Abstract
Sugarcane white leaf (SCWL) is one of the major sugarcane diseases in the Asian continent associated with phytoplasmas. It occurs in all sugarcane-growing areas of Sri Lanka, causing substantial economic losses. However, subgroup level identification has been not reported so far for the phytoplasma strains associated with this disease in Sri Lanka. In the present study, the geographical distribution of phytoplasma strains associated with SCWL in Sri Lanka, identification of their weed reservoirs and ribosomal subgroup analysis based on phytoplasma 16S rRNA gene were carried out. A total of 27 SCWL samples representing six main sugarcane-growing areas of Sri Lanka and six samples of two grass species (Cynodon dactylon and Brachiaria distachya) showing putative symptoms of phytoplasma were analyzed. A nested PCR product of 1.2 kb size was consistently amplified in all the symptomatic samples with primers amplifying the 16S rRNA gene. A higher level of sequence identity from 99.03 to 100% was shared among the 16S rRNA gene sequences of all tested SCWL phytoplasma strains. Both Bermudagrass white leaf (BGWL) and Brachiaria grass white leaf (BraWL) phytoplasma strains shared 97.5–98.1% identity of their 16S rRNA gene with SCWL strains. The 16S rRNA gene sequence comparisons and virtual RFLP analysis of SCWL, BGWL, and BraWL phytoplasma isolates allowed their affiliations with ‘Candidatus Phytoplasma sacchari’ (16SrXI-B subgroup) with SCWL and ‘Ca. P. cynodontis’ (16SrXIV-A subgroup) with the grasses samples. The phytoplasma subgroup 16SrXI-B was identified as the most widespread SCWL phytoplasma subgroup in commercial sugarcane varieties in Sri Lanka.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytoplasmas are plant pathogens that affect economically important annual perennial crops and natural floras worldwide (Rao et al. 2017, 2018). Sugarcane is one of the crucial crops infected by phytoplasma diseases, such as white leaf (SCWL), grassy shoot (SCGS), yellow leaf (SCLYP) and green grassy shoot (SCGGS) (Rao et al. 2012, 2017, 2018). The sugarcane white leaf disease has been identified as the most destructive disease. It has a serious impact on sugarcane yield and is regarded as one of the major threats to sugarcane industries in Sri Lanka (Kumarasinghe and Jones 2001; Keerthipala 2016).
Different phytoplasma subgroups belonging to the 16SrXI group were reported to be associated with SCWL and SCGS diseases in all over Asian countries. The phytoplasma subgroups, 16SrXI-B and 16SrXI-F in India (Viswanathan and Rao 2011; Rao et al. 2014; Yadav et al. 2017) and 16SrXI-D in China (Zhang et al. 2016), were, identified in the infected plants associated with SCWL and SCGS diseases. Among these, the 16SrXI-B is the major phytoplasma subgroup reported associated with SCWL and SCGS disease (Viswanathan and Rao 2011; Jung et al. 2003; Yadav et al. 2017; Nithya et al. 2020; Kirdat et al. 2020a, b). Earlier the SCWL phytoplasma had been classified under ‘Candidatus Phytoplasma oryzae’ species (Jung et al. 2003). However, recently a new ‘Candidatus Phytoplasma’ species has been published as ‘Candidatus Phytoplasma sacchari’ for the phytoplasma strains associated with SCWL and SCGS diseases infecting sugarcane crops in Asia (Kirdat et al. 2020b). So far, no information is available on subgroup level-based identification of phytoplasma strains associated with SCWL or SCGS disease in Sri Lanka.
Several weed species, including various grasses, act as natural reservoirs of phytoplasmas worldwide (Rao et al. 2008, 2017). Some of those weed species are also present in sugarcane plantations of Sri Lanka. Among those weeds, phytoplasma association symptoms are showing in Bermuda grass (Cynodon dactylon) and signal grass (Brachiaria distachya). However, no attempt has been made to identify phytoplasma association with these weed species to check their role as a natural reservoir for SCWL or SCGS phytoplasma in Sri Lanka. Therefore, our objectives were to study the geographical distribution of SCWL phytoplasma strains in Sri Lanka, detect the possible weed grass species acting as a reservoir of SCWL phytoplasmas, and to identify the ribosomal subgroup taxonomy of SCWL and grass phytoplasma strains based on the analysis of 16S rRNA gene sequences.
Materials and Methods
A survey was made of sugarcane fields in Sri Lanka at six locations, including sugar factories areas of Hingurana, Pelwatte, Sevanagala and Ethimale with more than 2500 ha of sugarcane plantation and established sugarcane nurseries at Kantale and Uda Walawe areas during 2019–2020 (Fig. 1; Table 1). Symptomatic samples of sugarcane, Brachiaria grass white leaf (BraWL) from signal grass (B. distachya) and Bermuda grass white leaf (BGWL) from Bermuda grass (C. dactylon) were collected from the surveyed sugarcane fields from Hingurana and Uda Walawe areas. Asymptomatic sugarcane and grass samples were also collected from the same fields and used as positive controls. Eighteen sugarcane fields and three sugarcane nurseries were surveyed to inspect the symptoms of SCWL. A total of 33 samples (27 sugarcane and six grass samples), which represent two symptomatic and one asymptomatic sample from nine locations, were collected in the surveyed locations for DNA extraction (Table 1).
DNA Extraction, PCR Assay and Sequence Analysis
DNA extraction was done by following the CTAB genomic DNA extraction protocol suggested by Doyle and Doyle (1990). The quantity and quality of the DNA were measured by using a Nanodrop spectrophotometer (Implant NP80, USA). PCR amplification was performed by using phytoplasma specific universal primer pairs P1/P7 (Deng and Hiruki 1991; Schneider et al. 1995) followed by nested PCR assays with R16F2n/R16R2 primer pair (Gundersen and Lee 1996). The amplified nested PCR products of the 16S rRNA gene from each location were purified using Wizard®SV gel and PCR clean-up system (Promega, USA), and direct sequencing was done for both directions using R16F2n/R16R2 primer pairs specific for 16S rRNA gene (ABI Genetic Analyzer 3500 series). Both forward and reverse sequencing data were checked and assembled using the BioEdit application (version 7.0) to prepare the contig sequences. The resulting contig sequences were submitted to the GenBank database as representing one sequence from each location.
Multiple sequence alignment was done to determine the similarities of the 16S rRNA gene in different sugarcane and grass phytoplasma strains by using the Clustal Omega program. Phylogenetic analysis was done using the MEGA 7.0 application (Kumar et al. 2016) to find the phylogenetic relationship between the 16S rRNA gene sequences of the SCWL, SCGS, BGWL, and BraWL phytoplasma strains. The phytoplasma sequences reported in Asian countries were retrieved from GenBank and the neighbor-joining (NJ) method with 1000 bootstrap value was used to construct the phylogenetic tree. Acholeplasma laidlawii (Acc No. AB680603) strain was used as an out group for the phylogenetic tree analysis. In silico RFLP analysis was done for all the SCWL and grass phytoplasma strains based on the virtual RFLP gel patterns of 17 restriction endonucleases by iPhyClassifier online tool (Zhao et al. 2009) to identify the group and subgroup of the phytoplasma strains.
Results and Discussion
Typical symptoms of the sugarcane white leaf disease such as the presence of chlorotic stripes on the leaves, leaf chlorosis with a varying degree from pale green to complete white leaf, production of whitish dwarf new tillers and dead of those without producing millable stalks and the proliferation of the axillary shoots in the mature cane were recorded in different sugarcane varieties during the survey. Similar symptoms were recorded on different sugarcane varieties in all the surveyed locations. Leaf chlorosis, a proliferation of axillary shoots, bushy growth habit, small leaves and shortened stolons symptoms were observed in C. dactylon in Sevanagala, Pelwatte, Hingurana and Kantale and B. distachya in Uda Walawe, Sevanagala, Pelwatte and Hingurana locations. Amplification of approximately 1250 bp PCR products was achieved in all the symptomatic sugarcane and two grass samples in nested PCR assay, while no amplified products could be seen in any of the asymptomatic sugarcane and grass samples (data not shown).
The sequence identity of the 16S rRNA gene varied from 99.03 to 100% among the SCWL phytoplasma strains identified from all the surveyed locations (Table 2). The results revealed that there was no significant genetic diversity among the SCWL phytoplasma strains infecting different commercial sugarcane varieties in Sri Lanka. The SCWL phytoplasma strain reported in SL 96 128 sugarcane variety from Galmaduwa (Acc. No. MT811046), Kantale (Acc. No. MT860714) and Uda Walawe (Acc. No. MT811809) shared 100% sequence identity among each other. This is the most cultivated sugarcane variety at present in Sri Lanka. The SCWL phytoplasma strains reported from Galmaduwa, Kantale and Uda Walawe (Acc. Nos. MT811046, MT860714 and MT811809) showed 100% sequence identity with SCWL Indian strain (Acc. No. JX862179). Furthermore, there was a 100% identity of the 16S rRNA gene sequence between SCWL Pelwatte strains (Acc. No. MT860722) and SCWL China strain (Acc. No. MT649297). The results confirmed almost similar genetic information in their 16S rRNA gene is being shared by different SCWL strains infecting the sugarcane crops in the Asian region. The identified SCWL strains in Sri Lanka shared a close identity from 99.2 to 100%, with earlier reported SCGS phytoplasma strain from India (Acc. No. MN889545). A very close identity of 16S rRNA gene sequence in the range of 99.2–99.9% between the SCGS and SCWL strains in the Asian countries has also been reported in previous studies (Nasare et al. 2007; Rao et al. 2008; Yadav et al. 2017; Kumar et al. 2018; Quan et al. 2020). However, comparatively low identity of 16S rRNA gene sequence was shared among SCWL Vietnam strain (Acc. No. KC295286) and Sri Lanka strains, which ranged from 96.93 to 99.61% (Table 2).
The 16S rRNA gene sequence of the Sri Lankan BGWL phytoplasma strains shared 97.51–98.19% sequence identity with SCWL phytoplasma Sri Lankan strains. The result revealed the close identity of the 16S rRNA gene sequence between BGWL and SCWL phytoplasma strains present in Sri Lanka. Similar confirmations were done by Kirdat et al. (2020a) revealed 98.07-99.59 % identity and Nasare et al. (2007) revealed 98.0% identity among BGWL and SCGS phytoplasma strains from India. Firrao et al. (2005) reported the 16S rRNA gene of SCGS phytoplasma shared 98.3–98.5% identity with the BGWL strains while the SCWL phytoplasma shared 98.2–98.4% identity with the BGWL strains. Further, the BraWL Sri Lanka strain shared 97.51–98.15% identity with SCWL phytoplasma strains reported in Sri Lanka (Table 2). Therefore, our results also suggested that the SCWL phytoplasma strains in Sri Lanka shared a significantly close genetic relationship with the BraWL strains in Sri Lanka.
Kirdat et al. (2020b) suggested a novel phytoplasma species for the phytoplasma strains identified associated with SCGS disease, namely ‘Ca. P. sacchari.' The phytoplasma strains reported in the present study are showing more than 99% of the identity of their 16S rRNA gene with SCGS strains reported from India. Few previous studies also confirmed both SCWL and SCGS phytoplasmas have a very close sequence identity of their 16S rRNA gene, and both strains have been categorized under the RYD phytoplasma group (Nakashima et al. 1996; Lee et al. 1997; Tran-Nguyen et al. 2000; Jung et al. 2003; Nasare et al. 2007; Kirdat et al. 2020b). Hence, the results of the present study proposed the Sri Lankan SCWL strains under the new ‘Ca. P. sacchari’ species introduced by Kirdat et al (2020b).
In the present study, SCWL/SCGS phytoplasma strains shared more than 97.5% identity of 16S rRNA gene sequence with BGWL and BraWL phytoplasma and they were categorized under two different ‘Ca. Phytoplasma species, viz. ‘Ca. P. sacchari’ and ‘Ca. P. cynodontis.' There are two different vector species such as Deltocephalus menoni transmitting SCWL in Sri Lanka (Seneviratne 2008) and Exitianus capicola transmitting BGWL phytoplasma strains (Salehi et al. 2009). Hence, further studies will be needed to confirm whether the same vector species is transmitting both SCWL and BGWL phytoplasmas. Furthermore, studies based on the multilocus genes of the phytoplasma genome other than the 16S rRNA gene will be required to confirm whether the phytoplasma strains in C. dactylon and B. distachya cause for SCWL disease.
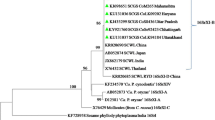
The phylogenetic analysis presented that all of the Sri Lankan SCWL strains clustered together with SCWL and SCGS phytoplasma strains reported from India, Thailand, Vietnam, China and Pakistan. Therefore, the results further suggested that all the SCWL Sri Lankan strains are phylogenetically closer with other SCWL and SCGS phytoplasma strains reported from different Asian countries and related to ‘Ca. P. sacchari’ species. Sri Lankan BGWL and BraWL strains clustered with earlier reported BGWL and BraWL phytoplasma strains (Fig. 2), and close phylogenetic relationship is shown among them. Furthermore, a few previous studies have been reported the evolutionary closest relatives of the rice yellow dwarf (RYD) phytoplasma group are the SCWL, SCGS, BGWL and BraWL phytoplasmas (Nakashima et al. 1996; Lee et al. 1997; Wongkaew et al. 1997; Tran-Nguyen et al. 2000).
Phylogenetic tree was constructed by neighbor-joining methods with 1000 bootstrap replications to show the phylogenetic relationship of the 16S rRNA gene sequence of different SCWL, SCGS (‘Ca Phytoplasma sacchari’) and BGWL, BraWL (‘Ca Phytoplasma cynodontis’) reported in different countries of the world. 16S rRNA gene sequence of Acholeplasma laidlawii was used as an out-group
Computer-simulated RFLP analyses were carried out with R16F2n/R16R2 sequences corresponding to the 16S rRNA gene sequence of SCWL, BGWL and BraWL phytoplasma strains for 16Sr group/subgroup assignment. Nine Sri Lankan SCWL phytoplasma strains (Acc. Nos. MT811809, MT811046, MT811811, MT811813, MT860714, MT860721, MT860722, MT862368 and MT872412) produced restriction patterns identical to the 16SrXI subgroup B with similarity coefficient 1.0 that confirmed the phytoplasma subgroup 16SrXI-B is associated with the SCWL in all sugarcane growing areas of Sri Lanka (Fig. 3). This is the first study performed to subgroup level identification of phytoplasma strains associated with SCWL in Sri Lanka. The previous studies reported that the phytoplasma subgroup 16SrXI-B was the dominant subgroup reported associated with SCWL and SCGS diseases in Asian countries (Viswanathan and Rao 2011; Rao et al. 2014; Zhang et al. 2016; Kumar et al. 2018; Quan et al. 2020). Further studies are required to analyze more SCWL samples in Sri Lanka to check the possibility of association of other phytoplasma subgroups with SCWL.
Virtual gel images created by iPhyClassifier after In silico analysis of 16S rRNA gene sequence of SCWL and grass white leaf phytoplasmas. 01. Representative virtual gel of 16SrXI-B subgroup for SCWL Sri Lankan strains. 02. iPhyClassifier reference virtual gel for the 16SrXI-B subgroup. 03. Representative virtual gel image of 16SrXIV-A subgroup for Sri Lankan grass white leaf strains. 04. iPhyClassifier reference virtual gel for 16SrXIV-A subgroup
The 16S rRNA sequences of the C. dactylon and B. distachya (Acc. Nos MT862162 and MT872407) phytoplasma strains showed 99% sequence identity with ‘Ca. P. cynodontis’ strains which belong to the 16SrXIV-A subgroup (Fig. 3). The present study confirms the 16SrXIV-A phytoplasma subgroup association with the white leaf symptoms in these two grass species in Sri Lanka. These two grass species were also reported to be the host of the BGWL and BraWL phytoplasma strains in other parts of the world (Lee et al. 1998, 2000; Seemuller et al. 1998; Sdoodee et al. 1999; Duduk et al. 2013). The results also confirmed that phytoplasma strains associated with two weed grass species were different from the SCWL phytoplasma strain in Sri Lanka.
Conclusion
No significant genetic diversity was found among the 16S rRNA gene sequences of SCWL strains reported in Sri Lanka. The 16S rRNA gene sequence of Sri Lankan SCWL strains is closer to SCGS and SCWL strains reported from other Asian countries. The 16SrXI-B subgroup is a dominant phytoplasma subgroup associated with SCWL in all sugarcane growing areas of Sri Lanka. On the basis of recent sequence analysis, the SCWL strains have been described under new phytoplasma species ‘Ca. P. sacchari.'
References
Deng, S., and C. Hiruki. 1991. Amplification of 16S rRNA genes from culturable and non-culturable mollicutes. Journal of Microbiology Methods 14: 53–61.
Doyle, J.J., and J.L. Doyle. 1990. Isolation of plant DNA from fresh tissue. Focus 12: 13–15.
Duduk, B., S. Paltrinieri, I.-M. Lee, and A. Bertaccini. 2013. Nested PCR and RFLP analysis based on the 16S rRNA gene. In Phytoplasma methods in molecular biology (methods and protocols), vol. 938, ed. M. Dickinson and J. Hodgetts. Totowa, NJ: Humana Press. https://doi.org/10.1007/978-1-62703-089-2_14.
Firrao, G., K. Gibb, and C. Streten. 2005. Short Taxonomic guide to the genus ‘Candidatus Phytoplasma’ . Journal of Plant Pathology 87(4): 249–263.
Gundersen, D., and I.-M. Lee. 1996. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathologia Mediterranea 35(3): 144–151.
Jung, H.Y., T. Sawayanagi, P. Wongkaew, S. Kakizawa, H. Nishigawa, W. Wei, K. Oshima, S. Miyata, M. Ugaki, T. Hibi, and S. Namba. 2003. Candidatus Phytoplasma oryzae’, a novel phytoplasma taxon associated with rice yellow dwarf disease. International Journal of Systematic and Evolutionary Microbiology 53(6): 1925–1929.
Keerthipala, A.P. 2016. Development of sugar industry in Sri Lanka. Sugar Tech 18: 612–626.
Kirdat, K., B. Tiwarekar, V. Thorat, N. Narawade, D. Dhotre, S. Sathe, Y. Shouche, and A. Yadav. 2020a. Draft genome sequences of two phytoplasma strains associated with sugarcane grassy shoot (SCGS) and bermuda grass white leaf (BGWL) diseases. Molecular Plant-Microbe Interactions 33(5): 715–717.
Kirdat, K., B. Tiwarekar, V. Thorat, S. Sathe, Y. Shouche, and A. Yadav. 2020b. Candidatus Phytoplasma sacchari’, a novel taxon-associated with Sugarcane Grassy Shoot (SCGS) disease. International Journal of Systematic and Evolutionary Microbiology 71(1): 004591.
Kumar, S., G.P. Rao, J.V. Singh, and V.K. Baranwal. 2018. Genetic diversity of phytoplasmas associated with sugarcane grassy shoot and leaf yellows diseases in India. Phytopathogenic Mollicutes 8 (2): 74–88.
Kumar, S., G. Stecher, and K. Tamura. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Molecular Biology and Evolution 33(7): 1870–1874.
Kumarasinghe, N.C., and P. Jones. 2001. Identification of white leaf disease of sugarcane in Sri Lanka. Sugar Tech 3: 55–58.
Lee, I.M., D.E. Gundersen, R.E. Davis, and I.M. Bartoszyk. 1998. Revised classification scheme of phytoplasma based on RFLP analysis of 16S rRNA and ribosomal protein gene sequences. International Journal of Systematic Bacteriology 48: 1153–1169.
Lee, I.M., M. Pastore, M. Vibio, A. Danielli, S. Attathorn, R.E. Davis, and A. Bertaccini. 1997. Detection and characterization of a phytoplasma associated with annual blue grass (Poa annua) white leaf disease in southern Italy. European Journal of Plant Pathology 103: 251–254.
Lee, I.M., R.E. Davis, and D.E. Gundersen. 2000. Phytoplasma: phytopathogenic mollicutes. Annual Review of Microbiology 54: 221–255.
Nakashima, K., T. Hayashi, W. Chaleeprom, P. Wongkaew, and P. Sirithorn. 1996. Complex phytoplasma flora in northeast Thailand as revealed by 16S rDNA analysis. Annals of the Phytopathological Society of Japan 62: 57–60.
Nasare, K., A. Yadav, A.K. Singh, K.B. Shivasharanappa, Y.S. Nerkar, and V.S. Reddy. 2007. Molecular and symptoms analysis reveal the presence of new phytoplasmas associated with sugarcane grassy shoot disease in India. Plant Disease 91: 1413–1418. https://doi.org/10.1094/PDIS-91-11-1413.
Nithya, K., B. Parameswari, A. Bertaccini, G.P. Rao, and R. Viswanathan. 2020. Grassy shoot: The destructive disease of sugarcane. Phytopathogenic Mollicutes 10(1): 10–24.
Quan, M.V., V.L. Nguyen, M.L. Quang, N.Q. Huy, V.L. Xuan, C.H.H. Viet, D.T. Nguyen, H.T.B. Thao, V.H. Nguyen, D.H. Nguyen, P.G. Weintraub, C. Keswani, L.T. Hang, M.H. Nguyen, and T.H. Hoat. 2020. A new phytoplasma strain associated with the sugarcane white leaf disease in Vietnam. Phytopathogenic Mollicutes 10(1): 60–68.
Rao, G.P., E. Alvarez, and A. Yadav. 2018. Phytoplasma diseases of industrial crops. In Phytoplasma: Plant Pathogenic Bacteria—I, ed. G.P. Rao, A. Bertaccini, N. Fiore, and L.W. Liefting, 91–122. Singapore: Springer. https://doi.org/10.1007/s12355-008-0013-1.
Rao, G.P., S. Mall, and C. Marcone. 2012. Recent biotechnological approaches in diagnosis and management of sugarcane phytoplasma diseases. Functional Plant Science and Biotechnology 6: 19–29.
Rao, G.P., S. Srivastava, P.S. Gupta, A. Singh, M. Singh, and C. Marcone. 2008. Detection of sugarcane grassy shoot phytoplasma infecting sugarcane in India and its phylogenetic relationships to closely related phytoplasmas. Sugar Tech 10: 74–80.
Rao, G.P., V. Madhupriya, A.K. Kumar, S. Tiwari, and V. Baranwal. 2014. Identification of sugarcane grassy shoot-associated phytoplasma and one of its putative vectors in India. Phytoparasitica 42: 349–354. https://doi.org/10.1007/s12600-013-0366-1.
Rao, G.P., V. Madhupriya, R. Thorat, A.K. Tiwari. Manimekalai, and A. Yadav. 2017. A century progress of research on phytoplasma diseases in India. Phytopathogenic Mollicutes 7(1): 1–38.
Salehi, M., K. Izadpanah, M. Siampour, and M. Taghizadeh. 2009. Molecular characterization and transmission of Bermuda grass white leaf phytoplasma in Iran. Journal of Plant Pathology 91: 655–661.
Schneider, B., E. Seemueller, C.D. Smart, and B.C. Kirkpatrick. 1995. Phylogenetic classification of plant pathogenic mycoplasma-like organisms or phytoplasmas. In Molecular and diagnostic procedures in mycoplasmology, ed. S. Razin and J.G. Tully, 369–380. San Diego, CA: Academic Press. https://doi.org/10.1016/B978-012583805-4/50040-6.
Sdoodee, R., B. Schneider, A. Padovan, and K. Gibb. 1999. Detection and genetic relatedness of phytoplasmas associated with plant diseases in Thailand. The Journal of Biochemistry, Molecular Biology and Biophysics 3: 133–140.
Seemuller, E., C. Marcone, U. Lauer, A. Ragozzino, and M. Göschl. 1998. Current status of molecular classification of the phytoplasma. Journal of Plant Pathology 80: 3–26.
Seneviratne, J.A.U.T. 2008. An investigation of the secondary transmission of sugarcane white leaf disease in Sri Lanka, PhD Thesis, University of Peradeniya, Sri Lanka.
Tran-Nguyen, L., K.R. Blanche, B. Egan, and K.S. Gibb. 2000. Diversity of phytoplasmas in northern Australian sugarcane and other grasses. Plant Pathology 49: 666–679.
Viswanathan, R., and G.P. Rao. 2011. Disease scenario and management of major sugarcane diseases in India. Sugar Tech 13: 336–353.
Wongkaew, P., Y. Hanboonsong, P. Sirithorn, C. Choosai, S. Boonkrong, T. Tinnangwattana, R. Kitchareonpanya, and S. Damak. 1997. Differentiation of phytoplasma associated with sugarcane and gramineous weed white leaf disease and sugarcane grassy shoot disease by RFLP and sequencing. Theoretical and Applied Genetics 95: 660–663.
Yadav, A., V. Thorat, S. Deokule, Y. Shouche, and D.T. Prasad. 2017. New subgroup 16SrXI-F phytoplasma strain associated with sugarcane grassy shoot (SCGS) disease in India. International Journal of Systematic and Evolutionary Microbiology 67: 374–378.
Zhang, R.Y., W.F. Li, Y.K. Huang, X.Y. Wang, H.L. Shan, Z.M. Luo, and Z.Y. Yin. 2016. Group 16SrXI phytoplasma strains, including subgroup 16SrXI-B and a new subgroup, 16SrXI-D, are associated with sugar cane white leaf. International Journal of Systematic and Evolutionary Microbiology 66: 487–491.
Zhao, Y., W. Wei, I.M. Lee, J. Shao, X. Sou, and R.E. Davis. 2009. Construction of an interactive online phytoplasma classification tool, iPhyClasifier, and its application in analysis of peach X-disease phytoplasma group (16SrIII). International Journal of Systematic and Evolutionary Microbiology 59: 2582–2593.
Acknowledgements
The authors wish to acknowledge the Dr. M.S. Perera Director (acting) and members of the paper reviewing committee of Sugarcane Research Institute (SRI) Sri Lanka, Prof. P.G.C. Bandaranayake, Mrs. A.G.M.L.K. Dayananda from the University of Peradeniya, Mr. L.M.J.R. Wijayawardhna and other supported members of SRI and sugar industries of Sri Lanka.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dayasena, Y.A.P.K., Panda, P., Thushari, A.N.W.S. et al. Geographical Distribution and Identification of Phytoplasma Strain Associated with Sugarcane White Leaf Disease in Sri Lanka. Sugar Tech 23, 1351–1358 (2021). https://doi.org/10.1007/s12355-021-00980-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-021-00980-w