Abstract
Echinococcus granulosus as an etiologic agent of hydatid cyst is one of the most important zoonotic helminthes in the world that causing enormous economic and health losses. The aim of this study was to evaluate genotype of E. granulosus isolated from sheep using mitochondrial cytochrome c oxidase subunit 1 (cox1) gene and sequencing method in East Azerbaijan province, northwest of Iran. Nineteen sheep hydatid cyst samples were collected. Genomic DNA was extracted from protoscoleces using commercial DNA extraction kit. Mitochondrial cox1 region was amplified by polymerase chain reaction (PCR) and all isolates were sequenced. Afterward, sequences were analyzed for determination of genotypes by related software. G1 (94.73 %) and G3 (5.27 %) genotypes were identified from the isolates which out of 19 hydatid cysts, 17 samples were G1B, 1 sample G1D and the other one had G3 genotype. Results of this study indicate that common sheep strain (G1); especially G1B is the dominant subtype of E. granulosus in East Azerbaijan province.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Echinococcus granulosus, as one of the smallest tapeworms of the Taeniidae, infects dogs and wolves, whereas the larval stage (hydatid cyst) expands in several species of wild and domestic mammals and in humans, causing a zoonosis of great veterinary and medical importance, cystic echinococcosis (CE) (Daniel Mwambete et al. 2004). Liver and lung are the main sites to formation hydatid cyst (Ghabouli Mehrabani et al. 2014).
This disease has been reported from the Middle East, Russia, Australia, New Zealand, America and Africa (Schantz 1995). Hydatidosis, as a main public health concern, is endemic in many areas of the Iran. The prevalence of CE in livestock (sheep, cattle, camels and goats) in Iran was reported 6.7 % totally. Also based on serological studies, seroprevalence rate of the infection in human demonstrated from 1.2 to 21.4 % in different areas of Iran (Hajialilo et al. 2012). According to studies, the prevalence of hydatid cyst in sheep and cattle in different regions of the country is 5.1–74.4 and 3.5–38.3 % respectively (Rokni 2009).
This organism shows a vast intraspecific variation associated to host specificity, morphology, epidemiology, biology, physiology and genetic (Daniel Mwambete et al. 2004). Since, ten strains or genotypes (G1–G10) have been described of E. granulosus (Piccoli et al. 2013). Some authors have suggested that these genotypes should be clustered into four different species: E. granulosus sensu stricto (G1, G2 and G3, or G1–G3 complex), Echinococcus equinus (G4), Echinococcus ortleppi (G5), and Echinococcus canadensis (G6 to G10, or G6–G10 complex) (Knapp et al. 2011). These genotypes are included common strains of sheep (G1); Tasmanian sheep strain (G2), buffalo strain (G3), horse strain (G4), cattle strain (G5), camel strain (G6), pig strain (G7 & G9) and deer strain (G8 & G10) (Youssefi et al. 2013).
Different techniques have been applied to the study on genetic variability of E. granulosus. Lately, researches have focused their analysis based on the parasite’s mitochondrial cytochrome c oxidase subunit 1 (cox1) region as suitable genetic marker (Pour et al. 2011). It was felt necessary to study due to hyperendemicity of disease in Iran and East Azerbaijan region and multiplicity of intermediate hosts which carry the disease. The results about E. granulosus’ genotyping in different region can be used in studies on the prevention and control, epidemiology, vaccine design, drug sensitivity, life cycle analysis, transmission and disease progression (Ergin et al. 2010).
The aim of this study was to evaluate genotype of E. granulosus isolated from sheep using (cox1) gene and sequencing method in East Azerbaijan province, northwest of Iran.
Material and methods
Sample collection
In this study, 19 E. granulosus cysts were collected from sheep. The animals originated from various locations within East Azerbaijan province that slaughtered in abattoirs located in the Tabriz. Each animal cyst was processed as an E. granulosus isolate. Cyst fluid containing protoscoleces were aspirated from cysts by a separate syringe under sterile conditions and were washed three times with normal saline and stored at −20 °C.
DNA extraction
Genomic DNA was extracted from protoscoleces using commercial DNA extraction kit (AccuPrep ® Genomic DNA Extraction kit Cat.No: K-3032) according to the manufacturer’s instructions and then stored at −20 °C until polymerase chain reaction (PCR) amplification.
Polymerase chain reaction (PCR)
To amplification of the cox1 gene fragment we used primers (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) and (5′TAAAGAAAGAACATAATGAAAATG-3′) as forward and reverse respectively (Ergin et al. 2010). Twenty microliter reaction volumes containing Taq DNA polymerase (1 U), from each dNTP (dATP, dCTP, dGTP, dTTP) 250 µM, Tris–HCL (pH 9.0) 10 mM, KCL (30 mM), Mgcl2 (1.5 mM), Template DNA (50 ng) 7 µl and (10 Pmol) 1 µl of each primer were used and amplified by PCR under the following temperature conditions: initial denaturation 94 °C (5 min) and then denaturation 94 °C (30 s), annealing 56 °C (45 s), extension 72° (35 s), in 35 cycles and final extension 72° (10 min). PCR products were electrophoresed on 1.5 % agarose gel after staining with safe stain and then 440 bp band of cox1 gene was seen under UV light using transilluminator.
Sequencing and phylogenetic analysis
Nineteen PCR products were purified by gel extraction kit (AccuPrep ® Gel Purification Kit Cat No: K-3035) according to the manufacturer’s instructions and sequenced by Genetic Analyzer 3130 ABI. Sequences were compared with each other and available reference sequences in GenBank using Chromas and Sequencher softwares and BLAST program. Reference sequences of E. granulosus genotypes (G1–G10) and Echinococcus vogeli (as outgroup) were inferred from previous publications (Schneider et al. 2010; Bowles and McManus 1993) and the National Center for Biology Information (http://www.ncbi.nlm.nih.gov/). After multiple alignments by ClustalW, phylogenetic analyses of the sequences data were performed using cox1 sequences and phylogeny tree was drawn using sequences obtained in this study as well as reference sequences of all described E. granulosus genotypes by MEGA4 software. GenBank accession numbers for the sequences obtained from this study and the reference genotypes are shown in Table 1.
Results
PCR amplification and sequencing was successfully performed on hydatid cyst isolates. After electrophoresis on PCR product, 440 bp bands were seen in all samples under UV light clearly. G1 (94.73 %) and G3 (5.27 %) genotypes were identified from the isolates which out of 19 sheep hydatid cysts, 17 samples were G1B, 1 sample G1D and the other one had G3 genotype.
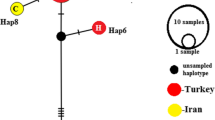
Three sequences (one of each subtype) were submitted to the GenBank and registered under following accession numbers KJ540227, KJ540229 and KJ540231. Phylogenetic tree based on analysis of sequences is shown in Fig. 1. Isolates were grouped into two distinct clusters related to the G1–G3 complex and the G6–G10 complex. In this study, most of the isolates (94.73 %) were identified as G1 (sheep strain) and were clustered with the G1 reference genotype. Also isolate identified as G3 (buffalo strain) (5.27 %) clustered with the G3 reference sequence.
Discussion
The results of this study indicated that the G1 genotype of E. granulosus (sheep strain) was the most commonly identified genotype from sheep in East Azerbaijan province, Iran. These finding suggest that sheep–dog cycle occur in this region. G1 is the dominant genotype found in world livestock (Moro and Schantz 2008). However in some North African countries, such as Sudan, G6 is the dominant genotype which found in sheep, goats and camels (Hajialilo et al. 2012).
In order to genotyping of sheep hydatid cysts using sequencing with cox1 gene, G1 is reported as predominant genotype in different countries such as Peru, China, Turkey, Italy and Tunisia that are consistent with this study (Sánchez et al. 2010; Ma et al. 2008; Utuk et al. 2008; Busi et al. 2007; Varcasia et al. 2006; M’rad et al. 2005).
In a study that was done by (Pezeshki et al. 2013) in the Ardabil province, out of 19 sheep hydatid cysts, 18 samples G1 and 1 sample G3 genotype were reported. The results of our study are quite similar with this study that could be due to the proximity of Ardabil with East Azerbaijan province in terms of parasite life cycle. This indicates that the G3 genotype could be as one of the etiologic factors in this area. Therefore, to determine transmission cycle, the study of G3 genotype reservoirs as intermediate hosts in this region is essential. In a study in Egypt (Abdel Aaty et al. 2012), 42 sheep hydatid cyst samples were genotyped that all samples were reported as G6 genotype. Also in Pakistan (Latif et al. 2010), one-third of samples were G1 genotype and in Indian study (Pednekar et al. 2009), out of 8 cysts, 6 samples were reported G3 genotype and 2 samples had G1 genotype. So far, 10 genotypes (G1–G10) have been reported from different intermediate hosts of E. granulosus species complex in the world (Thompsom 2008). Due to distribution of intermediate hosts in different parts of the world, there is probability to the difference of genotypes in different regions and their prevalence rate. In other words, the predominant or high prevalence of one genotype in an area indicates the significance and key role of its intermediate hosts in parasite life cycle.
Consequently, all prevention and control proceedings of disease are based on the dominant genotype in a region. PCR–RFLP, SSCP and Semi nested-PCR as other molecular methods for genotyping of E. granulosus were not able to separate correctly genotypes of G1–G3 (E. granulosus sensu stricto) and G6–G10 (E. Canadensis) (Simsek et al. 2011; Jabbar et al. 2011). In a Greek study in 2007 out of 20 sheep and goat cysts all sheep samples were G1 and goat samples have G6/G7 genotypes by Semi nested-PCR method, while in sequencing, 18 sheep samples, G1 and two samples were G3 genotype. The entire goat samples were reported as G7 genotype (Varcasia et al. 2007). In a study in Turkey, all bovine and sheep samples were reported G1–G3 genotype with SSCP method (Simsek et al. 2011). Also in study conducted in Iran on bovine, sheep and human samples, all samples were reported G1–G3 genotype using PCR–RFLP method (Dousti et al. 2013). As a result, because of exact separation between genotypes of sequencing method, this method is preferable than another methods for genotype determination.
In order to genotype of E. granulosus, there are used various molecular methods based on mitochondrial and nuclear DNA genes but according to results of conducted studies, mitochondrial genes are more preferable than other methods for E. granulosus genotyping. Information obtained from mitochondrial DNA can help researchers to solve parasite taxonomy problems (Gillespie and Pearson 2001; Schantz 2003).
In the present study, isolates were clustered in major group related to the G1–G3 genotype complex. One identified isolate as G3 genotype was grouped together with the G3 reference genotype (Fig. 1) and being the G1 isolates, providing further evidence that the G1 and G3 genotypes should be considered as E. granulosus sensu stricto (Hajialilo et al. 2012).
This study demonstrated that, G1 genotype is transmitted from sheep in Iran. Additionally, this study showed that G1 especially G1B subtype is the predominant among sheep in Northwest of Iran and sheep–dog cycle have interactions in this region.
References
Abdel Aaty H, Abdel-Hameed D, Alam-Eldin Y, El-Shennawy S, Aminou H, Makled S, Darweesh S (2012) Molecular genotyping of Echinococcus granulosus in animal and human isolates from Egypt. Acta Trop 121(2):125–128. doi:10.1016/j.actatropica.2011.10.014
Adwan G, Adwan K, Bdir S, Abuseir S (2013) Molecular characterization of Echinococcus granulosus isolated from sheep in Palestine. Exp Parasitol 134:195–199. doi:10.1016/j.exppara.2013.03.024
Bart JM, Abdukader M, Zhang YL, Lin RY (2006) Genotyping of human cystic echinococcosis in Xinjiang, PR China. Parasitology 133:571–579
Beato S, Parreira R, Calado M, Gracio M (2010) Apparent dominance of the G1-G3 genetic cluster of Echinococcus granulosus strains in the central inland region of Portugal. Parasitol Int 59:4. doi:10.1016/j.parint.2010.08.004
Bhattacharya D, Bera AK, Bera BC, Maity A, Das SK (2007) Genotypic characterisation of Indian cattle, buffalo and sheep isolates of Echinococcus granulosus. Vet Parasitol 143:371–374
Boufana B, Stidworthy MF, Bell S, Chantrey J (2012) Echinococcus and Taenia spp. from captive mammals in the United Kingdom. Vet Parasitol 190:95–103. doi:10.1016/j.vetpar.2012.05.023
Bowles J, McManus DP (1993) NADH dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus. Int J Parasitol 23:969–972
Bowles J, Blair D, McManus DP (1992) Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol 54:165–173
Busi M, Šnábel V, Varcasia A, Garippa G, Perrone V, De Liberato C, D’amelio S (2007) Genetic variation within and between G1 and G3 genotypes of Echinococcus granulosus in Italy revealed by multilocus DNA sequencing. Vet Parasitol 150:75–83. doi:10.1016/j.vetpar.2007.09.003
Daniel Mwambete K, Ponce-Gordo F, Cuesta-Bandera C (2004) Genetic identification and host range of the Spanish strains of Echinococcus granulosus. Acta Trop 91(2):87–93. doi:10.1016/j.actatropica.2004.04.001
Dousti M, Abdi J, Bakhtiyari S, Mohebali M, Mirhendi S, Rokni M (2013) Genotyping of hydatid cyst isolated from human and domestic animals in Ilam province, Western Iran using PCR-RFLP. Iran j Parasitol 8(1):47
Ergin S, Saribas S, Yuksel P, Zengin K, Midilli K, Adas G, Arikan S, Aslan M, Uysal H, Caliskan R (2010) Genotypic characterisation of Echinococcus granulosus isolated from human in Turkey. Afr J Microbiol Res 4:551–555
Ghabouli Mehrabani N, Kousha A, Khalili M, Mahami Oskouei M, Mohammadzadeh M, Alizadeh S, Maleksabet A, Hamidi F (2014) Hydatid cyst surgeries in patients referred to hospitals in East Azerbaijan province during 2009–2011. Iran J Parasitol 9(2):233–238
Gillespie SH, Pearson RD (2001) Principles and practice of clinical parasitology. Wiley, Chichester,UK, pp 585–612. doi:10.1002/0470842504.fmatter_indsub
Hajialilo E, Harandi MF, Sharbatkhori M, Mirhendi H, Rostami S (2012) Genetic characterization of Echinococcus granulosus in camels, cattle and sheep from the south-east of Iran indicates the presence of the G3 genotype. J Helminthol 86(3):263. doi:10.1017/S0022149X11000320
Jabbar A, Narankhajid M, Nolan MJ, Jex AR, Campbell BE, Gasser RB (2011) A first insight into the genotypes of Echinococcus granulosus from humans in Mongolia. MCP 25:49–54. doi:10.1016/j.mcp.2010.11.001
Kamenetzky L, Gutierrez AM, Canova SG, Haag KL, Guarnera EA, Parra A, García GE, Rosenzvit MC (2002) Several strains of Echinococcus granulosus infect livestock and humans in Argentina. Infect. Genet Evol 2:129–136
Knapp J, Nakao M, Yanagida T, Okamoto M, Saarma U, Lavikainen A, Ito A (2011) Phylogenetic relationships within Echinococcus and Taenia tapeworms (Cestoda: Taeniidae): an inference from nuclear protein-coding genes. Mol Phylogenet Evol 61:628–638. doi:10.1016/j.ympev.2011.07.022
Konyaev SV, Yanagida T, Nakao M, Ingovatova GM, Shoykhet YN, Bondarev AY (2013) Genetic diversity of Echinococcus spp. in Russia. Parasitology 140:1637–1647. doi:10.1017/S0031182013001340
Latif AA, Tanveer A, Maqbool A, Siddiqi N, Kyaw-Tanner M, Traub RJ (2010) Morphological and molecular characterisation of Echinococcus granulosus in livestock and humans in Punjab, Pakistan. Vet Parasitol 170:44–49. doi:10.1016/j.vetpar.2010.02.003
Lavikainen A, Lehtinen M, Meri T, Hirvelä-Koski V, Meri S (2003) Molecular genetic characterization of the Fennoscandian cervid strain, a new genotypic group (G10) of Echinococcus granulosus. Parasitology 127:207–215
M’rad S, Filisetti D, Oudni M, Mekki M, Belguith M, Nouri A, Sayadi T, Lahmar S, Candolfi E, Azaiez R (2005) Molecular evidence of ovine (G1) and camel (G6) strains of Echinococcus granulosus in Tunisia and putative role of cattle in human contamination. Vet Parasitol 129:267–272. doi:10.1016/j.vetpar.2005.02.006
Ma S, Maillard S, Zhao H, Huang X, Wang H, Geng P, Bart J-M, Piarroux R (2008) Assessment of Echinococcus granulosus polymorphism in Qinghai province, People’s Republic of China. Parasitol Res 102:1201–1206. doi:10.1007/s00436-008-0894-7
Monteiro DU, Botton SA, Tonin AA, Azevedo MI, Graichen DAS, Noal CB, de la Rue ML (2014) Echinococcus canadensis (G7) and Echinococcus granulosus sensu stricto (G1) in swine of southern Brazil. Vet Parasitol 28:335–338. doi:10.1016/j.vetpar.2014.01.023
Moro P, Schantz P (2008) Echinococcosis: a review. Int J Infect Dis 13:125–133. doi:10.1016/j.ijid.2008.03.037
Pednekar RP, Gatne ML, Thompson R, Traub RJ (2009) Molecular and morphological characterisation of Echinococcus from food producing animals in India. Vet Parasitol 165(1):58–65. doi:10.1016/j.vetpar.2009.06.021
Pezeshki A, Akhlaghi L, Sharbatkhori M, Razmjou E, Oormazdi H, Mohebali M, Meamar A (2013) Genotyping of Echinococcus granulosus from domestic animals and humans from Ardabil Province, northwest Iran. J Helminthol 10:10
Piccoli L, Bazzocchi C, Brunetti E, Mihailescu P, Bandi C, Mastalier B, Cordos I, Beuran M, Popa L, Meroni V (2013) Molecular characterization of Echinococcus granulosus in south-eastern Romania: evidence of G1–G3 and G6–G10 complexes in humans. Clin Microbiol Infect 19(6):578–582. doi:10.1111/j.1469-0691.2012.03993.x
Pour AA, Hosseini SH, Shayan P (2011) Comparative genotyping of Echinococcus granulosus infecting buffalo in Iran using cox1 gene. Parasitol Res 108(5):1229–1234. doi:10.1007/s00436-010-2170-x
Rokni MB (2009) Echinococcosis/hydatidosis in Iran. Iran J Parasitol 4:1–16
Sánchez E, Cáceres O, Náquira C, Garcia D, Patiño G, Silvia H, Volotão AC, Fernandes O (2010) Molecular characterization of Echinococcus granulosus from Peru by sequencing of the mitochondrial cytochrome C oxidase subunit 1 gene. Mem Inst Oswaldo Cruz 105:806–810. doi:10.1590/S0074-02762010000600013
Schantz P (1995) Epidemiology and control of hydatid disease. In Thompson RCA, Lymbery AJ (eds) Echinococcus and hydatid disease, CAB International, Oxon, pp 233–331
Schantz P (2003) Echinococcus species (agents of cystic, alveolar, and polycystic echinococcosis). Principles and practice of pediatric infectious diseases, 2nd edn. Churchill-Livingstone, New York, pp 1357–1361
Schneider R, Gollackner B, Schindl M, Tucek G, Auer H (2010) Echinococcus canadensis G7 (pig strain): an underestimated cause of cystic echinococcosis in Austria. Am J Trop Med Hyg 82(5):871. doi:10.4269/ajtmh.2010.09-0639
Sharifiyazdi H, Oryan A, Ahmadnia S, Valinezhad A (2011) Genotypic characterization of Iranian camel (Camelus dromedarius) isolates of Echinoccocus granulosus. J Parasitol 97:251–255. doi:10.1645/GE-2642.1
Sharma M, Sehgal R, Ahmad Fomda B, Malhotra A (2013) Molecular characterization of Echinococcus granulosus cysts in north Indian patients: identification of G1, G3, G5 and G6 genotypes. PLoS Negl Trop Dis 7:6. doi:10.1371/journal.pntd.0002262
Simsek S, Balkaya I, Ciftci AT, Utuk AE (2011) Molecular discrimination of sheep and cattle isolates of Echinococcus granulosus by SSCP and conventional PCR in Turkey. Vet Parasitol 178:367–369. doi:10.1016/j.vetpar.2011.01.033
Thompsom R (2008) The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol 119(4):439–446. doi:10.1016/j.exppara.2008.04.016
Umhang G, Richomme C, Boucher JM, Hormaz V, Boué F (2013) Prevalence survey and first molecular characterization of Echinococcus granulosus in France. Parasitol Res 112:1809–1812. doi:10.1007/s00436-012-3245-7
Utuk AE, Simsek S, Koroglu E, Mcmanus DP (2008) Molecular genetic characterization of different isolates of Echinococcus granulosus in east and southeast regions of Turkey. Acta Trop 107:192–194. doi:10.1016/j.actatropica.2008.05.026
Varcasia A, Canu S, Lightowlers MW, Scala A, Garippa G (2006) Molecular characterization of Echinococcus granulosus strains in Sardinia. Parasitol Res 98(3):273–277. doi:10.1007/S00436-005-0059-X
Varcasia A, Canu S, Kogkos A, Pipia AP, Scala A, Garippa G, Seimenis A (2007) Molecular characterization of Echinococcus granulosus in sheep and goats of Peloponnesus, Greece. Parasitol Res 101:1135–1139. doi:10.1007/s00436-007-0568-x
Vural G, Baca AU, Gauci CG, Bagci O (2008) Variability in the Echinococcus granulosus cytochrome C oxidase 1 mitochondrial gene sequence from livestock in Turkey and a re-appraisal of the G1-3 genotype cluster. Vet Parasitol 154:347–350. doi:10.1016/j.vetpar.2008.03.020
Youssefi MR, Tabaripour R, Omrani VF, Spotin A, Esfandiari B (2013) Genotypic characterization of Echinococcus granulosus in Iranian goats. Asian Pac J Trop Dis 3:362–366
Acknowledgments
This work was supported fully by Tabriz Infectious and Tropical Diseases Research Center (Grant no: 92-01), Tabriz University of Medical Sciences, Tabriz, Iran. This is a report of a database from thesis of Nader Ghabouli Mehrabani entitled “Genotyping of Echinococcus granulosus isolates from human and herbivores using mitochondrial cytochrome oxidase 1(cox1) gene by sequencing method in Tabriz” registered in Tabriz University of Medical Sciences.
Conflict of interest
The authors declare that there is no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mahami Oskouei, M., Ghabouli Mehrabani, N., Miahipour, A. et al. Molecular characterization and sequence analysis of Echinococcus granulosus from sheep isolates in East Azerbaijan province, northwest of Iran. J Parasit Dis 40, 785–790 (2016). https://doi.org/10.1007/s12639-014-0579-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12639-014-0579-3