Abstract
Seriola lalandi is an economically important species that is globally distributed in temperate and subtropical marine waters. Aquaculture production of this species has had problems associated with intensive fish farming, such as disease outbreaks or nutritional deficiencies causing high mortality. Intestinal microbiota are involved in many processes that benefit a host, such as disease control, stimulation of the immune response, and the promotion of nutrient metabolism. The aim of this study is to evaluate the in vitro probiotic properties of bacteria isolated from the intestinal content of wild Seriola lalandi. The probiotic potential was evaluated in terms of (i) the antimicrobial activity against vibrios causing outbreaks in farmed fish; (ii) the ability to stimulate genes related to an innate immune response in fish; and (iii) antibiotic resistance. Nineteen isolates identified as Pseudomonas, Shewanella, Psychrobacter, and Acinetobacter showed antimicrobial activity and significant relative expression of cytokines, serum amyloid A protein (SAA), hepcidin, and lysozyme. A positive correlation was observed between the levels of expression and the bacterial load after 24 h of exposure. Pseudomonas isolates showed a level of antibiotic resistance. In conclusion, isolates of the genera Shewanella, Psychrobacter, and Acinetobacter could serve as potential probiotics in S. lalandi culture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seriola lalandi (yellowtail kingfish) is a marine, pelagic, and carnivorous fish found globally in subtropical and temperate waters of the Pacific and Indian Oceans [1]. This species is mainly cultivated in Japan [2], Australia [1], and New Zealand [3]. In Chile, its cultivation is considered within the aquaculture diversification program [4]. This species has excellent attributes that promote its cultivation, including high rates of growth and market acceptance [5]. A problem associated with the cultivation of this species is the high mortality, up to 90%, in the stage of larvae and fry [6]. The disease outbreaks have increased with the intensification of aquaculture, especially during the early stages of fish development; an example of this principal problem in larval culture is vibriosis [7].
The control and prevention of diseases in aquaculture are antimicrobials; however, the excessive use of these has been questioned given the selection of bacteria resistant to antimicrobials [8, 9]. It has been described that the use of antimicrobials in farmed fish causes an imbalance in the intestinal microbiota known as dysbiosis [10,11,12,13]. This imbalance in the composition of the microbiota could cause an increase in the colonization of pathogenic bacteria, increasing host mortality [13]. In this sense, the use of probiotics is proposed as an alternative to overcome the problems associated with the excessive use of antimicrobials in the control of bacterial diseases in intensive culture systems. Among the reported benefits of the use of probiotics in aquaculture species are inhibition of colonization by pathogens [14,15,16]; the improvement in the immune response of the host [17,18,19,20]; enzymatic contribution, improving the digestion process, given by the extracellular production of enzymes, such as proteases, lipases, chitinases, phytases, and amylases [21]; and contribution with nutrients such as vitamins, amino acids, and fatty acids [22]. Desirable characteristics for the selection of probiotic potentials include (i) not causing host damage; (ii) having the ability to colonize and adhere to host epithelial surfaces; (iii) having the ability to reach target organs and exercise their function; (iv) lacking antimicrobial resistance genes [23, 24]; (v) bile and pH tolerance testing; and (vi) antagonistic activity. Previous studies have reported the use of autochthonous bacteria as an alternative to controlling bacterial diseases in fish culture [18, 25,26,27,28,29,30,31]. As Nayak [32] and Mills et al. [33] described, compared to the benefits provided by bacteria isolated from other sources, beneficial bacteria isolated from a host could be more beneficial if administered again in the same species or similar species. Currently, there is increasing attention on the composition of the gut microbiota of fish for their contribution to processes such as nutrition and defense of the host [32, 34].
Microbiota have been defined as the community of microorganisms present in most individuals of a population or a species that, despite continuous contact with different tissues, do not cause damage to their host [35]. Most of these microorganisms reside in the digestive tract, where they influence a wide range of biological processes that generate beneficial effects for the host. These microorganisms contribute to nutrition, digestion processes, and the provision of vitamins; in defense, microbiota play a protective role in preventing colonization by pathogens through competition for nutrients, competition for exclusion, and production of antimicrobial substances, and these microorganisms can modulate the host’s immune system [23, 24, 36, 37]. Many studies suggest that microbial communities have co-evolved with their respective hosts establishing mutualistic interactions for many physiological aspects, such as the synthesis of enzymes, vitamins, metabolites, and antimicrobial compounds and the development of the intestinal mucosa of the host and the immune system [32, 36, 38,39,40]. A recent study of Rosshart et al. [41] highlights the importance of the establishment of the microbiota in the response of the host to pathogenic microorganisms. These authors showed that wild and laboratory mice have different survival against a pathogen, 90% in wild mice and less than 20% in laboratory mice, attributing this response to the composition of the organism’s microbiota depending on the origin. This reflects that the animals in captivity, kept under controlled conditions, present a microbiota different from that of wild animals, which have been exposed to various environmental factors, pathogens, stress, etc. and that this microbiota selected by the host through natural selection confers benefits in the response to pathogens. Intensive culture systems maintain controlled conditions of various parameters, such as water quality, diet, and population density, which can lead to the establishment of a different intestinal microbiota compared to wild fish of the same species [4, 42,43,44,45]. A previous study by Ramírez and Romero [4] describes significant differences in the components of the intestinal microbiota of wild S. lalandi and aquaculture. At the genus level, those differentially represented in wild S. lalandi from that study are Psychrobacter, Shewanella, Pseudomonas, and Acinetobacter. The aim of this work was to study the probiotic potential of autochthonous bacteria isolated from wild S. lalandi by evaluating the antagonist activity against Vibrio sp. and the in vitro stimulation of genes related to the immune response.
Materials and Methods
Isolation of Bacterial Strains
A total of 12 fish of S. lalandi were collected from the Region of Coquimbo, Chile, specifically from latitude S 30.104; longitude W 71.377 to latitude S 30.302; longitude W 70.608 in February 2015. After capture, the fish were weighed, and the intestinal contents were aseptically removed, stored in ice, and transported to the laboratory. The intestinal contents were homogenized in a 10 mL sterile phosphate-buffered saline solution (PBS 1×) pH 7.2. The homogenates were disseminated in trypticase soy agar (Merck) supplemented with 2% NaCl (Oxoid) (TSA2), Luria–Bertani (LB), de MAN, ROGOSA, and SHARPE (MRS, Merck) and yeast extract-peptone-dextrose growth medium (YEPD) dishes supplemented with 2% NaCl. After incubation at 28 °C for 2 days, the predominant colonies were isolated in fresh media and examined for different morphologies. The isolates were identified based on 16S rRNA gene sequencing. Isolates from the bacterial genera Pseudomonas, Shewanella, Psychrobacter, and Acinetobacter were evaluated in this study. These isolates were selected according to previous results by Ramírez and Romero [4] in which these genera were found to be dominant in the gut microbiota of wild S. lalandi.
Antimicrobial Activity
The antimicrobial activity of the isolated strains was studied against Vibrio sp., a common pathogen in aquaculture, by the double-layer method of Dopazo et al. [46] modified according to Leyton and Riquelme [47]. On a plate count agar plate (prepared with 75% sea water and 25% distilled water) (PCAsw), 10 μL of an 18-h culture of the strain of interest grown in trypticase soy broth (TSB, Merck) supplemented with 2% NaCl (TSB2) was inoculated. The inoculum was incubated for a period of 36 h at 20 °C. Once the bacterial growth was verified, the macrocolony was inactivated by exposure to chloroform vapors for a period of 45 min. Subsequently, 4 mL of TSB2 soft agar media supplemented with 0.7% bacteriological agar (Oxoid) was used, which was inoculated with 100 μL of an 18-h culture of the three pathogenic bacteria of genus Vibrio cultivated in TSB2. Then, the medium was shaken gently and briefly by pouring on the plate with the macrocolony of the antagonist strain. Plates were incubated at 20 °C for 24 h. Subsequently, inhibition or growth of the pathogenic strain around the antagonist strain was verified.
Stimulation of Immune-Related Genes
Infection Kinetics in Cell Line
To determine the transcriptional activity of genes associated with the innate immune system during the infection process with probiotic potentials, an infection kinetic assay was performed using qRT-PCR in the SHK-1 cell line [48]. The cell line was maintained in L-15 medium (Leibovitz, Gibco) supplemented with 5% fetal bovine serum (FBS, Gibco), 4 mM l-glutamine, 50 μg mL−1 gentamicin sulfate, and 40 μM β-mercaptoethanol and was cultured at 18 °C in 75 cm2 tissue culture–treated flasks (Orange Scientific). The kinetic infection was measured at earlier stages of the infection (12 and 24 h. The cells used in this study corresponded to passages 45 and 48.
A single colony grown in TSA2 plates was used to inoculate 4 mL of TSB2 medium, incubated at 28 °C and 100 rpm for 24 h. Then, 1 mL of the medium of each isolate was used to quantify the bacterial concentration by means of a Petroff–Hausser chamber. First, it was centrifuged, and the pellet was washed twice with PBS 1×, to eliminate traces of culture medium. The volume corresponding to 1 × 105 bacteria was used to infect with a multiplicity of infection (MOI) of 1 in every 1.92 cm2 well with cells for every kinetic time point, including biological triplicates. SHK-1 cells were grown to 90% confluency in 24-well tissue culture plates under the conditions described above, but this time without addition of antibiotic. After infection, followed by an incubation period of 2 h at 18 °C, the cells were washed once with 1× PBS, and fresh L-15 medium was added to eliminate excessive bacterial growth [49]. Finally, the infected cells were incubated at 18 °C during the times described above.
RNA Isolation and cDNA Synthesis
Total RNA was isolated from cells using the TRIzol isolation reagent according to the manufacturer’s procedure (Roche, Canada). The concentration of extracted RNA was calculated at a wavelength of 260 nm using NanoQuant spectrophotometry (Tecan). To detect the purity of RNA, the optical density (OD) absorption ratio at 260/280 nm was determined, and only samples with a ratio of more than 1.8 were used for cDNA synthesis. cDNA was synthesized from 1 μg of RNA pre-treated with RNase-Free DNase RQ1 (Promega) for 1 h at 37 °C, using a kit ImProm-II™Reverse Transcription System (Promega) according to the manufacturer’s procedure.
Relative Expression of Immune-Related Genes
Triplicate qPCR reactions were carried out for each sample analyzed. The PCRs were performed with the SYBR green method. Arrays were run on a Corbett Rotor Gene 6000 instrument. The reactions were set for each sample with 1 μL of cDNA, 5 μL of 2× concentrated SYBR Green PCR Master Mix (Roche) containing SYBR Green as a fluorescent intercalating agent, 0.5 μM forward primer, and 0.5 μM of reverse primer (Table 1). The thermal profile for all reactions was 5 min at 95 °C, and then 40 cycles of 15 s at 95 °C, 10 s at 60 °C, and 10 s at 72 °C, except for the V3 region of the 16S rRNA, which had a profile of 40 cycles of 5 s at 95 °C and 2 s at 60 °C. Fluorescence monitoring occurred at the end of each cycle. Additional dissociation curve analysis was performed and showed in all cases one single peak. Relative quantification of cDNA was made using Elongation Factor 1a (EF-1a) as a reference gene (housekeeping gene). Modification of gene expression is represented with respect to the control sampled at the same time as the treatment. The graphics for the relative expression were performed as a heatmap in the R environment using the ggplot package.
Enzymatic Analysis
The enzymatic activities of the isolates were determined by using the API ZYM system (bioMérieux, Marcy-l’Etoile, France) according to the manufacturer’s guidelines. Briefly, isolated colonies grown on TSA plates were used to inoculate and were resuspended in a NaCl 0.85% solution (bioMérieux, Marcy-l’Etoile, France) to obtain a turbidity of 5–6 McFarland (1.5–1.8 × 109 bacteria mL−1), and 65 μL of this suspension was added to each cupule. Test strips were incubated for 4 h at 30 °C, and following incubation, 1 drop of ZYM A (API; Tris-hydroxymethyl-aminomethane, hydrochloric acid, sodium laurel sulfate, H2O) and ZYM B (API; fast blue BB, 2-methoxyethanol) were added to each cupule. Test strips were read after 5 min, and the results were scored using the following classification: 0 negative reaction; 1–2 weak activity; 3–5 strong activity.
Antimicrobial Resistance Patterns
The resistance/susceptibility profiles of the different isolates to five antimicrobial agents were determined by an agar disk diffusion method as described in the Clinical and Laboratory Standards Institute (CLSI) guideline M42-A [50], using Müeller–Hinton agar (Difco) supplemented with NaCl (2%). The antibacterial susceptibility patterns of resistant isolates were performed using disks containing the following antibacterial agents: florfenicol (FFC, 30 μg), oxytetracycline (OT, 30 μg), oxolinic acid (OA, 2 μg), flumequine (UB, 30 μg), and sulfamethoxazole–trimethoprim (SXT, 1.25 and 23.75 μg). All disks were obtained from Oxoid Ltd. (Basingstoke, Hampshire, England). Bacterial strains were suspended in sterile 0.85% saline to a turbidity to match a McFarland No. 2 standard (bioMerieux S.A.), diluted 1:20, and streaked on the used media. Plates were incubated for 24–48 h at 22 °C and E. coli ATCC 25922 was used as the control strain. Characterization of isolates as resistant was stated according to standards suggested by Miranda and Rojas [51].
Statistical Analysis
The analysis of the relative gene expression was conducted following the method described by Pfaffl [52]. A 5% level of significance was used for all statistical tests (P < 0.05). Statistical analysis was conducted using the Relative Expression Software Tool (REST©). The Pearson correlation test was used to find any correlation between load bacteria and the respective immune gene expression.
Results
Isolated Intestinal Microbiota of Seriola lalandi
From the intestinal content samples of 12 specimens of S. lalandi, 388 bacterial isolates were obtained, of which 59% corresponded to bacteria of phylum Proteobacteria, 16% to Firmicutes, 13% to Actinobacteria, and 10% to Bacteroidetes. The genera with the highest number of isolates were Alcaligenes (18%), Myroides (9%), Microbacterium (6%), and Pseudochrobactrum (5%). Among the genera of interest, Pseudomonas, Shewanella, Psychrobacter, and Actinobacteria, 39 isolates were obtained.
Antimicrobial Activity
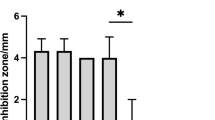
Direct antimicrobial activity of the selected isolates was carried out against three isolates obtained from outbreaks of mortality of fish in culture; these isolates were confirmed as Vibrio sp. Of the 39 isolates evaluated, nearly 50%, i.e., 19 isolates, registered growth inhibition of Vibrio sp. as shown in Table 2.
Relative Expression of Immune-Related Genes
First, the cytopathic effect of the 19 bacterial isolates selected above was evaluated for their antimicrobial activity against strains of Vibrio sp. using a multiplicity of infection (MOI) equal to 1. After 24 h post-infection (hpi), no cytopathic effect was observed in the monolayer on the part of the isolates evaluated. Subsequently, stimulation tests were performed up to 12 and 24 hpi. The results show that the expression of the evaluated genes is higher at 24 hpi than at 12 hpi. For example, at 24 hpi, citokynes (IL-8, TNFα, and IL-lβ) increased almost 30 times the expression observed at 12 hpi; the same tendency was observed in effectors such as (SAA), hepcidin and lysozyme, reaching 3 or 5 more expression (Fig. 1; Supplementary material). Different gene expression was also observed among bacterial genera. The isolates of the genera Pseudomonas and Shewanella present higher levels of expression at 12 and 24 hpi, reaching expression levels over 200–500 times the control, especially in citokynes. Isolates with higher expression levels were P21, P19, P14, P11, and P25 in the case of Pseudomonas (Fig. 1a), like those shown by the two isolates of Shewanella (Fig. 1b; Supplementary material), with expression levels over 100 times the control. The expression levels generated by the isolates of Psychrobacter and Acinetobacter (Fig. 1c, d) are lower than those previously described, with the exception of P29, P16, and P17, which were all significant with respect to the control, with the exception of P15 for the IL-1b and Hep genes (Fig. 1c). In addition, it was determined by Pearson correlation that the expression of the evaluated genes, although not significant, is associated with the bacterial load (Table 3) and has a positive correlation after 12 h of exposure (r 0.3–0.4). After 24 hpi, the correlation is positive (r 0.7–0.85) and significant (P < 0.001).
Enzymatic Activity
Nineteen enzymatic activities were tested with the API ZYM system. Some activities were shown by all or most strains and at high levels of esterase (C4), esterase lipase (C8), Leu-arylamidase, and naphthol-AS-BI-phosphohydrolase, highlighting the isolates of genus Shewanella that also showed high levels of activity of alkaline phosphatase, α-chymotrypsin, and acid phosphatase. In contrast, other activities such as those of α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mamnosidase, and α-fucosidase were detected at low levels and only in the case of isolates of Shewanella showed high to weak activity of α-chymotrypsin and N-acetyl-β-glucosaminidase. These results are shown in Table S1 (supplementary material).
Resistance of Antimicrobial Agents
The evaluated isolates belonging to the genera Shewanella, Psychrobacter, and Acinetobacter were susceptible to all the antimicrobial agents evaluated. On the other hand, among the isolates of the genus Pseudomonas, only two were susceptible to all the antimicrobials evaluated. Isolates P19, P14, P21, P11, P25, and P35 showed resistance to florfenicol; in addition, P21 and P11 showed resistance to sulfamethoxazole–trimethoprim as shown in Table 4.
Discussion
The microbiota plays an important role in the maintenance of the intestinal mucosal barrier, avoiding colonization by pathogenic microorganisms and contributing to immune homeostasis, this given by a close relationship between the microbiota and the host. In this way, the microbiota of the same species is presented as a source of probiotic potentials in aquaculture species. In the present study, 19 of 39 isolates evaluated showed antimicrobial activity against strains of Vibrio sp. derived from mortality outbreaks in aquaculture fish. It has been previously described that isolates of the genus Psychrobacter have an in vitro antagonistic effect against several pathogenic species, such as Staphylococcus aureus, Vibrio harveyi, V. metschnikovii, and V. alginolyticus [53]; V. anguillarum; and Aeromonas salmonicida [26]. The literature also describes a similar behaviour for some strains of Pseudomonas against V. anguillarum [14, 15]. On the other hand, strains of Acinetobacter have been described with antagonistic activity against pathogens such as Vibrio cholera, Flavobacterium sp., and A. hydrophila [54]. In the case of Shewanella, activity has been described against V. anguillarum, A. salmonicida [26], and Vibrio harveyi [37]. The antimicrobial activity produced by these strains could be attributed to the production of antimicrobial substances such as organic acids, bacteriocins, siderophores, and/or hydrogen peroxide, which have been described as mechanisms of pathogen inhibition [55]. For example, the study by Gram et al. [14] showed that the antimicrobial activity of Pseudomonas fluorescens against V. anguillarum is associated with the production of siderophores, similar to that described by Spanggaard et al. [15]. The antimicrobial activity of these strains against Vibrio can be considered relevant since vibriosis has been reported to generate significant mortality in marine fish cultures [7].
In this study, the levels of relative expression of genes associated with the innate immune system, by the isolates, revealed a positive tendency with respect to the bacterial load. This was corroborated by performing the Pearson correlation analysis, showing a positive (r between 0.70 and 0.85) and significant correlation (P < 0.001) after 24 hpi, demonstrating that the higher the bacterial load, the greater the relative expression of the genes. This reflects the need to evaluate the administered dose in the potential use as probiotics. Among the genes analyzed in this study, cytokines have been considered by various authors as a criterion for evaluating probiotic potential in fish [18, 19, 56,57,58]. The stimulation of cytokines initiates the response cascade of the immune system, where TNFα and IL-lβ play an essential role in the activation of prostaglandin, leukotriene, nitric oxide, and other cytokines, such as IL-6 and IL-8 [59]. IL-8 is produced mainly by macrophages, epithelial cells, and endothelial cells and acts as a neutrophil chemotactic factor that induces mostly the migration of neutrophils and other granulocytes to the site of an infection, thereby inducing phagocytosis [60]. In this study, the isolates evaluated showed significant overexpression compared to the control group after 24 h of exposure of the genes associated with cytokines and chemokine, with the exception of an isolate of the genus Psychrobacter (P15). Previously, Beck et al. [18] reported that the administration of a mix of autochthonous bacteria exerted significant stimulation of IL-8 and TNFα in addition to the substantial increase in survival of Paralichthys olivaceus challenged with Streptococcus iniae. On the other hand, Giri et al. [61] used heat-killed whole-cell products (HKWCP) from a probiotic strain of Pseudomonas aeruginosa (VSG2), demonstrating that it stimulates cytokine responses in head kidney macrophages (HK) of Labeo rohita. This report proposes a form of administration of beneficial bacteria that could be evaluated for the improvement of the Seriola culture. Previously, Lazado et al. [17] reported that strains of Pseudomonas (GP21) and Psychrobacter (GP12) stimulate the expression of IL-1β and IL-8 in head kidney leukocytes (HK) of Gadus morhua. Recent reports of Mohammadian et al. [19, 20] show that the administration of autochthonous probiotics generates significant stimulation of cytokines in the head kidney of Tor grypus. Also, it shows that probiotic strains generate significant decrease in mortality associated with infection with Aeromonas hydrophila [19].
In the early response of the innate immune system, an essential component is the serum amyloid A protein (SAA). It binds to gram-negative bacteria; acts as an opsonin; and, therefore, improves phagocytosis [62]. In addition, Bayne and Gerwick [59] describe that their function could help repair tissue damage. In this study, the 19 isolates evaluated showed significant stimulation of the SAA gene after 24 h of exposure. Given the described functionality of this protein, the stimulation of this gene by the evaluated isolates could be beneficial when faced with an infection with pathogenic bacteria. Previously, it has been described that SAA is strongly regulated during septicemia in Salvelinus alpinus [63]. Similar to that described by Kania et al. [64] demonstrating that the increase in the expression of the SAA gene in O. mykiss is directly related to the bacterial load post-infection, showing a positive Pearson correlation (r = 0.974; P < 0.01), similar to the results obtained in this study.
Another important component of host defense is the production of antimicrobial peptides (AMP). Recently, Muncaster et al. [65] characterized two antimicrobial peptides in S. lalandi, piscidin and hepcidin, describing that the latter has higher expression in the liver, with respect to spleen and gills. Álvarez et al. [66] describe the effect of synthetic hepcidin as a protector against a challenge with V. anguillarum in Dicentrarchus labrax. The cumulative mortality of D. labrax infected with V. anguillarum was 72.5% until day 7 post-injection; for its part in fish previously injected with hepcidin (Hep1), the cumulative mortality was 23.5%. Previously, Álvarez et al. [67] described that synthetic hepcidin presents antimicrobial activity against Piscirickettsia salmonis. On the other hand, Jiang et al. [68] showed that stimulation in hepcidin production prior to a challenge with pathogenic bacteria such as A. hydrophila and V. alginolyticus, increases the survival of challenged fish, from 46.7 to 76%. These authors propose a cooperative effect between the antibacterial and regulatory activities of iron, by hepcidin in the innate immune defense against bacterial infection. Given that the increase in hepcidin decreases the availability of iron in the serum, this is a major element required for bacterial growth. Among the isolates of Seriola evaluated, the isolates of the genera Pseudomonas and Shewanella emerged due to the robust induction in the expression of hepcidin, and therefore, they have advantages as candidates for probiotics for S. lalandi.
Several studies evaluating probiotic potential in fish have investigated the activity of serum lysozyme. Meidong et al. [69] reported a significant increase in serum lysozyme activity in Pangasius bocourti after administration of a Bacillus strain in the diet. Similar results have been published in different species such as Catla catla [70, 71], Labeo rohita [72, 73], Cyprinus carpio [74], Oreochromis niloticus [75], and O. mykiss [76]. The importance of the increase in serum lysozyme has to do with the bactericidal activity. Meidong et al. [69] reported this activity against A. hydrophila to be approximately 50% after 60 days of administration with a Bacillus strain. Notably, in this study, the activity of serum lysozyme was not evaluated, but the relative expression of this gene was evaluated, and the relative expression values were significant with respect to the control by the 19 isolates evaluated. In this sense, Caipang et al. [26] observed that the expression of lysozyme in the head kidney of Gadus morhua increased in the presence of a strain of Psychrobacter (GP11). Subsequently, Muñoz-Atienza et al. [57] described that the administration of a Weissella strain significantly regulates the expression of the lysozyme gene in the skin and intestine in addition to substantially decreasing mortality accumulated in Scophthalmus maximus larvae. In this study, the isolates that generated higher levels of lysozyme expression were those of the genus Pseudomonas and Shewanella, and those that generated lower levels belonged to Psychrobacter and Acinetobacter.
Our results show that the isolates evaluated, especially those of the genera Pseudomonas and Shewanella, could be beneficial for the host, helping in the response to Vibrio sp. infections. However, considering another judgment in this evaluation, such as resistance to antibiotics, it is observed that six of the eight isolates belonging to the genus Pseudomonas have resistance to florfenicol and/or sulfamethoxazole-trimethoprim. It has been described that Pseudomonas species harbor multiple intrinsic and acquired resistance genes [77, 78]. Intrinsic resistance is mainly conferred by mechanisms such as low permeability of the outer membrane, synthesis of β-lactamases and efflux systems. In addition, due to the plasticity of the genome, it is believed that members of Pseudomonas sp. can acquire almost all known mechanisms of resistance to antimicrobials [79]. Although this study was not evaluated at the level of genes associated with antimicrobial resistance, it has been described that resistance could be associated with mobile elements such as integrons, plasmids, and transposons, so that these would have the ability to transfer this resistance to other bacteria of the microbiota; therefore, isolates of this genus would be discarded as candidates for probiotics. In the case of sulfamethoxazole–trimethoprim resistance, Shin et al. [80] describe three genes associated with resistance to sulfonamides, sul1, sul2, and sul3. Of which, the sul1 gene is linked to other resistance genes in class 1 integrons and in large conjugative plasmids. On the other hand, sul2 is generally located in small plasmids, also in large transmissible multiresistant plasmids or through a common region of insertion elements (ISCR2) and sul3, a plasmid-borne sulfonamide resistance gene. For its part, resistance to trimethoprim is associated with the dfr gene, of which 30 different genes have been described, which are generally found in gene cassettes within integrons. In the case of resistance associated with florfenicol, three genes associated with the resistance of florfenicol, cfr, fexA, and floR, have been described, of which floR has been reported in several gram-negative bacteria [81]. The presence of these genes has been described in mobile elements such as the floR gene in plasmids [82] and transposons [81], similar to that described for the fexA and cfr genes [83].
According to the results obtained, 10 of the isolates of the genera Shewanella, Psychrobacter, and Acinetobacter are proposed as potential probiotics in cultures of S. lalandi; notably, the two isolates of Shewanella have higher levels of enzymatic activity of alkaline phosphatase, Leu-arylamidase, α-chymotrypsin, and N-acetyl-β-glucosaminidase. The presence of alkaline phosphatase is desired in probiotic candidates because it could potentially inhibit an inflammatory response in the intestine and give an immunomodulation benefit to the host [84]. The review by Ray et al. [85] also describes the importance of the contribution to nutrition in fish of enzymes of the microbiota, including proteases, lipases, amylase, chitinase, and cellulase.
Conclusion
The present study reveals that 11 isolates of the genera Shewanella, Psychrobacter, and Acinetobacter could serve as probiotic candidates in the culture of Seriola lalandi. However, more studies should be done to investigate the adhesion properties of these potential probiotic bacteria using animal models and to corroborate these results through in vivo challenges.
References
Fowler A, Ham J, Jennings P (2003) Discriminating between cultured and wild yellowtail kingfish (Seriola lalandi) in South Australia. Publication No. RD03/0159. SARDI Aquatic Sciences Publication, Adelaide
Nakada M (2002) Yellowtail culture development and solutions for the future. Rev Fish Sci 10:559–575
Moran D, Wells RMG, Pether SJ (2008) Low stress response exhibited by juvenile yellowtail kingfish (Seriola lalandi Valenciennes) exposed to hypercapnic conditions associated with transportation. Aquac Res 39:1399–1407
Ramírez C, Romero J (2017) The microbiome of Seriola lalandi of wild and aquaculture origin reveals differences in composition and potential function. Front Microbiol 8:1–10
Poortenaar CW, Hooker SH, Sharp N (2001) Assessment of yellowtail kingfish (Seriola lalandi) reproductive physiology, as a basis for aquaculture development. Aquaculture 201:271–286
Fielder S, Heasman M (2011) Hatchery manual for the production of Australian bass, mulloway and yellowtail kingfish Industry & Investment NSW, ISBN 978 1 74256 058 8. 176 pp
Toranzo AE, Magariños B, Romalde JL (2005) A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246:37–61
Dang H, Zhang X, Song L et al (2006) Molecular characterizations of oxytetracycline resistant bacteria and their resistance genes from mariculture waters of China. Mar Pollut Bull 52:1494–1503
Miranda CD, Godoy FA, Lee MR (2018) Current status of the use of antibiotics and the antimicrobial resistance in the Chilean salmon farms. Front Microbiol 9:1–14
Pindling S, Azulai D, Zheng B et al (2018) Dysbiosis and early mortality in zebrafish larvae exposed to subclinical concentrations of streptomycin. FEMS Microbiol Lett 365:1–9
Schmidt V, Gomez-Chiarri M, Roy C et al (2017) Subtle microbiome manipulation using probiotics reduces antibiotic-associated mortality in fish. mSystems 2:1–13
He S, Wang Q, Li S et al (2017) Antibiotic growth promoter olaquindox increases pathogen susceptibility in fish by inducing gut microbiota dysbiosis. Sci China Life Sci 60:1260–1270
Wang E, Yuan Z, Wang K et al (2019) Consumption of florfenicol-medicated feed alters the composition of the channel catfish intestinal microbiota including enriching the relative abundance of opportunistic pathogens. Aquaculture 501:111–118
Gram L, Melchiorsen J, Spanggaard B et al (1999) Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl Environ Microbiol 65:969–973
Spanggaard B, Huber I, Nielsen J et al (2001) The probiotic potential against vibriosis of the indigenous microflora of rainbow trout. Environ Microbiol 3:755–765
Seghouani H, Garcia-Rangel CE, Füller J et al (2017) Walleye autochthonous bacteria as promising probiotic candidates against Flavobacterium columnare. Front Microbiol 8:1–9
Lazado CC, Caipang CMA, Gallage S et al (2010) Expression profiles of genes associated with immune response and oxidative stress in Atlantic cod, Gadus morhua head kidney leukocytes modulated by live and heat-inactivated intestinal bacteria. Comp Biochem Physiol Part B 155:249–255
Beck BR, Kim D, Jeon J et al (2015) The effects of combined dietary probiotics Lactococcus lactis BFE920 and Lactobacillus plantarum FGL0001 on innate immunity and disease resistance in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol 42:177–183
Mohammadian T, Alishahi M, Tabandeh MR et al (2017) Changes in immunity, expression of some immune-related genes of shabot fish, Tor grypus, following experimental infection with Aeromonas hydrophila: effects of autochthonous probiotics. Probiotics Antimicrob Proteins 10:616–628
Mohammadian T, Alishahi M, Tabandeh MR et al (2018) Effects of autochthonous probiotics, isolated from Tor grypus (Karaman, 1971) intestine and Lactobacillus casei (PTCC 1608) on expression of immune-related genes. Aquac Int 27:239–260
Balcázar JL, de Blas I, Ruiz-Zarzuela I et al (2006) The role of probiotics in aquaculture. Vet Microbiol 114:173–186
Zorriehzahra MJ, Delshad ST, Adel M et al (2016) Probiotics as beneficial microbes in aquaculture: an update on their multiple modes of action: a review. Vet Q 36:228–241
Verschuere L, Rombaut G, Sorgeloos P, Verstraete W (2000) Probiotic bacteria as biological control agents in aquaculture. Microbiol Mol Biol Rev 64:655–671
Kesarcodi-Watson A, Kaspar H, Lategan MJ, Gibson L (2008) Probiotics in aquaculture: the need, principles and mechanisms of action and screening processes. Aquaculture 274:1–14
Makridis P, Martins S, Reis J, Dinis MT (2008) Use of probiotic bacteria in the rearing of Senegalese sole (Solea senegalensis) larvae. Aquac Res 39:627–634
Caipang CMA, Brinchmann MF, Kiron V (2010) Antagonistic activity of bacterial isolates from intestinal microbiota of Atlantic cod, Gadus morhua, and an investigation of their immunomodulatory capabilities. Aquac Res 41:249–256
Sharifuzzaman SM, Austin B (2010) Kocuria SM1 controls vibriosis in rainbow trout (Oncorhynchus mykiss, Walbaum). J Appl Microbiol 108:2162–2170
He S, Zhang Y, Xu L et al (2013) Effects of dietary Bacillus subtilis C-3102 on the production, intestinal cytokine expression and autochthonous bacteria of hybrid tilapia Oreochromis niloticus ♂×Oreochromis aureus ♀. Aquaculture 412–413:125–130
Sun YZ, Yang HL, Huang KP et al (2013) Application of autochthonous Bacillus bioencapsulated in copepod to grouper Epinephelus coioides larvae. Aquaculture 392–395:44–50
Tapia-Paniagua ST, Vidal S, Lobo C et al (2014) The treatment with the probiotic Shewanella putrefaciens Pdp11 of specimens of Solea senegalensis exposed to high stocking densities to enhance their resistance to disease. Fish Shellfish Immunol 41:209–221
Giri SS, Sukumaran V, Sen SS, Jena PK (2014) Effects of dietary supplementation of potential probiotic Bacillus subtilis VSG1 singularly or in combination with Lactobacillus plantarum VSG3 or/and Pseudomonas aeruginosa VSG2 on the growth, immunity and disease resistance of Labeo rohita. Aquac Nutr 20:163–171
Nayak SK (2010) Role of gastrointestinal microbiota in fish. Aquac Res 41:1553–1573
Mills S, Stanton C, Fitzgerald GF, Ross RP (2011) Enhancing the stress responses of probiotics for a lifestyle from gut to product and back again. Microb Cell Factories 10:S19
Romero J, Navarrete P (2014) Marine vertebrate animal metagenomics, Salmonidae: the microbiota of the digestive tract of salmonids. In: Highlander SK, Rodriguez-Valera F, White BA (eds) Encyclopedia of metagenomics. Springer, Boston, pp 1–7
Berg R (1996) The indigenous gastrointestinal microflora. Trends Microbiol 4:430–435
Rawls JF, Samuel BS, Gordon JI (2004) Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A 101:4596–4601
Chabrillón M, Rico RM, Arijo S et al (2005) Interactions of microorganisms isolated from gilthead sea bream, Sparus aurata L., on Vibrio harveyi, a pathogen of farmed Senegalese sole, Solea senegalensis (Kaup). J Fish Dis 28:531–537
Dethlefsen L, McFall-Ngai M, Relman DA (2007) An ecological and evolutionary perspective on humang-microbe mutualism and disease. Nature 449:811–818
Ley RE, Lozupone CA, Hamady M et al (2009) Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788
Hooper LV, Littman DR, Macpherson AJ (2012) Interactions between the microbiota and the immune system. Science 336(80):1268–1273
Rosshart SP, Vassallo BG, Angeletti D et al (2017) Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171:1015–1028
Bacanu GM, Lucian O (2013) Differences in the gut microbiota between wild and domestic Acipenser ruthenus evaluated by denaturing gradient gel electrophoresis. Rom Biotechnol Lett 18:8069–8076
Kim D-H, Kim D (2013) Microbial diversity in the intestine of olive flounder (Paralichthys olivaceus). Aquaculture 414–15:103–108
Dhanasiri AKS, Brunvold L, Brinchmann MF et al (2011) Changes in the intestinal microbiota of wild Atlantic cod Gadus morhua l. upon captive rearing. Microb Ecol 61:20–30
Ramírez C, Romero J (2017) Fine flounder (Paralichthys adspersus) microbiome showed important differences between wild and reared specimens. Front Microbiol 8:1–12
Dopazo CP, Lemos ML, Lodeiros C et al (1988) Inhibitory activity of antibiotic producing marine bacteria against fish pathogens. J Appl Bacteriol 65:97–101
Leyton Y, Riquelme C (2010) Marine Bacillus spp. associated with the egg capsule of Concholepas concholepas (common name “loco”) have an inhibitory activity toward the pathogen Vibrio parahaemolyticus. Microb Ecol 60:599–605
Dannevig B, Brudeseth B, Gjoen T et al (1997) Characterisation of a long-term cell line (SHK-1) developed from the head kidney of Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol 7:213–226
Vourc’h M, Roquilly A, Broquet A et al (2017) Exoenzyme T plays a pivotal role in the IFN-γ production after Pseudomonas challenge in IL-12 primed natural killer cells. Front Immunol 8:2–10
CLSI (2006) Methods for antimicrobial disk susceptibility testing of bacteria isolated from aquatic animals, approved guideline M42-A. Clinical and Laboratory Standards Institute, Wayne
Miranda CD, Rojas R (2007) Occurrence of florfenicol resistance in bacteria associated with two Chilean salmon farms with different history of antibacterial usage. Aquaculture 266:39–46
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Sun Y, Yang H, Ling Z et al (2009) Gut microbiota of fast and slow growing grouper. Afr J Microbiol Res 3:713–720
Balakrishna A, Keerthi TR (2012) Screening of potential aquatic probiotics from the major microflora of guppies (Poecilia reticulata). Front Chem Sci Eng 6:163–173
Hoseinifar SH, Sun YZ, Wang A, Zhou Z (2018) Probiotics as means of diseases control in aquaculture, a review of current knowledge and future perspectives. Front Microbiol 9:1–18
Lazado CC, Caipang CMA (2014) Mucosal immunity and probiotics in fish. Fish Shellfish Immunol 39:78–89
Muñoz-Atienza E, Araújo C, Magadán S et al (2014) In vitro and in vivo evaluation of lactic acid bacteria of aquatic origin as probiotics for turbot (Scophthalmus maximus L.) farming. Fish Shellfish Immunol 41:570–580
Cordero H, Morcillo P, Meseguer J et al (2016) Effects of Shewanella putrefaciens on innate immunity and cytokine expression profile upon high stocking density of gilthead seabream specimens. Fish Shellfish Immunol 51:33–40
Bayne CJ, Gerwick L (2001) The acute phase response and innate immunity of fish. Dev Comp Immunol 25:725–743
Mukaida N, Shiroo M, Matsushima K (1989) Genomic structure of the human monocyte-derived neutrophil chemotactic factor IL-8. J Immunol 143:1366–1371
Giri SS, Sen SS, Jun JW et al (2016) Heat-killed whole-cell products of the probiotic Pseudomonas aeruginosa VSG2 strain affect in vitro cytokine expression in head kidney macrophages of Labeo rohita. Fish Shellfish Immunol 50:310–316
Shah C, Hari-dass R, Raynes JG (2006) Serum amyloid A is an innate immune opsonin for Gram-negative bacteria. Blood 108:1–34
Jensen L, Hiney M, Shields D et al (1997) Acute phase proteins in salmonids. Evolutionary analyses and acute phase response. J Immunol 158:384–392
Kania PW, Chettri JK, Buchmann K (2014) Characterization of serum amyloid A (SAA) in rainbow trout using a new monoclonal antibody. Fish Shellfish Immunol 40:648–658
Muncaster S, Kraakman K, Gibbons O et al (2018) Antimicrobial peptides within the yellowtail kingfish (Seriola lalandi). Dev Comp Immunol 80:67–80
Álvarez CA, Acosta F, Montero D et al (2016) Synthetic hepcidin from fish: uptake and protection against Vibrio anguillarum in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol 55:662–670
Álvarez CA, Guzmán F, Cárdenas C et al (2014) Antimicrobial activity of trout hepcidin. Fish Shellfish Immunol 41:93–101
Jiang XF, Liu ZF, Lin AF et al (2017) Coordination of bactericidal and iron regulatory functions of hepcidin in innate antimicrobial immunity in a zebrafish model. Sci Rep 7:1–15
Meidong R, Khotchanalekha K, Doolgindachbaporn S et al (2018) Evaluation of probiotic Bacillus aerius B81e isolated from healthy hybrid catfish on growth, disease resistance and innate immunity of Pla-mong Pangasius bocourti. Fish Shellfish Immunol 73:1–10
Das A, Nakhro K, Chowdhury S, Kamilya D (2013) Effects of potential probiotic Bacillus amyloliquifaciens FPTB16 on systemic and cutaneous mucosal immune responses and disease resistance of catla (Catla catla). Fish Shellfish Immunol 35:1547–1553
Sangma T, Kamilya D (2015) Dietary Bacillus subtilis FPTB13 and chitin, single or combined, modulate systemic and cutaneous mucosal immunity and resistance of catla, Catla catla (Hamilton) against edwardsiellosis. Comp Immunol Microbiol Infect Dis 43:8–15
Giri SS, Sen SS, Sukumaran V (2012) Effects of dietary supplementation of potential probiotic Pseudomonas aeruginosa VSG-2 on the innate immunity and disease resistance of tropical freshwater fish, Labeo rohita. Fish Shellfish Immunol 32:1135–1140
Ramesh D, Vinothkanna A, Rai AK, Vignesh VS (2015) Isolation of potential probiotic Bacillus spp. and assessment of their subcellular components to induce immune responses in Labeo rohita against Aeromonas hydrophila. Fish Shellfish Immunol 45:268–276
Lin B, Chen S, Cao Z et al (2007) Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: striking similarities and obvious differences with mammals. Mol Immunol 44:295–301
Makled S, Hamdan A, El-Sayed A-F, Hafez E (2017) Evaluation of marine psychrophile, Psychrobacter namhaensis SO89, as a probiotic in Nile tilapia (Oreochromis niloticus) diets. Fish Shellfish Immunol 61:194–200
Ramos MA, Gonçalves JFM, Batista S et al (2015) Growth, immune responses and intestinal morphology of rainbow trout (Oncorhynchus mykiss) supplemented with commercial probiotics. Fish Shellfish Immunol 45:19–26
Pfeifer Y, Cullik A, Witte W (2010) Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 300:371–379
Kittinger C, Lipp M, Baumert R et al (2016) Antibiotic resistance patterns of Pseudomonas spp. isolated from the river Danube. Front Microbiol 7:1–8
Livermore DM (2002) Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640
Shin HW, Lim J, Kim S et al (2014) Characterization of trimethoprim-sulfamethoxazole resistance genes and their relatedness to class 1 integron and insertion sequence common region in gram-negative bacilli. J Microbiol Biotechnol 25:137–142
Doublet B, Schwarz S, Kehrenberg C, Cloeckaert A (2005) Florfenicol resistance gene floR is part of a novel transposon. Antimicrob Agents Chemother 49:2106–2108
Gordon L, Cloeckaert A, Doublet B et al (2008) Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J Antimicrob Chemother 62:65–71
Kehrenberg C, Schwarz S (2006) Distribution of florfenicol resistance genes fexA and cfr among chloramphenicol-resistant Staphylococcus isolates. Antimicrob Agents Chemother 50:1156–1163
Whitehead J (2009) Intestinal alkaline phosphatase: the molecular link between rosacea and gastrointestinal disease? Med Hypotheses 73:1019–1022
Ray AK, Ghosh K, Ringø E (2012) Enzyme-producing bacteria isolated from fish gut: a review. Aquac Nutr 18:465–492
Acknowledgments
CR would like to acknowledge the National Commission of Scientific and Technologic Research (CONICYT) for the funding through the National PhD funding program.
Funding
This project was funded by a research grant from FONDECYT 1140734 and 1171129 (Fondo Nacional de Desarrollo Científico y Tecnológico, Chile) and supported by project Aquapacifico 15PCTI-46284 from Corfo.
Author information
Authors and Affiliations
Contributions
CR performed microbial isolation, DNA and RNA extraction, and data analysis and interpretation and wrote the original draft. RR performed the antimicrobial activity and antibiotic resistance studies and reviewed and edited. JR designed the work and reviewed and edited manuscript. All authors have made intellectual contributions to the work and approved it for publication.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(XLSX 26 kb)
Rights and permissions
About this article
Cite this article
Ramírez, C., Rojas, R. & Romero, J. Partial Evaluation of Autochthonous Probiotic Potential of the Gut Microbiota of Seriola lalandi. Probiotics & Antimicro. Prot. 12, 672–682 (2020). https://doi.org/10.1007/s12602-019-09550-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-019-09550-9