Abstract
Bemisia tabaci is an important agriculture pests and vector of viruses. The MEAM1 species of B. tabaci, first described in Brazil in the 90s is now the most prevalent species and primary cause of the emergence of begomoviruses in tomatoes. The Mediterranean species (MED) was recently detected in Brazil and is a new concern for Brazilian agriculture. The potential impact of this species as a vector of economically important virus in Brazil is unknown. We therefore evaluated the ability of MED to transmit four whitefly transmitted viruses prevalent in Brazil, Cowpea mild mottle virus (CpMMV, carlavirus), Bean golden mosaic virus (BGMV, begomovirus) infecting beans; and the Tomato severe rugose virus (ToSRV, begomovirus), Tomato chlorosis virus (ToCV, crinivirus) infecting tomatoes. The colony of MED harbouring the secondary endosymbionts was tested: 14% positive for Hamiltonella and 29% positive for Rickettsia. After six months being maintained on cotton plants, this colony changed the frequency of endosymbionts (97% of Hamiltonella and 1% of Rickettsia) and was denominated as MEDH. Additionally, a colony of MEAM1 (98% positive for Hamiltonella and 91% positive for Rickettsia) was also tested. The viruses were efficiently transmitted by MED, but transmission efficiency varied among the MED and MEDH, being CpMMV, BGMV and ToCV better transmitted by MEDH. Moreover, transmission efficiency of ToSRV and ToCV by MEDH was even significantly better than MEAM1. We conclude that specimens from B. tabaci MED are good vectors of virus infecting tomato and beans in Brazil and populations with Hamiltonella prevalence increased the virus transmission.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is a phloem–feeding insect that inflicts a serious worldwide threat by direct feeding on plants and by the transmission of more than 300 plant virus species (Gilbertson et al. 2015). Bemisia tabaci transmits viruses of the genera Begomovirus, Crinivirus, Carlavirus, Ipomovirus and Torradovirus (Navas-Castillo et al. 2011), with the viruses of the genus Begomovirus highlighted, representing 90% of the viruses transmitted by whiteflies.
Bemisia tabaci is considered as a cryptic species complex (Boykin and De Barro 2014; De Barro et al. 2011; Dinsdale et al. 2010). In Brazil, only four species of the complex are reported to date: the indigenous species from the New World group, New World 1 (NW1) and New World 2 (NW2) (Marubayashi et al. 2013), and the exotic invasive species Middle East-Asia Minor 1 (MEAM1, also called B biotype) and Mediterranean (MED, also called Q biotype) (Barbosa et al. 2015). Among the species of this complex, MEAM1 and MED are the most important worldwide (Hadjistylli et al. 2016). The MEAM1 species was first recorded in Brazil in the early 1990’s (Lourencao and Nagai 1994) and is now the primary vector of plant viruses in the country (Ribeiro et al. 1998; Inoue-Nagata et al. 2016).
The Mediterranean species was first reported in Brazil in Rio Grande do Sul State in 2014 (Barbosa et al. 2015) and more recently in the states of Santa Catarina, Paraná, São Paulo and Minas Gerais (Moraes et al. 2017; Moraes et al. 2018). The dispersion of MED is continuing in Brazil and raising great concerns for farmers across the country mainly because this whitefly species is associated with low susceptibility to insecticides which hinders control (Horowitz et al. 2005; Yao et al. 2017).
Begomoviruses infecting vegetables and weeds are typically native to Brazil (Barreto et al. 2013), so the potential of MED as a vector of these viruses is unknown. Moreover, populations of B. tabaci harbour different symbiotic bacteria compositions, in which some endosymbionts are reported to enhance and reduce virus transmission capability. According to Ghosh et al. 2018, the Arsenophonus endosymbionts has negative effect in East African cassava mosaic virus transmission by B. tabaci. However, the endosymbiont Rickettisia in B. tabaci was reported to increase Tomato yellow leaf curl virus (TYLCV) transmission (Kliot et al. 2014), as well, Hamiltonella has been identified as responsible for high TYLCV transmission efficiency (Gottlieb et al. 2010; Su et al. 2013). Populations of B. tabaci MEAM1 in Brazil has been reported with high frequency of Hamiltonella and Rickettsia, while for MED different sets and frequency were observed (Moraes et al. 2018; Marubayashi et al. 2014), but there is not studies indicating the role of specific endosymbionts on virus transmission.
Therefore, the goal of this study was to evaluate the ability of MED with low and high Hamiltonella frequency to transmit the begomovirus, Bean golden mosaic virus (BGMV) and the carlavirus, Cowpea mild mottle virus (CPMMV) to beans, this later an emergent virus infecting beans and soybean in Brazil (Faria et al. 2016; Zanardo et al. 2014). We also tested the transmission of Tomato severe rugose virus (ToSRV), that is the prevalent begomovirus species infecting tomato in Brazil and the crinivirus Tomato chlorosis virus (ToCV), frequently found on mixed infection with ToSRV (Macedo et al. 2014). Viruses were tested in single and mixed infections.
Materials and methods
Establishing whitefly populations
For the transmission assays, one colony of MEAM1 and one colony of MED were established. The whiteflies were collected in the field and separately reared on cotton plants in controlled conditions (26 °C, 12 L:12D, light 06:00–18:00). MEAM1 was collected in Campinas/SP (Brassica oleracea) (22°52′14”S, 47°04′38”W) in 2015, and MED (Begonia spp.; 23°22′20”S, 46°10′35”W) was collected in Santa Isabel/SP in 2015.
For whitefly species identification, the DNA of adult whiteflies was extracted using the Chelex protocol (Walsh et al. 1991) and used as a template for a PCR with C1-J-2195 and TL2-N-3014 primers (Simon et al. 1994) that amplified the partial mtCOI fragment of B. tabaci followed by restriction fragment length polymorphism (RFLP) analysis of the amplicons utilizing the TaqI enzyme (Bosco et al. 2006). The mtCOI amplicons were then purified, sequenced and analysed using a global, curated dataset of mitochondrial COI (Boykin and De Barro 2014) to confirm the species.
Characterization of endosymbionts
For the characterization of endosymbionts, 100 insects were analysed per colony. The same DNA was used for the screening of Portiera aleyrodidarum and the six secondary endosymbionts Hamiltonella, Rickettsia, Wolbachia, Arsenophonus, Cardinium and Fritschea using genus-specific primers targeting the 16S or 23S rDNA gene. PCR cycling was performed as described by (Marubayashi et al. 2014). To confirm endosymbiont presence, the amplified sequences from representative individuals were sequenced. The analysis was repeated every six months to check the endosymbiont frequency. Colonies of MEAM1 an MED were maintained on cotton plants.
Transmission assays
Single-infected isolates were assessed for the viruses: BGMV, CpMMV, ToSRV and ToCV. Virus sources with mixed infections were also tested (BGMV + CpMMV and ToSRV + ToCV). CpMMV and BGMV were maintained in common bean cv. Jalo and ToSRV and ToCV were maintained in tomato plants of cv. Mariana, in whitefly-proof screened cages (Fig. 1).
Virus acquisition was performed by transferring whitefly MEAM1 and MED species with different endosymbiont frequency and compositions (MEDH) to cages containing plants infected or coinfected with the specific virus/es, for an acquisition access period (AAP) of 24 h. Following virus acquisition, whiteflies (10 adults per plant) were transferred to cages containing healthy tomato plants cv. Mariana with 3–4 true-leafs or beans cv. Jalo with the primary leaves-true, using a hand-held aspirator on which they remained for a 24 h inoculation access periods (IAPs) under controlled conditions at 30 °C. Thirty plants for each treatment were individually inoculated in separate cages. Following inoculation, the plants were sprayed with insecticides (Cartap® and Oberon®) to kill all the whitefly adults, nymphs and eggs. Plants were grown in whitefly-proof screened cages in greenhouses. Virus-free adult whiteflies collected from either rearing cages (MEAM1 and MED) were given inoculation access to 10 non-infected plants for 24 h at 30 °C as negative controls. Negative control plants for each treatment were also sprayed with insecticides at the same intervals as those in the assays and the presence of virus analysed at 30 days after the inoculation.
Virus detection
Thirty days after the IAP, plants were analysed for the presence of viruses. The begomoviruses (ToSRV and BGMV) were detected by DNA extraction (Dellaporta et al. 1983) and PCR using the degenerated primer set PAL1v1978/PAR1c496 (Rojas et al. 1993). For detection of ToCV, total RNA was extracted with Trizol® (Invitrogen) and submitted to a RT-PCR using primers HS-11/HS-12 (Dovas et al. 2002), which amplified a conserved region of a heat shock protein (HSP-70). The RT-PCR was used to make a Nested-PCR using ToC-5/ToC-6 specific primers of ToCV (Dovas et al. 2002),. CpMMV detection was conducted biologically by sap transmission into a susceptible plant (soybean BRS-132) followed by visualization of the symptoms. Plants showing symptoms were selected for RNA extraction with a “Total RNA Purification Kit (NORGEN)” followed by a RT-PCR One Step using AMV@ reverse transcriptase (Promega, Brazil) with the specific primers CpMMV 1280-F/1696-R (De Marchi et al. 2017).
Statistical analyses
Dates of frequency of endosymbionts and virus transmission efficiency by B. tabaci MEAM1 and MED were analyzed by chi-square test (p < 0.01) using a generalized linear model (GLM) with log link functions or just linear model. The analysis was performed using the software package R 3.1.0 (RDevelopment C. 2018).
Results
Whitefly colonies
Two colonies harbouring different endosymbiont compositions were established: the MEAM1 colony with 98% of individuals testing positive for Hamiltonella and 91% positive for Rickettsia and MED starting colony with 14% of individuals testing positive for Hamiltonella and 29% positive for Rickettsia. The endosymbionts constitution of MEAM1 and MED populations was re-evaluated after six months and one year from the beginning of the experiments, and an expressive increase of Hamiltonella (98%) and a decrease of Rickettsia (1%) in MED population was verified from the sixth month (Table 1). This population was named as MEDH.
Transmission assays for bean
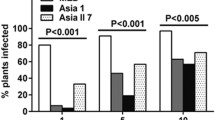
The transmission of the viruses changed significantly according to the colony of B. tabaci tested. In single infection, the MED population (14% of Hamiltonella and 29% of Rickettsia frequency) transmitted BGMV with 53.3% efficiency (16 positive out of 30 tested) while MEAM1 (98% of Hamiltonella and 91% of Rickettsia) with 93.3% efficiency (28 positive out of 30 tested). MEDH (98% of Hamiltonella and 1% of Rickettsia) transmitted BGMV with 100% of efficiency. CpMMV, in single infection was transmitted with 56.6% and 90% efficiency by MED and MEAM1, respectively, and an increase in CpMMV transmission (56.6 to 96.6%) was observed for MEDH. In mixed infection of BGMV and CpMMV, the MEAM1 colony transmitted both viruses with 100% of efficiency, while MED with 96.6% the CpMMV and 26.6% the BGMV, respectively. Population of MEDH was not tested for mixed infection of BGMV+CpMMV.
Transmission assays for tomato
In single-infection, the crinivirus ToCV and the begomovirus ToSRV were efficiently transmitted to tomatoes by the populations tested. Comparing MED and MEDH, an increasing in ToCV transmission could be observed (83.3% to 96.6%), respectively. An isolate of ToSRV in single infection was not available at the beginning of the experiments, so it was not possible to test this virus for MED colony. ToCV and ToSRV in mixed infection were more efficiently transmitted by MEAM1 than MED (Table 2).
Discussion
Our results indicate that the recently introduced B. tabaci Meditterranean species can be an important vector of viruses infecting tomato and beans in Brazil. Additionally, in this study, the frequency and composition of endosymbionts changed with the maintenance of the MED population on a specific host (cotton) and in controlled conditions (Table 1), as was also observed by Su et al. (2013). In our case, an expressive increase of Hamiltonella (from 14% to 98%) and a decrease of Rickettsia (29% to 1%) was observed. This population then called MEDH, transmitted ToCV to tomatoes, BGMV and CpMMV to beans better than the original population and also proved to be an excellent vector of ToSRV to tomatoes.
In Brazil, the variability of the secondary endosymbionts found in MED is very large, possibly explained by the recent and different introductions of the species into the country (Moraes et al. 2017, 2018). MED populations worldwide commonly harbour Hamiltonella, Rickettsia, Cardinium, Wolbachia and Arsenophonus while Hamiltonella and Rickettsia are commonly found in populations of MEAM1 (Gueguen et al. 2010; Czosnek and Ghanim 2016). Brazilian populations of MEAM1 presented high frequency of Hamiltonella and Rickettsia (Marubayashi et al. 2014). The colony of MEAM1 established in this work (98% positive for Hamiltonella and 91% for Rickettsia) was considered an excellent vector of the virus tested.
Bemisia tabaci Mediterranean species is known as an efficient vector of TYLCV, the most destructive begomovirus in the world (Pan et al. 2012). The influence of endosymbiont on virus transmission is very well documented for B. tabaci MED where the absence or low frequency of the Hamilltonella results in low transmission efficiency of TYLCV to tomatoes (Su et al. 2013; Gottlieb et al. 2010). TYLCV interacts with the “heat shock protein 70” (HSP 70), and after circulation in the insect filter chamber and midgut, the virus interacts with the GroEl protein (produced by Hamiltonella) in the haemolymph and crosses into the insect primary salivary glands (Ghanim 2014; Gottlieb et al. 2010). For the viruses tested in this study, there is no information about the contribution of each specific endosymbiont on virus transmission. Here we have evidence that frequency of Hamiltonella on MED populations could also contribute for transmission efficiency of BGMV, CpMMV and ToCV, but further tests should be performed to understand the influence of the endosymbiont Hamiltonella for each virus.
In Brazil, the B. tabaci MEAM1 is still the predominant species (Moraes et al. 2018; Marubayashi et al. 2014, 2013), however, there has been a notable shift in the whitefly dynamics since the first detection of the MED species and it dispersion to others regions of the country (Moraes et al. 2018, 2017; Barbosa et al. 2015). The Mediterranean B. tabaci species is less susceptible to insecticides compared to MEAM1 (Ghanim and Kontsedalov 2009; Horowitz et al. 2005). Additionally, the ability of MED to colonize bell pepper (Capsicum annuum) is significantly higher than that of MEAM1 (Sun et al. 2013). The combination of host and high insecticide use can lead for the displacement of indigenous and also the invasive MEAM1 species (Sun et al. 2013).
Furthermore, the emergence of new virus diseases in several regions can be related to an introduced species of whitefly. Until the 1990’s, only indigenous species of B. tabaci of the New World group, also called A biotype, were in Brazil (Barbosa et al. 2014; Marubayashi et al. 2013). Virus-related epidemics were sporadic, generally occurring in common beans, which are preferred by native whiteflies for colonization (Costa et al. 1977). With the report of the MEAM1 species in the early 1990’s (Lourencao and Nagai 1994), outbreaks of begomoviruses infecting tomatoes occurred in Brazil (Ribeiro et al. 1998) and are frequent until the present days, causing great damage to tomatoes (Inoue-Nagata et al. 2016; Macedo et al. 2014). The MEAM1 species is highly polyphagous and was the responsible for transferring native weed-virus to cultivated hosts (Navas-Castillo et al. 2011; Barreto et al. 2013).
In China the TYLCV became an emergent virus after the report of MED (Ning et al. 2015). There is evidence that the interaction between TYLCV and MED is mutually beneficial to the virus and its vector, because B. tabaci MED feeds more efficiently after acquisition of TYLCV (Moreno-Delafuente et al. 2013). Moreover, the B. tabaci MED can supress the host plant defences involving the jasmonic acid (JA) and proteinase inhibitor (PI) and TYLCV viruliferous MED whiteflies increases the longevity and fecundity of non-viruliferous whiteflies that subsequently feed on the same plant (Shi et al. 2014). In the same way, pre-infested tomato plants with viruliferous TYLCV MED whiteflies, but not with viruliferous MEAM1, promotes the subsequent whitefly infestation and induces plant volatile neophytadiene which recruits whiteflies (Shi et al. 2018). These outcomes have clear implications in the epidemiology and management of the TYLCV and whiteflies and help to explain the concurrent outbreaks of TYLCV and B. tabaci MED in China. The TYLCV has already been reported in our neighbouring country Venezuela, in South America (Zambrano et al. 2007) and actions should avoid its arrival in Brazil.
We can conclude that B. tabaci MED represent a great constraint for Brazilian agriculture, as pest and vector of of the main important viruses infecting tomatoes and beans and populations of MED with Hamiltonella prevalence, can increase the transmission efficiency and contribute for virus epidemic in different crops.
References
Barbosa, L. F., Marubayashi, J. M., De Marchi, B. R., Yuki, V. A., Pavan, M. A., Moriones, E., et al. (2014). Indigenous American species of the Bemisia tabaci complex are still widespread in the Americas. Pest Management Science, 70(10), 1440–1445.
Barbosa, L. B., Yuki, V. A., Marubayashi, J. M., De Marchi, B. R., Perini, F. L., Pavan, M. A., et al. (2015). First report of Bemisia tabaci Mediterranean (Q biotype) species in Brazil. Pest Management Science, 71(4), 501–504. https://doi.org/10.1002/ps.3909.
Barreto, S. S., Hallwass, M., Aquino, O. M., & Inoue-Nagata, A. K. (2013). A study of weeds as potential inoculum sources for a tomato-infecting Begomovirus in Central Brazil. Phytopathology, 103(5), 436–444. https://doi.org/10.1094/PHYTO-07-12-0174-R.
Bosco, D., Loria, A., Sartor, C., & Cenis, J. L. (2006). PCR-RFLP identification ofBemisia tabaci biotypes in the Mediterranean Basin. Phytoparasitica, 34(3), 243–251.
Boykin, L. M., & De Barro, P. J. (2014). A practical guide to identifying members of the Bemisia tabaci species complex: And other morphologically identical species. Frontiers in Ecology and Evolution, 2. https://doi.org/10.3389/fevo.2014.00045.
Costa, A. S., Oliveira, A. R., & Silva, D. M. (1977). Transmissao mecanica do agente causal do mosaico dourado do tomateiro [Lycopersicum esculentum]. Summa Phytopathologica (Brasil), 3(3), 194–200.
Czosnek, H., & Ghanim, M. (2016). Management of insect pests to agriculture: Lessons learned from deciphering their genome, transcriptome and proteome. Management of Insect Pests to Agriculture: Lessons Learned from Deciphering their Genome, Transcriptome and Proteome, 1–290. https://doi.org/10.1007/978-3-319-24049-7.
De Barro, P. J., Liu, S.-S., Boykin, L. M., & Dinsdale, A. B. (2011). Bemisia tabaci : A statement of species status. Annual Review of Entomology, 56(1), 1–19. https://doi.org/10.1146/annurev-ento-112408-085504.
De Marchi, B. R., Marubayashi, J. M., Favara, G. M., Yuki, V. A., Watanabe, L. F. M., Barbosa, L. F., et al. (2017). Comparative transmission of five viruses by Bemisia tabaci NW2 and MEAM1. Tropical Plant Pathology, 1, 495–499. https://doi.org/10.1007/s40858-017-0186-9.
Dellaporta, S. L., Wood, J., & Hicks, J. B. (1983). A plant DNA minipreparation: Version II. Plant Molecular Biology Reporter, 1(4), 19–21.
Dinsdale, A., Cook, L., Riginos, C., Buckley, Y. M., De Barro, P., & Barro, P. D. (2010). Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Annals of the Entomological Society of America, 103(2), 196–208. https://doi.org/10.1603/AN09061.
Dovas, C. I., Katis, N. I., & Avgelis, A. D. (2002). Multiplex detection of Criniviruses associated with epidemics of a yellowing disease of tomato in Greece. Plant Disease, 86(12), 1345–1349. https://doi.org/10.1094/PDIS.2002.86.12.1345.
Faria, J. C., Aragão, F. J. L., Souza, T. L. P. O., Quintela, E. D., Kitajima, E. W., & Ribeiro, S. G. (2016). Golden mosaic of common beans in Brazil : Management with a transgenic approach. APS Journal, 1–14. https://doi.org/10.1094/APSFeature-2016-10.Plant.
Ghanim, M. (2014). A review of the mechanisms and components that determine the transmission efficiency of tomato yellow leaf curl virus (Geminiviridae; Begomovirus) by its whitefly vector. Virus Research, 186, 47–54. https://doi.org/10.1016/j.virusres.2014.01.022.
Ghanim, M., & Kontsedalov, S. (2009). Susceptibility to insecticides in the Q biotype of Bemisia tabaci is correlated with bacterial symbiont densities. Pest Management Science, 65(9), 939–942. https://doi.org/10.1002/ps.1795.
Ghosh, S., Bouvaine, S., Richardson, S. C. W., Ghanim, M., & Maruthi, M. N. (2018). Fitness costs associated with infections of secondary endosymbionts in the cassava whitefly species Bemisia tabaci. Journal of Pest Science, 91(1), 17–28. https://doi.org/10.1007/s10340-017-0910-8.
Gilbertson, R. L., Batuman, O., Webster, C. G., & Adkins, S. (2015). Role of the insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the emergence and global spread of plant viruses. Annual Review of Virology, 2(1), 67–93. https://doi.org/10.1146/annurev-virology-031413-085410.
Gottlieb, Y., Zchori-Fein, E., Mozes-Daube, N., Kontsedalov, S., Skaljac, M., Brumin, M., Sobol, I., Czosnek, H., Vavre, F., Fleury, F., & Ghanim, M. (2010). The transmission efficiency of tomato yellow leaf curl virus by the whitefly Bemisia tabaci is correlated with the presence of a specific symbiotic bacterium species. Journal of Virology, 84(18), 9310–9317. https://doi.org/10.1128/JVI.00423-10.
Gueguen, G., Vavre, F., Gnankine, O., Peterschmitt, M., Charif, D., Chiel, E., et al. (2010). Endosymbiont metacommunities, mtDNA diversity and the evolution of the Bemisia tabaci (Hemiptera: Aleyrodidae) species complex. Molecular Ecology, 19(19), 4365–4376.
Hadjistylli, M., Roderick, G.K., & Brown, J.K. (2016). Global population structure of a worldwide Pest and virus vector : Genetic diversity and population history of the Bemisia tabaci sibling species group. https://doi.org/10.5061/dryad.h7s57.
Horowitz, A. R., Kontsedalov, S., Khasdan, V., & Ishaaya, I. (2005). Biotypes B and Q of Bemisia tabaci and their relevance to neonicotinoid and pyriproxyfen resistance. Archives of Insect Biochemistry and Physiology, 58(4), 216–225. https://doi.org/10.1002/arch.20044.
Inoue-Nagata, A. K., Lima, M. F., & Gilbertson, R. L. (2016). A review of geminivirus diseases in vegetables and other crops in Brazil: Current status and approaches for management. Horticultura Brasileira, 34(1), 8–18. https://doi.org/10.1590/S0102-053620160000100002.
Kliot, A., Cilia, M., Czosnek, H., & Ghanim, M. (2014). Implication of the bacterial endosymbiont rickettsia spp. in interactions of the whitefly Bemisia tabaci with tomato yellow leaf curl virus. Journal of Virology, 88(10), 5652–5660. https://doi.org/10.1128/JVI.00071-14.
Lourencao, A. L., & Nagai, H. (1994). Surtos populacionais de Bemisia tabaci no Estado de Sao Paulo. Bragantia, 53(1), 53–59. https://doi.org/10.1590/S0006-87051994000100006.
Macedo, M., Barreto, S., Hallwass, M., & Inoue-nagata, A. (2014). High incidence of Tomato chlorosis virus alone and in mixed infection with begomoviruses in two tomato fields in the Federal District and Goiás state , Brazil. Tropical Plant Pathology, 39(6), 449–452. https://doi.org/10.1590/S1982-56762014000600005.
Marubayashi, J. M., Yuki, V. A., Rocha, K. C. G., Mituti, T., Pelegrinotti, F. M., Ferreira, F. Z., Moura, M. F., Navas-Castillo, J., Moriones, E., Pavan, M. A., & Krause-Sakate, R. (2013). At least two indigenous species of the Bemisia tabaci complex are present in Brazil. Journal of Applied Entomology, 137(1–2), 113–121. https://doi.org/10.1111/j.1439-0418.2012.01714.x.
Marubayashi, J. M., Kliot, A., Yuki, V. A., Rezende, J. A. M., Krause-Sakate, R., Pavan, M. A., & Ghanim, M. (2014). Diversity and localization of bacterial endosymbionts from whitefly species collected in Brazil. PLoS One, 9(9), e108363. https://doi.org/10.1371/journal.pone.0108363.
Moraes, L. A., Marubayashi, J. M., Yuki, V. A., Ghanim, M., Bello, V. H., De Marchi, B. R., et al. (2017). New invasion of Bemisia tabaci Mediterranean species in Brazil associated to ornamental plants. Phytoparasitica, 45, 1–525. https://doi.org/10.1007/s12600-017-0607-9.
Moraes, L. A., Muller, C., Bueno, R. C. O. F., Santos, A., Bello, V. H., De Marchi, B. R., et al. (2018). Distribution and phylogenetics of whiteflies and their endosymbiont relationships after the Mediterranean species invasion in Brazil. Scientific Reports, 8(1), 14589. https://doi.org/10.1038/s41598-018-32913-1.
Moreno-Delafuente, A., Garzo, E., Moreno, A., & Fereres, A. (2013). A plant virus manipulates the behavior of its whitefly vector to enhance its transmission efficiency and spread. PLoS One, 8(4), e61543. https://doi.org/10.1371/journal.pone.0061543.
Navas-Castillo, J., Fiallo-Olivé, E., & Sánchez-Campos, S. (2011). Emerging virus diseases transmitted by whiteflies. Annual Review of Phytopathology, 49, 219–248. https://doi.org/10.1146/annurev-phyto-072910-095235.
Ning, W., Shi, X., Liu, B., Pan, H., Wei, W., Zeng, Y., Sun, X., Xie, W., Wang, S., Wu, Q., Cheng, J., Peng, Z., & Zhang, Y. (2015). Transmission of tomato yellow leaf curl virus by Bemisia tabaci as affected by whitefly sex and biotype. Scientific Reports, 5(1), 10744. https://doi.org/10.1038/srep10744.
Pan, H., Chu, D., Yan, W., Su, Q., Liu, B., Wang, S., & Wu, Q. (2012). Rapid Spread of Tomato Yellow Leaf Curl Virus in China Is Aided Differentially by Two Invasive Whiteflies, 7(4), e34817. https://doi.org/10.1371/journal.pone.0034817.
RDevelopment, C. (2018). R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Ribeiro, S. G., De Ávila, A. C., Bezerra, I. C., Fernandes, J. J., Faria, J. C., Lima, M. F., et al. (1998). Widespread occurrence of tomato geminiviruses in Brazil, associated with the new biotype of the whitefly vector. Plant Disease, 82(7), 830.
Rojas, M. R., Gilbertson, R. L., Russell, D. R., & Maxwell, D. P. (1993). Use of degenerate primers in the polymerase chain reaction to detect whitefly-transmitted Geminiviruses. Plant Disease, 77, 340. https://doi.org/10.1094/PD-77-0340.
Shi, X., Pan, H., Xie, W., Jiao, X., Fang, Y., Chen, G., Yang, X., Wu, Q., Wang, S., & Zhang, Y. (2014). Three-Way Interactions Between the Tomato Plant, Tomato Yellow Leaf Curl Virus, and Bemisia tabaci (Hemiptera: Aleyrodidae) Facilitate Virus Spread. Journal of Economic Entomology, 107(3), 920–926. https://doi.org/10.1603/EC13476.
Shi, X., Chen, G., Pan, H., Xie, W., Wu, Q., Wang, S., et al. (2018). Plants pre-infested with viruliferous MED/Q cryptic species promotes subsequent Bemisia tabaci infestation. Frontiers in Microbiology, 9(JUN), 1–8. https://doi.org/10.3389/fmicb.2018.01404.
Simon, C., Frati, F., Beckenbach, A., Crespi, B., Liu, H., & Flook, P. (1994). Evolution, weighting, and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Annals of the Entomological Society of America, 87(6), 651–701.
Su, Q., Pan, H., Liu, B., Chu, D., Xie, W., Wu, Q., Wang, S., Xu, B., & Zhang, Y. (2013). Insect symbiont facilitates vector acquisition, retention, and transmission of plant virus. Scientific Reports, 3, 1–6. https://doi.org/10.1038/srep01367.
Sun, D.-B., Liu, Y.-Q., Qin, L., Xu, J., Li, F.-F., & Liu, S.-S. (2013). Competitive displacement between two invasive whiteflies: Insecticide application and host plant effects. Bulletin of Entomological Research, 103(03), 344–353. https://doi.org/10.1017/S0007485312000788.
Walsh, P. S., Metzger, D. A., & Higuchi, R. (1991). Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques, 10(4), 506–513.
Yao, F. L., Zheng, Y., Huang, X. Y., Ding, X. L., Zhao, J. W., Desneux, N., He, Y. X., & Weng, Q. Y. (2017). Dynamics of Bemisia tabaci biotypes and insecticide resistance in Fujian province in China during 2005-2014. Scientific Reports, 7(December 2016), 1–12. https://doi.org/10.1038/srep40803.
Zambrano, K., Carballo, O., Geraud, F., Chirinos, D., Fernández, C., & Marys, E. (2007). First report of tomato yellow leaf curl virus in Venezuela. Plant Disease, 91(6), 768.
Zanardo, L. G., Silva, F. N., Lima, A. T. M., Milanesi, D. F., Castilho-Urquiza, G. P., Almeida, A. M. R., Zerbini, F. M., & Carvalho, C. M. (2014). Molecular variability of cowpea mild mottle virus infecting soybean in Brazil. Archives of Virology, 159(4), 727–737. https://doi.org/10.1007/s00705-013-1879-0.
Acknowledgments
This study was financed in party by the Coordenaçao de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. BRM and BRS received a CNPq/Brazil scholarship. Financial support was received from FAPESP 2017/21588-7, 2017/50222, -2014/047289-4 and CNPq479101/2013-2. RKS and MAP received CNPq fellowships.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that the work is in compliance with ethical standards.
Conflict of interest
The authors declare no conflict of interests.
Research involving human participants and/or animals
The authors declare that the manuscript does not contain research involving Human Participants and/or Animals.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bello, V.H., Watanabe, L.F.M., Santos, B.R. et al. Evidence for increased efficiency of virus transmission by populations of Mediterranean species of Bemisia tabaci with high Hamiltonella prevalence. Phytoparasitica 47, 293–300 (2019). https://doi.org/10.1007/s12600-019-00729-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12600-019-00729-y