Abstract
Trypanosoma cruzi is the etiological agent of Chagas disease, a neglected tropical infection with great public health importance. This protozoan has triatomine insects as vector but may also be transmitted through blood transfusion, organ transplants, ingestion of contaminated food, or congenitally. It has a heterogeneous population classified into Discrete Typing Units (DTUs), TcI–TcVI and TcBat. The aim of this study was to molecularly characterize the DTUs of T. cruzi in triatomines from a Chagas disease endemic area in Northeastern Brazil. Triatomines were collected and the gut content was microscopically analyzed to investigate the presence of trypanosomatid flagellates. In addition, digestive tracts of some specimens were dissected and molecularly analyzed through PCR for Trypanosoma spp. and sequencing. PCR positive samples were further submitted to a multiplex PCR for DTUs of T. cruzi. A total of 117 triatomines were collected, 93.16% being in intradomicile and 6.84% in peridomicile environments. Insects were identified as Panstrongylus lutzi (37.60%), Triatoma pseudomaculata (26.50%), Triatoma brasiliensis (23.08%) and Panstrongylus megistus (12.82%). The specimens herein analyzed presented infection rates by T. cruzi of 5.49% and 12.09% in parasitological and molecular examinations, respectively. Multiplex PCR screening revealed 70.59% of the TcI genotype, detected in all triatomine species identified in this study and 29.41% of the DTU TcIII/TcIV detected in P. megistus and P. lutzi. T. cruzi infect triatomines in intradomicile and peridomicile environments, which brings attention to the risk of human infections and to the importance of the implementation of surveillance and entomological control actions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protozoa of the Trypanosomatidae Family have been causing important diseases of public health concern for a long time. Amongst these parasites, Trypanosoma cruzi (Kinetoplastida: Trypanosomatidae) has been identified as the etiological agent of Chagas disease or American trypanosomiasis, a neglected tropical zoonosis still presents in Latin America. Kissing-bugs (Hemiptera: Reduviidae: Triatominae) act as vectors of this protozoan, mainly in developing countries where poor housing conditions of the population facilitate the infestation of insect vectors (Barbosa-Silva et al. 2019). It may also be transmitted through blood transfusion, organ transplant, ingestion of contaminated food, and congenitally. Currently, it is believed that approximately 8 million people are infected worldwide, with over 10,000 deaths every year (WHO 2021).
It is known that T. cruzi has a heterogeneous population with a genetic diversity currently classified into Discrete Typing Units (DTUs), denoted TcI, TcII, TcIII, TcIV, TcV, TcVI and TcBat (Marcili et al. 2009a; Zingales et al. 2009, 2012). In Brazil, all DTUs have been reported in different biomes (Jansen et al. 2018) and isolated from wild and domestic hosts (Marcili et al. 2009b; Bezerra et al. 2014; Jansen et al. 2018), as well as from triatomines recovered from domestic or sylvatic habitats (Cominetti et al. 2014; Martins et al. 2015; Ribeiro et al. 2016; Barbosa-Silva et al. 2016; Bezerra et al. 2018; Jansen et al. 2018; Lima-Oliveira et al. 2020).
TcI has a wide geographical distribution throughout the American continent and different mammalian (e.g., opossums, rodents, primates, and anteaters) and triatomine species [e.g., Panstrongylus megistus (Burmeister, 1835), Triatoma brasiliensis Neiva, 1911, Triatoma pseudomaculata Corrêa & Espínola, 1964, Rhodnius nasutus Stål, 1859, Rhodnius pictipes Stål, 1872] are involved in the sylvatic cycle (Zingales et al. 2012; Zingales 2018). It has also been isolated from domestic dogs (Canis lupus familiaris) (Bezerra et al. 2014). Conversely, TcII is predominantly found in southern and central regions of South America, being rarely reported in North America (Zingales 2018). This DTU has been detected especially in bats, primates, rodents, marsupials, coatis (Jansen et al. 2015, 2018); and triatomine vectors (Lilioso et al. 2017; Dario et al. 2018).
TcIII is geographically distributed from northeastern Venezuela to Argentina, being predominantly associated with the sylvatic cycle (Zingales 2018). This DTU is commonly found in terrestrial and fossorial ecotopes, having armadillos of the genera Dasypus, Chaetophractus and Euphractus acting as main reservoirs (Zingales et al. 2012; Zingales 2018), but they can also be found in marsupials (e.g., Didelphis spp., Monodelphis spp.), rodents (e.g., Galea spixii), and humans (Abolis et al. 2011; Zingales et al. 2012; Martins et al. 2015). Similarly, TcIV is mostly related with the sylvatic cycle, being reported in North and South America. The main hosts for this DTU in South America are wild primates and coatis (Nasua nasua), whereas raccoons act as important reservoirs in North America (Zingales 2018).
TcV and TcVI are rare in the wild cycle and data about their host range is scant. In fact, some reports have been performed in mammalian hosts of the genera Dasypus, Euphractus and Octodon (Zingales et al. 2012). In Brazil, TcVI has been isolated in T. brasiliensis (Lima-Oliveira et al. 2020), and both DTUs (TcV and TcVI) have been associated with cardiomyopathy and mega syndromes in humans (Zingales et al. 2012). Lastly, TcBat has been isolated from Myotis spp., Noctilio sp., and humans (Marcili et al. 2009a; Ramírez et al. 2014), and it was also detected in Triatoma sordida (Stål, 1859) (Cominetti et al. 2014).
Despite of all efforts of Brazilian Health Service, Chagas disease is still a real trouble for indigenous populations living in endemic areas. In some regions the domiciliation of triatomine species increase the risk of human and animal infection as it has been observed with the increase of reports of T. cruzi infection in dogs over the last five years. Therefore, investigations on naturally infected T. cruzi vectors, as well as the genotypic characterization of this protozoan contribute to the understanding of the eco-epidemiology of Chagas disease, facilitating decisions on preventive measures to reduce the risk for human and animal infections. The aim of this study was to detect different DTUs of T. cruzi in triatomines from a Chagas disease endemic area in Northeastern Brazil.
Methods
Study area
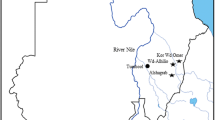
The study was performed in the state of Pernambuco, Northeastern region of Brazil (Fig. 1). The area is comprised of 21 municipalities belonging to the micro region of Garanhuns (Latitude 8°53′27″ South and Longitude 36°29′48″ West), and it is featured by a semi-arid climate with average annual temperature of 22 °C (ranging from 17 °C to 30 °C), rainfall mean of 147 mm (ranging from 25 to 295 mm) and air relative humidity of 90%.
All rural communities in this region have similar landscapes, with residences located near forest fragments and the presence of native palm trees. Dogs, cats and chickens are frequently reported in these domiciles.
Triatomine collection and morphological identification
From July 2018 to June 2019, triatomine specimens were actively collected with the aid of tweezers in intradomicile (e.g., bed frames, stored objects, boxes, walls, pictures stuck on walls) and peridomicile (e.g., chicken coops, pigpens, cattle sheds, piles of tiles, wood and bricks) areas that could serve as natural shelter for these insects. Two operators performed each sampling between 8 and 12 am for a period of 30 min. Afterwards, samples were placed in plastic vials and transported to laboratory for morphological identification (Lent and Wygodzinsky 1979). Information about vector species, life stage, site of capture, and municipality were recorded.
Detection of T. cruzi infection in triatomines
Microscopic examination
The direct parasitological detection was made by abdominal compression of each insect in 50μL saline solution (0.9% NaCl), that was then examined in an optical microscopic at 400× magnification to investigate the presence of trypanosomatid flagellates.
Molecular analysis
Triatomines were individually dissected and digestive tracts were separated for genomic DNA extraction, which was performed following a previously described protocol (Ramos et al. 2015).
DNA samples were individually tested for Trypanosoma spp. through Polymerase Chain Reaction (PCR) using the primers 18ST nF2 (5′-CAACGATGACACCCATGAATTGGGGA-3′) and 18ST nR3 (5′-TGCGCGACCAATAATTGCAATAC-3′), which amplify a product of 700-800 bp of the 18S rRNA gene (Geysen et al. 2003). Positive (DNA of T. cruzi from a triatomine) and negative (DNA of an uninfected triatomine) controls were used in all reactions. PCR products were subjected to electrophoresis in 1.5% agarose gel, stained with GelRed™ (Biotium) and visualized under an UV transilluminator. The amplified products were purified using ExoSAP-IT® (Thermo Fisher Scientific), according to manufacturer’s instructions, and sequenced in both directions in an automatic sequencer ABI 3130 Genetic Analyzer (Applied Biosystems), using the Sanger’s method (Sanger et al. 1977). DNA sequences were compared with sequences from the GenBank database using the BLASTn search tool (Altschul et al. 1990).
PCR positive samples were further submitted to a multiplex PCR, based on the non-transcribed spacer of the miniexon gene (Fernandes et al. 2001). For this, the set of primers TC1 (5′-ACACTTTCTGTGGCGCTGATCG-3′), TC2 (5′-TTGCTCGCACACTCGGCTG-CAT-3′) and TC3 (5′-CCGCGWACAACCCCTMATAAAAATG-3′) from the intergenic region of T. cruzi miniexon, and a common oligonucleotide downstream from the most conserved part of the miniexon gene Me (5′-TACCAATATAGTACAGAAACTG-3′) were used. These primers amplify products with 200 bp (TcI), 250 bp (TcII/ TcV/TcVI) and 150 bp (TcIII/TcIV) (Fernandes et al. 2001; Aliaga et al. 2011). Positive (DNA of T. cruzi from a triatomine) and negative (DNA of an uninfected triatomine) controls were used in all reactions, which were also subjected to electrophoresis as previously described, but in 3% agarose gel.
Results
Out of 117 triatomines, 93.16% (109/117) were collected in intradomicile areas and 6.84% (8/117) in peridomicile areas. All insect samples were classified as adults and identified as Panstrongylus lutzi (37.60%; 44/117), Triatoma pseudomaculata (26.50%; 31/117), Triatoma brasiliensis (23.08%; 27/117) and Panstrongylus megistus (12.82%, 15/117) from 57.14% (12/21) of municipalities of the study area. The direct parasitological detection rate was 9.40% (11/117). The infection rate observed in triatomines was 13.33% (2/15) for P. megistus, 12.90% (4/31) for T. pseudomaculata, 7.41% (2/27) for T. brasiliensis and 6.82% (3/44) for P. lutzi (Neiva and Pinto 1923).
The digestive tracts of 91 specimens (34 P. lutzi, 30 T. pseudomaculata, 15 T. brasiliensis and 12 P. megistus) were molecularly analyzed because in some samples (n = 26) the material was insufficient to perform both analyses. The microscopical examination detected an overall infection rate by T. cruzi of 5.49% (5/91), whereas Trypanosoma spp. DNA was detected in 26.37% (24/91) of the samples. In particular, 26.47% (9/34) were detected in P. lutzi, 20.00% (6/30) in T. pseudomaculata, 13.33% (2/15) in T. brasiliensis and 58.33% (7/12) in P. megistus. Homologies varying from 96.5% to 99.8% were detected with T. cruzi sequences available at the GenBank database. The molecular assessment revealed an infection rate by T. cruzi of 12.09% (11/91), being 8.82% (3/34) in P. lutzi, 10.00% (3/30) in T. pseudomaculata and 41.67% (5/12) in P. megistus. The DNA sequences herein obtained were deposited in the GenBank under the accession numbers: MN721297, MN721298, MN721299, MN721300, MN721302, MN721303, MN721304, MN721305, MN721306, MN721308, MN721309. It is worth mentioning that the difference between the number of positive samples for PCR (24) and the number of sequences deposited in GenBank (11), was due to the quality of the sequences obtained.
Molecular typing for the miniexon gene of T. cruzi was performed for 17 samples, in which Trypanosoma spp. DNA was detected. Twelve samples were classified as TcI and the remaining five were classified as DTU TcIII/TcIV (Table 1). All samples of TcI were obtained from triatomines captured from the intradomicile environment. Most of the samples that amplified for TCIII primer (DTU TcIII/TcIV) were triatomines from intradomicile and only one sample from peridomicile environment.
Discussion
This study revealed for the first time the presence of T. cruzi DTUs TcI and TcIII/TcIV group in triatomine species collected from a Chagas disease endemic area in Northeastern Brazil (state of Pernambuco), with predominance of the genotype TcI strain detected exclusively in samples collected in intradomicile environment. The genotyping of T. cruzi based on a single genetic target had been considered a limitation of the research due to the potential influence of genetic exchange (Zingales et al. 2012). Even though, data herein presented are important and contributes to the epidemiological knowledge of T. cruzi genotypes in Brazil.
Different triatomine species (i.e., P. lutzi, T. pseudomaculata, T. brasiliensis and P. megistus), most of them captured in intradomicile areas, were evaluated in this study. This data is supported by the results obtained in a recent research in the Northeastern region of Brazil, which demonstrated that 94.5% of the triatomine specimens were captured in indoor environments (Silva et al. 2019). In general, species of both genera herein detected (i.e., Panstrongylus and Triatoma) are found in burrows, tree cavities, terrestrial rocky habitats and rodent lairs (Gaunt and Miles 2000). However, they may search for refuge or food sources in artificial environments close to domestic animal shelters, increasing the risk of human and animal infection (Ribeiro et al. 2014; Barbosa-Silva et al. 2019).
In previous study conducted in the same area, P. lutzi was also the most frequent species (Silva et al. 2012). This insect has a promiscuous feeding behavior using domestic and synanthropic animals, as well as human as source of blood (Silva et al. 2017). Additionally, it presents high infection rates by T. cruzi, demonstrating its epidemiological importance in endemic areas for CD (Silva et al. 2012). On the other hand, T. pseudomaculata is found predominantly in tree trunks, and feed mainly on blood of birds. Another important species herein detected, T. brasiliensis, has been considered the main vector of T. cruzi in Northeastern Brazil. Although, this species had been predominantly associated with rodents (e.g., Galea spixii and Kerodon rupestris) (Lilioso et al. 2020; Ferreira et al. 2020), it has been proved that they may use a wide variety of blood food sources (e.g., bird, skunk, dog, goat and human) (Silva et al. 2017). The area of study is an important producer of milk in Brazil. Accordingly, the expansion of areas of bovine rearing increase the deforestation, which may be associated reduction of natural habitats of vectors, resulting in an intense invasion of domiciles (Parente et al. 2017; Santos et al. 2020).
The specimens herein analyzed presented infection rates by T. cruzi of 5.49% and 12.09% at microscopic and molecular examinations, respectively. Microscopical studies performed in other endemic areas demonstrated lower infection rates in the states of Rio Grande do Norte (2.5%) (Barbosa-Silva, 2019), Ceará (1.4%) (Fidalgo et al. 2018) and Piauí (0.8%) (Gurgel-Gonçalves et al. 2010). Similarly, a molecular investigation performed in the state of Bahia showed an infection rate of 10% (Ribeiro-Junior et al. 2019), also lower than what was observed in the present study. The higher infection rate herein detected in the molecular analyses was an expected finding, since the sensitivity of PCR is very high when compared to microscopic techniques (Dworak et al. 2017). The microscopical analysis cannot be disregarded as it is pivotal in differentiating the stages of protozoan development and consequently the metacyclogenesis rate, which is an important feature related to the dispersion ability of the parasite. Accordingly, the combination of microscopical analysis and molecular tools is advisable to increase accuracy in diagnosis and avoid false negative results (Dworak et al. 2017; Herrera et al. 2021).
The TcI genotype was predominant in all species of triatomines herein identified. It is known that this DTU is commonly found in these invertebrates (Brenière et al. 2016), being highly prevalent in anthropic environments (Lima-Oliveira et al. 2020). In Brazil, TcI has been isolated from P. megistus (Ribeiro et al. 2016), Triatoma petrochiae Pinto and Barreto, 1925 (Lima-Oliveira et al. 2020), T. brasiliensis (Bezerra et al. 2018; Costa et al. 2018; Lilioso et al. 2017; Lima-Oliveira et al. 2020), Triatoma vitticeps (Stål, 1859) (Dario et al. 2018), Triatoma sordida (Cominetti et al. 2014), T. pseudomaculata, Rhodnius nasutus (Brito et al. 2008) and Rhodnius pictipes (Xavier et al. 2014). Though detected only in single infections in this study, this genotype has been found in mixed infections with TcIV in triatomine species collected across United States, suggesting that the vectors take blood meal from different hosts species, or from a single vertebrate host species co-infected with distinct DTUs (Curtis-Robles et al. 2018). In Brazil, TcI co-infections have been reported in T. brasiliensis (TcI + TcII/VI and TcI + Trypanosoma rangeli genotype A) (Lima-Oliveira et al. 2020); in açaí samples (TcI + TcIII + TcV + TcVI) (Ferreira et al. 2018) and cardiac tissue from a fatal case of acute oral Chagas disease (TcI + TcII + TcIII + TcIV + Trypanosoma dionisii) (Dario et al. 2016). In experimental conditions, TcI presented the highest rate of infection in macrophages, followed by TcII and TcIII (Ribeiro et al. 2018). This demonstrates the complexity of the Chagas disease pathogenesis and the influence of the heterogeneity of different T. cruzi strains in the physiopathology of the disease (Ribeiro et al. 2018). Moreover, TcI has been isolated in humans presenting different clinical evolution of Chagas disease (i.e., asymptomatic, severe cardiomyopathy, and in chronic and fatal acute infections) (Abolis et al. 2011; Ramírez et al. 2010; Santana et al. 2014; Oliveira et al. 2017; Calvopina et al. 2020).
Five specimens of Panstrongylus spp. amplified for the primer TCIII, which is specific for DTUs (TcIII and TcIV), related to the sylvatic cycle (Zingales 2018). TcIII has already been isolated from P. lutzi and from a chronically infected human in Brazil (Abolis et al. 2011). Although predominant in the sylvatic cycle, TcIV has already been isolated in triatomines from the domestic cycle and in humans in Venezuela (Carrasco et al. 2012) and Brazil (Monteiro et al. 2012).
Overall, the genetic diversity of T. cruzi is underestimated (Jansen et al. 2020) and the real importance of this knowledge has been neglected in endemic regions of Latin America. In fact, the molecular characterization of these strains is pivotal to better understand the eco-epidemiology of the infection (Brenière et al. 2016). This approach provides important information on host-parasite interactions (Ribeiro et al. 2018). However, the association of these DTUs with vertebrate hosts and different biological cycles should be carefully interpreted (Jansen et al. 2020), since it has not been possible to unequivocally associate T. cruzi genotypes with any biological response variable (biome and environment) or host species (Jansen et al. 2020).
In the state of Pernambuco, Brazil, in a previous study, blood samples from patients with chronic Chagas disease presented TcII and TcVI (Rodrigues-dos-Santos et al. 2018), however, to the best of our knowledge, this is the first molecular analysis of infections and genotyping of T. cruzi in triatomines in the studied area, demonstrating a higher infection rate compared to the technique (microscopic examination) commonly used by the National Program of Control of Chagas Disease (PNCDCh). The detection of positive vectors inside or close to human dwellings suggests that people living in this Chagas disease endemic area have potential risks of becoming infected by T. cruzi. Additionally, it is an alert for the need of implementing preventive measures such as entomological surveillance to reduce the risk of human and animal infection.
References
Abolis NG, Araújo SM, Toledo MJO, Fernandez MA, Gomes ML (2011) Trypanosoma cruzi I-III in southern Brazil causing individual and mixed infections in humans, sylvatic reservoirs and triatomines. Acta Trop 120:167–172
Aliaga C, Brenière SF, Barnabé C (2011) Further interest of miniexon multiplex PCR for a rapid typing of Trypanosoma cruzi DTU groups. Infec Gen Evol 11:1155–1158
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment 23 search tool. J Mol Biol 215:403–410
Barbosa-Silva AN, Câmara ACJ, Martins K, Nunes DF, Oliveira PI, Azevedo PRM et al (2016) Characteristics of triatomine infestation and natural Trypanosoma cruzi infection in the State of Rio Grande do Norte, Brazil. Rev Soc Bras Med Trop 49:57–67
Barbosa-Silva AN, Souza RCM, Diotauiti L, Aguiar LMA, Câmara ACJ, Galvão LMC et al (2019) Synanthropic triatomines (Hemiptera: Reduviidae): infestation, colonization, and natural infection by trypanosomatids in the State of Rio Grande do Norte Brazil. Rev Soc Bras Med Trop 52:1–8
Bezerra CM, Barbosa SE, Souza R, Barezani CP, Gürtler RE, Ramos ANJr, et al (2018) Triatoma brasiliensis Neiva, 1911: food sources and diversity of Trypanosoma cruzi in wild and artificial environments of the semiarid region of Ceará, northeastern Brazil. Parasit Vectors 11:1–14
Bezerra CM, Cavalcanti LP, Souza R, Barbosa SE, Xavier SC, Jansen AM et al (2014) Domestic, peridomestic and wild hosts in the transmission of Trypanosoma cruzi in the Caatinga area colonised by Triatoma brasiliensis. Mem Inst Oswaldo Cruz 109:887–898
Brenière SF, Waleckx E, Barnabé C (2016) Over six thousand Trypanosoma cruzi strains classified into Discrete Typing Units (DTUs): attempt at an inventory. PLoS Neg Trop Dis 10:1–19
Brito CMM, Lima MM, Sarquis O, Pires MQ, Coutinho CFS, Duarte R et al (2008) Genetic polymorphism in Trypanosoma cruzi I isolated from Brazilian Northeast triatomines revealed by low-stringency single specific primer–polymerase chain reaction. Parasitol Res 103:1111–1117
Calvopina M, Segovia G, Cevallos W, Vicuña Y, Costales JA, Guevara A (2020) Fatal acute Chagas disease by Trypanosoma cruzi DTU TcI, Ecuador. BMC Infec Dis 20:1–5
Carrasco HJ, Segovia M, Llewellyn MS, Morocoima A, Urdaneta-Morales S, Martínez C et al (2012) Geographical distribution of Trypanosoma cruzi genotypes in Venezuela. PLoS Neg Trop Dis 6:1–9
Cominetti MC, Csordas BG, Cunha RC, Andreotti R (2014) Geographical distribution of Trypanosoma cruzi in triatomine vectors in the State of Mato Grosso do Sul, Brazil. Rev Soc Bras Med Trop 47:747–755
Costa AP, Ferreira JIGS, Silva RE, Tonhosolo R, Araújo AC, Guimarães MF et al (2018) Trypanosoma cruzi in triatomines and wild mammals in the National Park of Serra das Confusões, Northeastern Brazil. Rev Soc Bras Med Trop 51:445–451
Curtis-Robles R, Auckland LD, Snowden KF, Hamer GL, Hamer SA (2018) Analysis of over 1500 triatomine vectors from across the US, predominantly Texas, for Trypanosoma cruzi infection and Discrete Typing Units. Infec Gen Evol 58:171–180
Dario MA, Andrade T, Dos Santos CB, Fux B, Brandão AA, Falqueto A (2018) Molecular characterization of Trypanosoma cruzi samples derived from Triatoma vitticeps and Panstrongylus geniculatus of the Atlantic rainforest, southeast Brazil. Parasite 25:1–9
Dario MA, Rodrigues MS, Barros JH, Xavier SC, D’Andrea PS, Roque AL et al (2016) Ecological scenario and Trypanosoma cruzi DTU characterization of a fatal acute Chagas disease case transmitted orally (Espírito Santo state, Brazil). Parasit Vectors 9:1–14
Dworak ES, Araújo SM, Gomes ML, Massago M, Ferreira ÉC, Toledo MJO (2017) Sympatry influence in the interaction of Trypanosoma cruzi with triatomine. Rev Soc Bras Med Trop 50:629–637
Fernandes O, Santos SS, Cupoulo E, Mendonga B, Derre R, Junqueira ACV et al (2001) A mini-exon multiplex polymerase chain reaction to distinguish the major groups of Trypanosoma cruzi and T. rangeli in the Brazilian Amazon. Trans Royal Soc Trop Med Hyg 95:97–99
Ferreira ALS, Santana MA, Santos LVB, Monteiro DP, Campos JHF, Sena LLJ et al (2020) Triatoma brasiliensis Neiva, 1911 and Triatoma pseudomaculata Corrêa and Espínola, 1964 (Hemiptera, Reduviidae, Triatominae) in rural communities in Northeast Brazil. Rev Inst Med Trop S Paulo 62:1–8
Ferreira RTB, Cabral ML, Martins RS, Araujo PF, Silva SA, Britto C et al (2018) Detection and genotyping of Trypanosoma cruzi from açai products commercialized in Rio de Janeiro and Pará, Brazil. Parasit Vectors 11:1–11
Fidalgo ASOBV, Costa AC, Silva Filho JD, Cândido DS, Freitas EC, Pereira LS et al (2018) Insect vectors of Chagas disease (Trypanosoma cruzi) in Northeastern Brazil. Rev Soc Bras Med Trop 51:174–182
Gaunt M, Miles M (2000) The ecotopes and evolution of triatomine bugs (Triatominae) and their associated trypanosomes. Mem Inst Oswaldo Cruz 95:557–565
Geysen D, Delespaux V, Geerts S (2003) PCR–RFLP using Ssu-rDNA amplification as an easy method for species-specific diagnosis of Trypanosoma species in cattle. Vet Parasitol 110:171–180
Gurgel-Gonçalves R, Pereira FCA, Lima IP, Cavalcante RR (2010) Distribuição geográfica, infestação domiciliar e infecção natural de triatomíneos (Hemiptera: Reduviidae) no Estado do Piauí, Brasil, 2008. Rev Pan-Amaz Saude 1:57–64
Herrera L, Aguilar CM, Morocoima A, Viettri M, Lares M, Ferrer E (2021) Detection of Trypanosoma cruzi DNA in false negative samples of collected triatomines, xenodiagnosis material, and biopsies of experimentally infected animals. Int Microbiol 24:141–147
Jansen AM, Xavier S, Roque A (2020) Landmarks of the knowledge and Trypanosoma cruzi biology in the wild environment. Front Cel Infec Microbiol 10:1–15
Jansen AM, Xavier SCC, Roque ALR (2015) The multiple and complex and changeable scenarios of the Trypanosoma cruzi transmission cycle in the sylvatic environment. Acta Trop 151:1–15
Jansen AM, Xavier SCC, Roque ALR (2018) Trypanosoma cruzi transmission in the wild and its most important reservoir hosts in Brazil. Parasit Vectors 11:1–25
Lent H, Wygodzinsky P (1979) Revision of the Triatominae (Hemiptera, Reduviidae), and their significance as vectors of Chagas’ disease. Bul Am Mus Nat Hist 163:127–520
Lilioso M, Folly-Ramos E, Rocha FL, Rabinovich J, Capdevielle-Dulac C, Harry M et al (2017) High Triatoma brasiliensis densities and Trypanosoma cruzi prevalence in domestic and peridomestic habitats in the State of Rio Grande do Norte, Brazil: The Source for Chagas Disease Outbreaks? Am J Trop Med Hyg 96:1456–1459
Lilioso M, Reigada C, Pires-Silva D, Fontes FVHM, Limeira C, Monsalve-Lara J et al (2020) Dynamics of food sources, ecotypic distribution and Trypanosoma cruzi infection in Triatoma brasiliensis from the northeast of Brazil. PLoS Neg Trop Dis 14:1–18
Lima-Oliveira TM, Fontes FVHM, Lilioso M, Pires-Silva D, Teixeira MMG, Meza JGV et al (2020) Molecular eco-epidemiology on the sympatric Chagas disease vectors Triatoma brasiliensis and Triatoma petrocchiae: Ecotopes, genetic variation, natural infection prevalence by trypanosomatids and parasite genotyping. Acta Trop 201:1–6
Marcili A, Lima L, Cavazzana M, Junqueira A, Veludo H, Maia Da Silva F et al (2009a) A new genotype of Trypanosoma cruzi associated with bats evidenced by phylogenetic analyses using SSU rDNA, cytochrome b and Histone H2B genes and genotyping based on ITS1 rDNA. Parasitol 136:641–655
Marcili A, Valente V, Valente A, Junqueira ACV, Maia da Silva F, Naiff R et al (2009b) Trypanosoma cruzi in Brazilian Amazonia: lineages TcI and TcIIa in wild primates, Rhodnius spp. and in humans with Chagas disease associated with oral transmission. Int J Parasitol 39:615–623
Martins K, Andrade CM, Barbosa-Silva AN, Nascimento GB, Chiari E, Galvão LMC et al (2015) Trypanosoma cruzi III causing the indeterminate form of Chagas disease in a semi-arid region of Brazil. Int J Infec Dis 39:68–75
Monteiro WM, Magalhães LK, Sá AR, Gomes ML, Toledo MJ, Borges L et al (2012) Trypanosoma cruzi IV causing outbreaks of acute Chagas disease and infections by different haplotypes in the Western Brazilian Amazonia. PLoS ONE 7:1–9
Oliveira TSF, Santos BN, Galdino TS, Hasslocher-Moreno AM, Bastos OMP, Sousa MA (2017) Trypanosoma cruzi I genotype among isolates from patients with chronic Chagas disease followed at the Evandro Chagas National Institute of Infectious Diseases (FIOCRUZ, Brazil). Rev Soc Bras Med Trop 50:35–43
Parente CC, Bezerra FSM, Parente PI, Dias-Neto RV, Xavier SCC, Ramos NA Jr et al (2017) Community-based entomological surveillance reveals urban foci of Chagas disease vectors in Sobral, State of Ceará, Northeastern Brazil. PLoS ONE 12:1–11
Ramírez JD, Guhl F, Rendón LM, Rosas F, Marin-Neto JA, Morillo CA (2010) Chagas cardiomyopathy manifestations and Trypanosoma cruzi genotypes circulating in chronic chagasic patients. PLoS Neg Trop Dis 4:1–9
Ramírez JD, Hernández C, Montilla M, Zambrano P, Flórez AC, Parra E et al (2014) First report of human Trypanosoma cruzi infection attributed to Tcbat genotype. Zoon Pub Health 61:477–479
Ramos RAN, Campbell BE, Whittle A, Lia RP, Montarsi F, Parisi A et al (2015) Occurrence of Ixodiphagus hookeri (Hymenoptera: Encyrtidae) in Ixodes ricinus (Acari: Ixodidae) in Southern Italy. Ticks Tick-Borne Dis 6:234–236
Ribeiro AR, Lima L, Almeida LA, Monteiro J, Moreno C, Nascimento JD et al (2018) Biological and molecular characterization of Trypanosoma cruzi strains from four states of Brazil. Am J Trop Med Hyg 98:453–463
Ribeiro AR, Mendonça VJ, Alves RT, Martinez I, Araújo RF, Mello F et al (2014) Trypanosoma cruzi strains from triatomine collected in Bahia and Rio Grande do Sul, Brazil. Rev Saude Pub 48:295–302
Ribeiro AR, Oliveira RC, Ceretti Junior W, Lima L, Almeida LA, Nascimento JD et al (2016) Trypanosoma cruzi isolated from a triatomine found in one of the biggest metropolitan areas of Latin America. Rev Soc Bras Med Trop 49:183–189
Ribeiro-Júnior G, dos Santos C, Lanza F, Reis J, Vaccarezza F, Diniz C et al (2019) Wide distribution of Trypanosoma cruzi-infected triatomines in the State of Bahia, Brazil. Parasit Vectors 12:1–10
Rodrigues-dos-Santos Í, Melo MF, de Castro L, Hasslocher-Moreno AM, do Brasil PEAA, Silvestre de Sousa A et al (2018) Exploring the parasite load and molecular diversity of Trypanosoma cruzi in patients with chronic Chagas disease from different regions of Brazil. PLoS Negl Trop Dis 12:1–19
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Nat Acad Sci United States of America 74:5463–5467
Santana RA, Magalhães LK, Magalhães LK, Prestes SR, Maciel MG, Silva GA et al (2014) Trypanosoma cruzi strain TcI is associated with chronic Chagas disease in the Brazilian Amazon. Parasit Vectors 7:1–7
Santos JP, Guimarães LM, Lima IP, Batista FMA, Carvalho-Costa FA, Santos-Mallet JR (2020) Spatial distribution of synanthropic triatomines in Piaui State, Northeastern Brazil. Rev Inst Med Trop S Paulo 62:1–13
Silva MBA, Barreto AVMS, Silva HA, Galvão C, Rocha D, Jurberg J et al (2012) Synanthropic triatomines (Hemiptera, Reduviidae) in the state of Pernambuco, Brazil: geographical distribution and natural Trypanosoma infection rates between 2006 and 2007. Rev Soc Bras Med Trop 45:60–65
Silva MBA, Menezes KR, Farias MCG, Andrade MS, Victor CCA, Lorosa ES et al (2017) Description of the feeding preferences of triatominae in the Chagas disease surveillance study for the State of Pernambuco, Brazil (Hemiptera: Reduviidae). Rev Soc Bras Med Trop 50:543–546
Silva TRM, Barros GMMR, Lima TARF, Giannelli A, Silva GMda, Alves KML et al (2019) Spatial distribution of triatomine bugs in a Chagas disease endemic region in Brazil. Rev Soc Bras Med Trop 52:1–5
World Health Organization (WHO) (2021) Chagas disease (American trypanosomiasis): Epidemiology. https://www.who.int/chagas/epidemiology/en/ Accessed 26 June 2021
Xavier SC, Roque AL, Bilac D, Araújo VA, Costa Neto SF, Lorosa ES et al (2014) Distantiae transmission of Trypanosoma cruzi: a new epidemiological feature of acute Chagas disease in Brazil. PLoS Neg Trop Dis 8:1–9
Zingales B (2018) Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity. Acta Trop 184:38–52
Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O et al (2009) A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz 104:1051–1054
Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MM et al (2012) The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infec Gen Evol 12:240–253
Acknowledgements
The authors would like to thank Adeji Maria do Carmo and Jeane Cristina O. L. Silva from the Endemic Laboratory (V Gerência Regional de Saúde) for the direct microscopic examination assistance. This article is based on the PhD thesis (Postgraduate Program in Animal Bioscience) of the first author, developed at the Federal Rural University of Pernambuco, supported by a grant fellowship from the Coordination for the Improvement of Personnel of Higher Education (CAPES).
Author information
Authors and Affiliations
Contributions
Conceptualization: TRMS, RANR, Methodology: TRMS, TGR, CAdNR, TARFL, Formal analysis and investigation: TRMS, TGR, CAdNR, Writing—original draft preparation: TRMS, RANR, Writing—review and editing: TRMS, TGR, CAdNR, AS, TARFL, LCA, RANR, GAdC.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics
The Ethics Committee on Animal Experimentation of the Federal Rural University of Pernambuco approved all procedures herein performed (approval number 12/2019).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Silva, T.R.M., Rios, T.G., do Nascimento Ramos, C.A. et al. Molecular characterization of Trypanosoma cruzi DTUs of the triatomine species in a Chagas disease endemic area. J Parasit Dis 46, 64–71 (2022). https://doi.org/10.1007/s12639-021-01418-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12639-021-01418-6