated overnight at 4 ℃, followed by a secondary antibody (CST, USA) bound to horseradish peroxidase for 1 h at 37 ℃. Color development was done with the ECL kit (ultrassignal, China).
2.14 Animal experiment
The animal experiments were approved by the Animal Protection and Use Committee of Xiangtan Central Hospital (No.20201206N). Four-week-old male BALB/c nude mice (Laboratory Animal Resources, Chinese Academy of Sciences) were housed in an SPF-grade laminar flow chamber, with 12/12-h light/dark cycles, and enough foodstuff and water. HONE1 cells (1 × 106) stably knocking down circ_ROBO1 were constructed using circ_ROBO1-targeted shRNA sequence lentiviruses (HanBio, Shanghai, China). Mice were given a subcutaneous injection with HONE1 cells to observe tumor growth or an intravenous injection via the tail vein to detect lung metastasis (10 mice per group). Tumor volume = length × width2 × 0.5. Lungs were harvested from euthanized mice 4 weeks later. The collected samples were analyzed for HE staining, and lung metastatic nodules were observed under an inverted microscope. Severe: ≥ 50% metastatic nodules in the visual field; moderate: 25–50%; mild: less than 25%.
2.15 Statistical analysis
All the experiments were done at least three times. SPSS 19.0 software was adopted for statistical analysis. All measurements were represented as mean ± standard deviation. Data comparisons were done with student t-test or one-way ANOVA and post hoc test. Pearson method was utilized to analyze data correlation. P < 0.05 indicates statistical significance.
3 Results
3.1 circ_ROBO1 expression in NPC
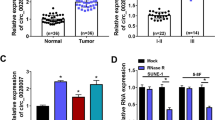
Based on the GSE143797 dataset, high-throughput sequencing was performed to identify circRNA expression profiles in NPC patients. A total of 93 up-regulated circRNAs and 77 down-regulated circRNAs were identified. circ_ROBO1 was significantly up-regulated in NPC (Fig. 1A). It was found by RT-qPCR that circ_ROBO1 was upregulated in cancer tissues of NPC patients (Fig. 1B). Overexpressed circ_ROBO1 was also found in the 4 NPC cell lines compared to NP69 cell line (Fig. 1C). Moreover, circ_ROBO1 expression was associated with lymph node metastasis and clinical stage in NPC patients (Table 1).
circ_ROBO1 expression in NPC. A. Volcano plots of the differentially expressed circRNAs. Horizontal dotted line: P = 0.05 (–log10 scaled); red points: upregulated circRNAs with statistical significance; green points: downregulated circRNAs with statistical significance. B. RT-qPCR detected circ_ROBO1 in 35 pairs of NPC tissues and normal tissues. C. RT-qPCR detected circ_ROBO1 in CNE2, HK1, HONE1, HNE2 and NP69 cells. The data were presented as mean ± SD (P < 0.01). ***
3.2 Expression characteristics of circ_ROBO1 in NPC cells
According to data from CircBase, circ_ROBO1 (hsa_circ_0001946) was derived from exons 3 to 5 of the ROBO1 gene located on chromosome Xq27.1 via reverse folding, with a length of 1485 bp (Fig. 2A). RNAs from HONE1 and HK1 cells were reverse-transcribed using random hexamers or oligomers (dT) 18 primers. The expression of circ_ROBO1 was determined by RT-qPCR in the presence of random hexamer primers (Fig. 2B), demonstrating the absence of poly A tails in circ_ROBO1.circ_ROBO1 and ROBO1 were detected in HONE1 and HK1 cells treated with actinomycin D. The results showed that circ_ROBO1 was more stable than ROBO1 (Fig. 2C). Moreover, circ_ROBO1 was resistant to RNAse R degradation, while ROBO1 mRNA was not (Fig. 2D). Circ_ROBO1 was mainly localized in the cytoplasm of HONE1 and CNE-2 cells (Fig. 2E).
Expression characteristics of circ_ROBO1 in NPC cells. A. circ_ROBO1 gene information. B. RT-qPCR detected circ_ROBO1 and ROBO1 expression. C, D. RT-qPCR detected circ_ROBO1 and ROBO1 expression in cells treated with actinomycin D or RNAse R. E. RT-qPCR detected circ_ROBO1 subcellular location. The data were presented as mean ± SD (P < 0.01). ***
3.3 Downregulating circ_ROBO1 represses NPC cell growth
circ_ROBO1 level was the highest in HONE1 and HK1 cells. Thus, siRNAs targeting circ_ROBO1 (si-circ_ROBO1#1/2) were transfected into HONE1 and HK1 cells. si-circ_ROBO1#2 with high knockout efficiency was utilized in the following cellular assays (Fig. 3A and Supplementary Fig. 1). Data from CCK-8 assay and BrdU analysis showed that HONE1 and HK1 cell proliferation was impaired after circ_ROBO1 silencing (Fig. 3B, C). Transwell test data validated that circ_ROBO1 silencing weakened cellular migration and invasion (Fig. 3D, E). Western blot analysis detected that circ_ROBO1 knockdown inhibited Vimentin and elevated E-cadherin expression in cells (Fig. 3F). Flow cytometry demonstrated that downregulating circ_ROBO1 enhanced apoptosis of NPC cells (Fig. 3G).
Elimination of circ_ROBO1 expression represses NPC cell growth. A. RT-qPCR detected circ_ROBO1 after si-circ_ROBO1#1#2 transfection. B, C. CCK-8 and BrdU assays detected cell proliferation. D, E. Transwell assays detected cell migration and invasion. F. Western blot assayed E-cadherin and Vimentin expression. G. Flow cytometry measured cell apoptosis. The data were presented as mean ± SD (P < 0.01).***
3.4 circ_ROBO1 competes with miR-324-3p
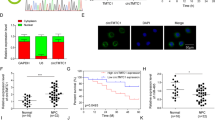
miR-324-3p may be a downstream target of circ_ROBO1 (Fig. 4A), and the binding sites between them were found (Fig. 4B). When detecting luciferase activity, it was measured that miR-324-3p upregulation caused a reduction in luciferase activity of circ_ROBO1-WT, but not affected that of circ_ROBO1-MUT (Fig. 4C). RIP experiments detected the enrichment of circ_ROBO1 and miR-324-3p in the microspheres containing Ago2, but not in those containing IgG (Fig. 4D). In HONE1 and HK1 cells, circ_ROBO1 was pulled down by Bio-miR-324-3p-WT (Fig. 4E). miR-324-3p was lower in NPC tissues than in normal tissues (Fig. 4F). Pearson correlation analysis proved the existence of a negative correlation between miR-324-3p and circ_ROBO1 expression in NPC samples (Fig. 4G).
circ_ROBO1 competes with miR-324-3p. A. Bioinformatics analysis predicted the targeted miRNA downstream of circ_ROBO1. B. The schematic diagram showed the presumed binding site between miR-324-3p and circ_ROBO1. C–E. Luciferase reporter gene assay, RIP, and RNA pull-down verified the interaction between miR-324-3p and circ_ROBO1. F. RT-qPCR detected miR-324-3p expression in 35 pairs of NPC tissues and normal tissues. G. Pearson correlation analysis determined the correlation between circ_ROBO1 and miR-324-3p in NPC. The data were presented as mean ± SD (P < 0.01). ***
3.5 miR-324-3p downregulation reverses the effect of knockdown of circ_ROBO1 on NPC cellular progression
miR-324-3p inhibitor lowered miR-324-3p expression in HONE1 and HK1 cells (Fig. 5A). Next, HONE1 and HK1 cells were co-treated with si-circ_ROBO1 and miR-324-3p inhibitor (Fig. 5B). Although knockdown of circ_ROBO1 blocked cellular proliferation, inhibition of miR-324-3p partially offset these effects (Fig. 5C, D). Additionally, inhibition of miR-324-3p expression partially mitigated the effect of circ_ROBO1 knockdown on migration, invasion, and apoptosis of HONE1 and HK1 cells (Fig. 5E–G).
miR-324-3p reduction after knockdown of circ_ROBO1 promotes NPC cellular progression. A. RT-qPCR detected miR-324-3p. B. RT-qPCR detected miR-324-3p after co-transfection. C, D. CCK-8 and BrdU assays detected cell proliferation. E, F. Transwell assays detected cell migration and invasion. G. Flow cytometry measured cell apoptosis. The data were presented as mean ± SD (P < 0.01).***; (P < 0.01).##
3.6 circ_ROBO1 regulates NME1 expression by miR-324-3p
Potential targets of miR-324-3p were identified by bioinformatics databases, and NME1 was finally focused (Fig. 6A, B). NME1 level was higher in NPC tissues than in normal tissues (Fig. 6C). Based on the binding site shared by miR-324-3p and NME1 (Fig. 6D), luciferase activity was determined, showing that miR-324-3p mimic significantly weakened NME1-WT luciferase activity (Fig. 6E). Downregulation of circ_ROBO1 significantly inhibited NME1 protein expression, while inhibiting miR-324-3p partially offset this impact (Fig. 6F).
circ_ROBO1 regulates NME1 by miR-324-3p. A. Venn diagram showed the target of miR-324-3p. B. Predicted target of miR-324-3p. C. RT-qPCR detected NME1 expression in 32 pairs of NPC tissues and normal tissues. D. Binding site between miR-324-3p and NME1 3’UTR. E. Luciferase reporter gene assay verified the interplay between miR-324-3p and NME13. F. Western blot assayed NME1 expression after regulating circ_ROBO1 and miR-324-3p. The data were presented as mean ± SD (P < 0.05; P < 0.01).***
3.7 NME1 upregulation reversed the circ_ROBO1 shRNA-induced inhibition of NPC cell invasion, proliferation and migration
To elucidate the role of NME1, in circ_ROBO1 shRNA-mediated NPC progression, NPC cells were transfected with pcDNA3.1-NME1,. As shown in Fig. 7A, B, the NME1, levels in NPC cells were decreased by circ_ROBO1 shRNA, while the effect of sh-circ_ROBO1 on NME1, expression was restored by NME1, overexpression. In addition, the circ_ROBO1 shRNA-induced decrease in NPC cell viability was significantly abolished by NME1, (Fig. 7C). Consistently, the migration and invasion of circ_ROBO1 shRNA-treated NPC cells were obviously increased by the upregulation of NME1, (Fig. 7D). In summary, overexpression of NME1, reversed the circ_ROBO1 shRNA-induced downregulation of NPC cell invasion, proliferation and migration.
Overexpression of NME1 reversed the circCRIM1 shRNA-induced inhibition of NPC cell invasion, proliferation and migration. A. the circ_ROBO1, miR-324-3p and NME1 levels in NPC cells were determined by RT–qPCR. B. The NME1 levels in NPC cells were determined by Western blot. GAPDH was used for normalization. C. NPC cell viability was assessed by the MTT assay. D. NPC cell invasion and migration were assessed by the transwell assay. **P < 0.01. All data are expressed as the mean ± standard deviation (SD)
3.8 circ_ROBO1 stimulates xenograft tumor growth and metastasis
Xenotransplantation experiments confirmed that circ_ROBO1 knockout reduced tumor weight and volume (Fig. 8A, B). In addition, downregulating circ_ROBO1 inhibited circ_ROBO1 and NME1 levels in tumor tissues while elevating miR-324-3p expression (Fig. 8C–E). HE staining validated that knocking down circ_ROBO1 repressed lung metastasis of NPC cells (Fig. 8F).
4 Discussion
circRNAs are related to tumorigenesis [17, 18] and regulate the activities of cells [19]. In recent years, new circRNAs have been confirmed to be abnormally expressed in NPC and are involved in NPC pathogenesis [20,21,22,23,24]. circRNAs act as gene expression regulators and are involved in cancer progression. However, their functions have not been sufficiently investigated in nasopharyngeal carcinoma (NPC). In this work, circ_ROBO1 was upregulated in NPC, and circ_ROBO1 knockdown inhibits NPC cell activities, suggesting that circ_ROBO1 plays a carcinogenic role in NPC.
circRNAs can bait miRNAs and participate in tumor progression by regulating downstream targets of miRNAs. For example, circ_0000215 silencing blocks NPC cell metastasis via competing with miR-512-5p to mediate phosphoinositide-3-kinase regulatory subunit 1 [25]. circ-ZNF609 accelerates NPC cell metastasis through miR-150-5p to upregulate Sp1 [20]. This study indicates that miR-324-3p was the direct downstream target of circ_ROBO1. miR-324-3p has been studied to suppress tumor growth in ovarian cancer [26]. Also, miR-324-3p is associated with tumorigenesis, including NPC [27,28,29]. Additionally, miR-324-3p increases the sensitivity of lung adenocarcinoma cells to cisplatin by induction of ferroptosis [30]. In gastric cancer, miR-324-3p is a stimulating factor for the colonization and growth of tumor cells [31]. In this study, miR-324-3p downregulation pattern was determined in NPC, and suppressing miR-324-3p impaired the effect of circ_ROBO1 knockdown on NPC cells.
The NME gene family was the first gene to be discovered as being related to tumor metastasis inhibition. At present, 10 members of the NME gene family have been found, including NME1 ~ NME10 [32]. The NME gene family has conserved domains of nucleoside diphosphokinase function [33] and modulates some physiological and pathological processes [34]. NME1, the most studied member of the NME gene family, is differentially expressed in cancers and indicates an association with tumor metastasis [35]. For example, the interaction of linc00261 with NME1 protects NME1 from degradation and increases NME1 levels, thereby inhibiting tumor metastasis in breast cancer [36]. NME1 expression is upregulated in HCC, and knocking down NME1 inhibits cancer cell malignant biological behaviors [37]. In addition, NME1 also acts as an oncogene in breast cancer and neuroblastoma [38, 39]. This work identified NME1 as a target of miR-324-3p. Downregulating circ_ROBO1 inhibited NME1, while downregulating miR-324-3p restored NME1 expression, implying that circ_ROBO1/miR-324-3p/NME1 network is involved in NPC development.
In summary, circ_ROBO1 level is elevated in NPC and modulates NPC malignancy through the miR-324-3p/NME1 axis. Useful information can be obtained from this work to explain circ_ROBO1-related mechanism in NPC, thereafter confirming circ_ROBO1 as a potential target for NPC management. There are still some shortcomings in this study. NPC cells are used for functional experiments and mechanism studies in the later stage about NME1, and other NPC cell lines can be added for similar experiments in the later stage to further confirm the research conclusion. In addition, whether circ_ROBO1 plays a role in the clinicopathological progression of tumor TNM staging in nasopharyngeal carcinoma patients needs further clinical studies and in vivo experiments to verify. Finally, because of circ_ROBO1’s covalent ring structure, circ_ROBO1 is not easily degraded by exocentase, indicating that circ_ROBO1 is structurally stable. Therefore, it is expected to be a molecular marker for the study of early diagnosis and prognosis of nasopharyngeal carcinoma. Due to the lack of relevant literature support and data reference, we will continue to verify its functional relationship with the occurrence and development of nasopharyngeal carcinoma.
Data availability
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
References
Shah, A.B. and S. Nagalli, Nasopharyngeal Carcinoma, in StatPearls. 2023: Treasure Island (FL).
Wong Y, et al. Estimating the global burden of Epstein-Barr virus-related cancers. J Cancer Res Clin Oncol. 2022;148(1):31–46.
Chen YP, et al. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80.
Huang SCM, Tsao SW, Tsang CM. Interplay of viral infection, host cell factors and tumor microenvironment in the pathogenesis of nasopharyngeal carcinoma. Cancers. 2018. https://doi.org/10.3390/cancers10040106.
Luo Y, Ye X. Baicalein exerts anti-tumor effects on nasopharyngeal carcinoma by mediating the FOXO-1/NF-κB signaling pathway. J Biol Regul Homeost Agents. 2023;37(12):6781–90.
Li T, et al. Establishment and validation of a two-step screening scheme for improved performance of serological screening of nasopharyngeal carcinoma. Cancer Med. 2018;7(4):1458–67.
Mahmoudi E, Cairns MJ. CircRNA and ageing. Subcell Biochem. 2023;102:249–70.
Huang G, et al. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/beta-catenin pathway. Cell Death Dis. 2020;11(12):1065.
Meng X, et al. CircPTK2/PABPC1/SETDB1 axis promotes EMT-mediated tumor metastasis and gemcitabine resistance in bladder cancer. Cancer Lett. 2023;554:216023.
Zhou WY, et al. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172.
Ma C, et al. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19(1):157.
Zhang H, et al. CircLIFR synergizes with MSH2 to attenuate chemoresistance via MutSalpha/ATM-p73 axis in bladder cancer. Mol Cancer. 2021;20(1):70.
Liu H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. 2021;11(3):1396–411.
Dong J, et al. Challenges and opportunities for circRNA identification and delivery. Crit Rev Biochem Mol Biol. 2023;58(1):19–35.
Wang S, et al. RNA-binding proteins and cancer metastasis. Semin Cancer Biol. 2022;86(Pt 2):748–68.
Song J, et al. A novel protein encoded by ZCRB1-induced circHEATR5B suppresses aerobic glycolysis of GBM through phosphorylation of JMJD5. J Exp Clin Cancer Res. 2022;41(1):171.
Hu F, et al. Circular RNAs: implications of signaling pathways and bioinformatics in human cancer. Cancer Biol Med. 2023;20(2):104–28.
Zhang Y, Qi W, Wu Y. EIF4A3-induced circular RNA SCAP facilitates tumorigenesis and progression of non-small-cell lung cancer via miR-7/SMAD2 signaling. Environ Sci Pollut Res Int. 2023;30(24):65237–49.
Luo YY, et al. Hsa_Circ_0098181 suppresses hepatocellular carcinoma by sponging miR-18a-3p and targeting PPARA. Front Pharmacol. 2022;13:819735.
Zhu L, et al. CircRNA ZNF609 promotes growth and metastasis of nasopharyngeal carcinoma by competing with microRNA-150-5p. Eur Rev Med Pharmacol Sci. 2019;23(7):2817–26.
Shuai M, et al. Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett. 2018;16(5):6495–501.
Mo Y, et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20(1):112.
Hong X, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19(1):33.
Fang X, et al. CircRNA circTRAF3 promotes nasopharyngeal carcinoma metastasis through targeting miR-203a-3p/AKT3 axis. Pathol Res Pract. 2021;221:153438.
Chen X, et al. Circ_0000215 exerts oncogenic function in nasopharyngeal carcinoma by targeting miR-512-5p. Front Cell Dev Biol. 2021;9:688873.
Geng L, Wang Z, Tian Y. Down-regulation of ZNF252P-AS1 alleviates ovarian cancer progression by binding miR-324-3p to downregulate LY6K. J Ovarian Res. 2022;15(1):1.
Wang X, et al. Long non-coding RNA SLC25A21-AS1 promotes multidrug resistance in nasopharyngeal carcinoma by regulating miR-324-3p/IL-6 Axis. Cancer Manag Res. 2020;12:3949–57.
Liu C, et al. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer Cell Int. 2017;17:2.
Xu J, et al. MiR-185-3p and miR-324-3p predict radiosensitivity of nasopharyngeal carcinoma and modulate cancer cell growth and apoptosis by targeting SMAD7. Med Sci Monit. 2015;21:2828–36.
Deng SH, et al. miR-324-3p reverses cisplatin resistance by inducing GPX4-mediated ferroptosis in lung adenocarcinoma cell line A549. Biochem Biophys Res Commun. 2021;549:54–60.
Sun GL, et al. miR-324-3p promotes gastric cancer development by activating Smad4-mediated Wnt/beta-catenin signaling pathway. J Gastroenterol. 2018;53(6):725–39.
Desvignes T, et al. Nme protein family evolutionary history, a vertebrate perspective. BMC Evol Biol. 2009;9:256.
Puts GS, et al. Nuclear functions of NME proteins. Lab Invest. 2018;98(2):211–8.
Boissan M, et al. The mammalian Nm23/NDPK family: from metastasis control to cilia movement. Mol Cell Biochem. 2009;329(1–2):51–62.
Banerjee S, Jha HC, Robertson ES. Regulation of the metastasis suppressor Nm23-H1 by tumor viruses. Naunyn Schmiedebergs Arch Pharmacol. 2015;388(2):207–24.
Guo G, Dai S, Chen Q. Long noncoding RNA LINC00261 reduces proliferation and migration of breast cancer cells via the NME1-EMT pathway. Cancer Manag Res. 2020;12:3081–9.
Yang J, et al. High expression of NME1 correlates with progression and poor prognosis in patients of hepatocellular carcinoma. Int J Clin Exp Pathol. 2017;10(8):8561–8.
Lodillinsky C, et al. Metastasis-suppressor NME1 controls the invasive switch of breast cancer by regulating MT1-MMP surface clearance. Oncogene. 2021;40(23):4019–32.
Adam K, et al. The potential functional roles of NME1 histidine kinase activity in neuroblastoma pathogenesis. Int J Mol Sci. 2020. https://doi.org/10.3390/ijms21093319.
Acknowledgements
Not applicable.
Tumor size in experimental animals
The maximum tumour size/burden permitted by the Ethics Committee is a volume of 2000 mm3. No mouse in this study has violated this standard and did not exceed the maximum tumour size/burden.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
FenFeng Liang designed the research study. Hai Zhao and GengChun Liu performed the research. MeiLing Huang and ZhengJia Peng provided help and advice. FenFeng Liang and ZhengJia Peng analyzed the data. FenFeng Liang wrote the manuscript. All authors contributed to editorial changes in the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Xiangtan Central Hospital (No.202010HN93) and written informed consent was provided by all patients prior to the study start. All procedures were performed in accordance with the ethical standards of the Institutional Review Board and The Declaration of Helsinki, and its later amendments or comparable ethical standards. And all animal experiments were complied with the ARRIVE guidelines and performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The experiments were approved by Xiangtan Central Hospital (No.20201206N).
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liang, F., Zhao, H., Liu, G. et al. Hsa_circ_ROBO1 competes with miR-324-3p and upregulates NME1 to promote tumor metastasis in nasopharyngeal carcinoma. Discov Onc 15, 398 (2024). https://doi.org/10.1007/s12672-024-01220-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-024-01220-9