Abstract
Gastric cancer is one of the most common malignancy worldwide. The various genetic and epigenetic events have been found to be associated with its carcinogenesis. The epigenetic is a heritable and transient/reversible change in the gene expression that is not accompanied by modification in the DNA sequence. This event is characterized by the alteration in the promoter CpG island of the gene or histone modification. These events are associated with silencing of critical tumor suppressor gene and activation of oncogenes leading to carcinogenesis. The DNA methylation is a chemical change in the DNA sequence that most commonly occurs at cytosine moiety of CpG dinucleotide and histone, primarily on N- terminal tail that ultimately effect the interaction of DNA with chromatin modifying protein.
Hypermethylation of tumor suppressor genes and global hypomethylation of oncogenes are widely studied epigenetic modifications. There are large number of publish reports regarding epigenetic events involving gastric cancer. These changes are potentially useful in identifying markers for early diagnosis and management of this lethal malignancy. Also, role of specific miRNAs and long non coding RNAs in regulation of gene expression is gaining interest and is a matter of further investigation. In this review, we aimed to summarize major epigenetic events (DNA methylation) in gastric cancer along with alteration in miRNAs and long non coding RNAs which plays an important role in pathology of this poorly understood malignancy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric carcinoma (GC) is the second most common cancer in the world and a leading cause of cancer related mortality [1]. Gastric carcinoma is classified into intestinal and diffuse type and there are differences in the pathways leading to these two types. Intestinal tumors are progressed through a number of sequential steps. This step begins with atrophic gastritis followed by intestinal metaplasia, dysplasia and carcinoma. Diffuse gastric carcinoma are not characterized by preceding steps other than the chronic gastritis associated with the Helicobacter pylori infection [2, 3].
The pathogenesis of gastric cancer represents a classic example of gene-environment interaction. Among the environmental factors, diet and infection with Helicobacter pylori are the most common suspects. Genetic factors play an important role in gastric carcinogenesis due to aberrant gene expression leading to malignant phenotype [3]. Oncogenic activation of β-catenin (17–27% in intestinal type) and K-ras (0–18% in both histological types) have been found in GC [4, 5]. In addition, amplifications of the c-erbB2 and c-met genes have each been observed in approximation of about 10% in both histological types. Among tumor suppressor genes, p53 mutations are reported in both diffuse (0–21%) and intestinal type (36–43%) [6]. Mutations in APC are found frequently in gastric adenomas, but rarely in gastric carcinoma [1]. Somatic mutations of E- cadherin are observed specifically in sporadic diffuse type gastric cancer (33–50%) [7, 8]. Microsatellite instability (MSI) is observed in 5–10% of diffuse type GC and in 15–40% of intestinal type gastric carcinoma [1]. In addition to these well characterized genetic changes, epigenetic alterations including promoter CpG island hypermethylation are the most common molecular alteration in human neoplasia [9]. Broadly, epigenetics refer to alteration in gene expression which are not regulated by the changes in the DNA sequences. DNA methylation and histone modifications are commonly studied epigenetic events. Promoter hypermethylation of mismatch repair gene hMLH1 is the major mechanism for MSI in gastric carcinoma. Similarly, while, hypermethylation of p16 is common in gastric cancer with higher incidence in intestinal type, mutation of the p16 gene is infrequent [10].
In this review, we aim to summarize major epigenetic changes (DNA methylation) and its known mechanism. We reviewed DNA methylation profile of genes, miRNAs and long non coding RNAs, their roles as oncogenes and tumor suppressors and its clinical utility. To make this review specific, we do not include other epigenetic events i.e. histone modification and nucleosome positioning.
Epigenetics
The term “epigenetics” was first used by Conrad Waddington in 1939 to describe “the casual interaction between the genes and their products”. Later Riggs and colleagues defined epigenetics as “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” [9]. In the present era the term epigenetics has broadened to include heritable and transient/reversible changes in gene expression that is not accompanied by a change in the DNA sequence.
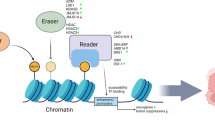
The critical role of epigenetic modifications in human diseases is coming to the fore. Comprehensive understanding of different biological activity like DNA methylation, chromatin structure, transcriptional activities and histone modification has contributed in the development of epigenetics. Two major epigenetic modifications include, DNA methylation and chromatin remodeling. While DNA methylation is a chemical change in the DNA sequence that most commonly occurs at cytosine moiety of CpG dinucleotides, chromatin remodeling occurs via histone modifications (primarily on the N- terminal tails) that ultimately affect the interaction of DNA with chromatin modifying proteins. Both DNA methylation and histone modifications are associated with silencing critical tumor suppressor genes and activating oncogenes involved in cancer [9,10,11].
The overall 5 year survival of GC is about 10–20% [12]. The poor prognosis of gastric carcinoma is because of its late detection. Thus search for new molecular markers becomes imperative for early detection and prediction of prognosis. The use of novel chemotherapeutic drugs against the identified new molecular targets may further improve the prognosis of the gastric carcinoma. Various genetic markers has been predicted for early tumor detection, prognostic prediction and explaining the genetic pathway of gastric carcinogenesis [1, 13]. But the epigenetic marker has gained popularity in recent years particularly the promoter hypermethylation, which has various advantages over a genetic marker. First, promoter hypermethylation is much more common than genetic alteration in cancer. Second, promoter hypermethylation occurs in the same defined region of that gene in all forms of cancer in comparison to wide range of mutational variations in a specific gene. Thus epigenetic detection of promoter hypermethylation will be both efficient and cost effective method of tumor detection. Third promoter hypermethylation constitute a “positive signal” that can be easily detected against a background of normal cells, where as some genetic marker such as loss of heterozygosity, homozygous deletion and MSI represent “negative signals” and would be difficult to detect in background of normal cells [14].
DNA Methylation and Epigenetic Gene Silencing
DNA methylation is a reversible chemical modification of the cytosine in the CpG islands of promoter sequences, catalyzed by a family of DNA methyltransferases. DNA methylation does not change the genetic information but it just alters the readability of the DNA and results in the inactivation of gene by subsequent transcript repression [15]. DNA methylation plays a critical role in the control of cellular process including embryonic development, transcription, X- chromosome inactivation and genomic imprinting [16]. The CpG dinucleotides are not found frequently throughout the human genome. Approximately half of the human gene promoter regions have CpG rich regions of 0.5 to 2 kb in length where CpG dinucleotides frequency is higher than expected. These CpG rich sequences are often known as CpG islands [17]. The majority (94%) of CpG islands remain unmethylated in normal cell. However, particular subgroups of promoter CpG are methylated such as in tissue and germ line specific genes. In general, CpG island methylation is associated with gene silencing. The methylated CpG island also recruits histone deacetylases and other factors involved in transcriptional silencing [9].
The changes in DNA methylation status in cancer cells is complicated by global hypomethylation and localized hypermethylation. Global hypomethylation of the genome was initially thought to be an exclusive event in cancer development [14]. The loss of methylation in cancer is mainly due to hypomethylation of repetitive DNA sequences. During the development of neoplasm, the degree of hypomethylation of genomic DNA increases as the lesion progress from a benign disease to metastatic [9, 18]. Three mechanisms of DNA hypomethylation have been proposed in the development of cancers. First, it increases the genomic instability, second by reactivation of transposable elements, and third by loss of imprinting. Demethylation of DNA can favor mitotic recombination, leading to deletions, translocations and chromosomal instability [9]. The aberrant activation of oncogenes due to promoter demethylation (hypomethylation) has yet not been established [18]. The exact relationship between global hypomethylation and development of cancer remain to be consolidated [9].
Paradoxically, in parallel to global hypomethylation, the genomes of cancer cells are also characterized by localized regions of de novo hypermethylation, typically in the CpG island of tumor suppressor genes and microRNAs (miRNAs) genes. Inactivation of tumor suppressor genes through hypermethylation of CpG islands within promoter regions is a major event in carcinogenesis [9]. Hypermethylation of CpG island also has silencing effect on miRNAs. Micro RNA are short, 18–22 nucleotide, noncoding RNAs that regulate many cellular functions including cell proliferation, apoptosis and differentiation by silencing specific target genes through translational repression or mRNA degradation [19, 20]. In recent past, a number of genes that are critical in tumorigenesis have been identified that undergo epigenetic silencing. It includes various genes involved in different cellular process like cell cycle regulation (p16NK4a, p15INK4b and p14ARF), DNA repair (hMLH 1 and MGMT), cell- cell/cell matrix adhesion (E- cadherin, H- cadherin and adenomatous polyposis coli), apoptosis (DAPK, TMS1 and Caspase-8) and angiogenesis (THBS-1 and p73) [14].
Histone Modification
Histones are evolutionarily highly conserved proteins characterized by an accessible amino terminal tail and a histone fold domain that mediates interactions between histones to form the nucleosome scaffold [9]. The N-terminal of histone polypeptides are extensively modified by more than 60 different posttranslational modifications including methylation, acetylation, phosphorylation, ribosylation, unbiquitinylation, sumoylation, carbonylation and glycosylation [9, 11, 21].
In normal cell, a precise balance maintains nucleosomal DNA in either an active/acetylated or an inactive/deacetylated form. This adequate balance is controlled by acetylating enzymes (histone acetyltransferases, HATs) and deacetylating enzymes (histone deacetylases, HDACs). The other modification includes methylation of arginine and lysine residues of histones. This methylation is catalyzed by histone methyltranferase (HMT) and the process is involved in the regulation of a wide range of gene activities and chromatin structures. In general, lysine methylation at H3K9, H3K27 and H4K20 is associated with gene silencing where as methylation at H3K4, H3K36 and H3K79 are associated with gene activation [11]. The genomic changes in the pattern of CpG methylation may in turn lead to global changes in histone modification patterns in many human cancers. Changes in histone modification pattern independent of the CpG methylation has also been directly linked to the cancer development. In addition to its roles in transcriptional regulation, histone modifications have been implicated in DNA replication, repair and condensation [9].
Mechanism of Epigenetic Activity
The epigenetic regulation by DNA methylation is possible through DNA methyltransferase (DNMT). In human, various DNMT has been described having different vital functions. DNMT3A and DNMT3B are implicated in establishing the de novo methylation pattern where as DNMT1 was thought to be responsible for maintaining the DNA methylation patterns [14, 22]. DNMT2 has an unknown biological function. Its strong binding to DNA suggests that it may mark specific sequence in the genome [11].
There are three main mechanisms by which DNA methylation can suppress gene transcription [23]. First is the methyl CpG binding domain (MBD) - mediated gene silencing. The various methyl-CpG- binding proteins (MBPs) have been identified like methyl CpG binding protein 2 (MeCP2) and methyl CpG biding domains 1–4 (MBD1, MBD2, MBD3 and MBD4). All these proteins posses a transcriptional repressor domain thus directly repress transcription. In addition, MBDs can recruit transcriptional corepressors such as histone deacetylases (HDACs) and Sin3A, to methylated DNA [14, 23]. The deacetylation of chromatin histone result in closed or repressed chromatin configuration, which in turn leads to exclusion of transcription factors and allele specific gene silencing. The second mechanism of gene silencing is mediated through DNMTs. All three DNMTs (DNMT1, DNMT3A and DNMT3B) have a transcription repressor domain and can thus directly suppress transcription. In addition, these DNMTs can recruit co transcriptional repressors such as HDACs to methylated DNA identical to MBDs. In third mechanism, CpG island hypermethylation can interrupt the binding of activating transcription factors to gene promoters [14]. The detail about the histone modifications are discussed above.
Polycomb group (PcG) proteins are epigenetic chromatin modifiers involved in carcinogenesis. Two distinct multiprotein PcG complexes have been identified [24, 25]. Polycomb Repressive Complex 2 (PRC2), which is involved in the initiation of gene repression, and Polycomb Repressive Complex 1 (PRC1) that act as maintenance complex. EZH2 (Enhancer of Zeste homolog 2), a member of PRC2, catalyses the addition of methyl groups to H3K27 (H3K27 trimethylation). EZH2 also interacts with DNMTs and is essential for DNA methylation of EZH2 target promoters suggesting that there is a direct link between PcG mediated gene repression and DNA methylation [21].
Epigenetic Alternations in Gastric Carcinoma
Most of the traditional molecular studies on gastric carcinoma are based on identifying the genetic mutation. Recently, studies are now focused in discovering new biomarkers that are epigenetically silenced in early carcinogenesis. It is also seen that almost half of the tumor suppressor genes that causes familial cancers through mutations can also get inactivated by promoter hypermethylation in sporadic cancers [21]. Increasing evidence suggests that epigenetic changes play a key role in cancer development including GC. It has become apparent that different tumor types pose different spectrum, profile or clustering of gene hypermethylation referred to as CpG island methylator phenotype (CIMP). The CIMP group was defined as having concordant tumor specific DNA methylation and clearly distinguished due to exhibiting higher methylation index in comparison to non CIMP tumors that shows only low levels of tumor specific methylation [14].
Tumors with concurrent hypermethylation in multiple loci have been defined on basis of CpG island methylation phenotype as high CIMP (CIMP-H). CIMP play an important role in gastric carcinoma progression. An et al. (2005) [26] has shown that concurrent hypermethylation of gene promoters in associated with microsatellite instability in CIMP high phenotype gastric carcinoma. They showed that concordant methylation of multiple genes/loci found in 31% of the tumors was associated with better survival but it was not an independent predictor of prognosis in gastric cancer. Thus the prognostic role of CIMP status in gastric cancer is unclear and a matter of further investigation [10]. Table 1 illustrate commonly methylated gene associated with gastric cancer.
The genes such as p16INK4a, CDKN2B/p151NK2b and p14ARF are hypermethylated in human cell lines and primary tumors. Silencing of p16 (INK4a) by promoter hypermethylation has also been reported in gastric carcinoma [27]. The hypermethylation of p16 may predict the malignant potential of dysplasia and early diagnosis of carcinoma. CDKN2A promoter methylation was reported in 30% of gastric cancer [10, 28]. CDKN2A hypermethylation may contribute to the malignant transformation of gastric precursor lesions [29]. Promoter hypermethylation of hMLH1 is a frequent event in gastric cancer and is associated with the loss of hMLH1 expression in the majority of gastric cancers exhibiting MSI [10, 30,31,32]. Methylation of hMLH1 is present in 71% of MSI-H tumor, but only 8% of MSI- low tumors and 13% of microsatellite stable tumors has been observed [29]. The DNA alkyl repair gene O′ (6) methyl guanine DNA methyltransferase (MGMT) hypermethylation is reported in 31% cases of gastric cancer [33].
The capacity of cancer cells to migrate and invade other organs using vascular channels is characterized by the various genes like APC, E-Cadherin (CDH1), H-Cadherin (CDH13) and FAT tumor suppressor cadherin [11]. CDH1 promoter hypermethylation has been shown in 54.8% of the analyzed sporadic gastric carcinoma [34] and down regulation of E-cadherin, might be associated with poor prognosis [35]. Interestingly, CDH1 promoter hypermethylation was more frequent in diffuse histological type (18 out of 20 cases) [35]. CDH1 methylation has been shown to be significantly higher in gastric tissue with lymph node (LN) metastasis than in those without LN metastasis, and has been associated with serosal invasion [36]. It was reported that H. pylori infection was associated with E-cadherin methylation leading to down regulation of E-cadherin. Interestingly, H. pylori eradication therapy could reverse methylation in patients with chronic gastritis only which may halt the process of gastric carcinogenesis [10, 37,38,39]. CDH4 gene methylation has also been seen in high frequency of gastric carcinoma cases and may be an early event in tumor progression.
Hypermethylation of Death associated protein kinase (DAPK) was observed in intestinal, diffuse and mixed type of gastric cancer and correlated with the presence of LN metastasis, advanced stage and poor survival [40]. Methylation associated inactivation of RASSF1A; a tumor suppressor gene is frequently seen in lung and breast cancer [41]. Loss or down regulation of RASSF1A correlates with stage and grade of the gastric tumor. RASSF1A non-expressing primary gastric carcinoma showed methylation at CpG sites in 95% cases [41]. Methylation pattern of RASSF1A may be use as a potential diagnostic and therapeutic target for gastric cancer [42]. Also, RAS pathway genes such as hDAB2IP, HRASLS, RASSF2, RKIP are found to be methylated in gastric carcinoma and associated with cancer risk [43, 44].
X–linked inhibitor of apoptosis (XIAP) is the most potent member of IAP family that exerts anti-apoptotic efforts by interfering with the activities of Caspases. Recently, XIAP associated factor 1 (XAF1) have been identified to negatively regulate the Caspase inhibiting activity of XIAP. The epigenetic silencing of XAF1 gene by aberrant promoter methylation is reported in gastric cancer [45]. On the other hand, Caspase-1 (interleukin 1 beta converting enzyme), a member of the cysteine protease family, show loss of expression in 19.3% cases of gastric carcinoma [46] and the expression is reversed on 5-aza 2’deoxycytidine and/or trichostatin treatments in gastric cancer cell line. The expression of TSPYL5 mRNA was frequently down regulated and inversely correlated with DNA methylation in seven out of nine gastric cancer cell lines [47]. In primary GC, methylation specific PCR of TSPYL5 showed hypermethylation at CpG island in 23 out of the 36 (63.9%) cases [47]. hSRBC is a putative tumor suppressor gene located at 11p15.4 and frequent genomic loss has been observed in several malignancies. hSRBC increases the protein stability of p53 and expression of p53 target genes such as p21 (WAF1), PUMA and NOXA, while hSRBC mediated cell cycle arrest and apoptosis were abolished by blockade of p53 function. Loss or reduction of hSRBC expression has been demonstrated in 73% of cancer cell lines and 41% of primary gastric tumor [48]. While the allelic loss or somatic mutation of this gene is infrequent, its expression was restored in tumor cells by 5-aza 2′ deoxycytidine (DNA methyltransferase inhibitor) treatment [48]. Activation of Wnt signaling has been implicated in tumorigenesis. Secreted frizzled related proteins (SFRP) are identified as possible negative modulators of the Wnt signal transduction pathway. DICKKOPF (DKK) family genes are identified as the Wnt antagonist [49, 50]. The downregulated expression of SFRP2 has been correlated with promoter hypermethylation in 73.3% cases of primary gastric cancer tissue [49]. Nojima et al. (2007) [51] showed high frequency of CpG methylation in SFRP1, SFRP2, and SFRP5 in both gastric cell line and primary gastric cancer. Hypermethylation and loss of β-catenin (CTNNB1) expression, an integral component of the Wnt signaling pathway, has been reported in a subgroup of primary gastric cancer, cell lines and in metastases [52]. DKK methylation is also reported in gastric cancer cell line [50]. Apoptosis pathway genes (BNIP3, CACNA2D3, DAPK, GPX3, PCDH17) hypermethylation are found to be associated with gastric cancer [44]. Recently, PCDH10 was found to be methylated at early stages of gastric cancer and is an independent prognostic factor for this malignancy [53].
Homozygous loss of very low density lipoprotein receptor (VLDLR) gene and epigenetic silencing by DNA methylation been reported in gastric cancer cell line by Takada et al. (2006) [54]. Suppressor of cytokine signaling (SOCS) – 1 inhibits signaling of the Janus kinase (JAK)/signal transducers and activators of transcription (STAT) pathway by several cytokines and has tumor suppressor activity. AKAP12/Gravin is one of the A-kinase anchoring proteins (AKAPs) which functions as a kinase scaffold protein and dynamic regulator of beta 2 adrenergic receptor complex. Hypermethylation of two form of AKAP12 gene (AKAP12A and AKAP12B) has been demonstrated in gastric carcinoma [55]. Promoter hypermethylation of RIZ1, the retinoblastoma protein interacting zinger finger gene which is involved in chromatin mediated gene expression and is also a target for frame shift mutation in microsatellite-unstable cancers, has been shown in 69% cases of gastric carcinoma and 21% cases of non-neoplastic mucosa [56]. An essential epigenetic regulator of the mammalian SWI/SNF chromatin remodeling complex contains Brm molecule as its catalytic submit. Frequent loss of Brm expression has been shown in gastric cancer cell lines and primary gastric tumour which is rescued by treatment with histone deacetylase inhibitor in gastric cancer cell lines suggesting epigenetic regulation of this gene [57]. Recent studies have shown that members of the SWI/SNF superfamily can function as tumor suppressor genes. Methylation of HLTF, a homologue of the SWI/SNF, is reported in 50% cases of gastric carcinoma [58]. Among RUNX gene family, RUNX3 is often involved in gastric carcinogenesis. Hypermethylation of CpG island of RUNX3 was reported in 64% cases of gastric carcinoma [10]. Kim et al. (2004) [59] also showed RUNX3 methylation in 8% case of chronic gastritis, 28% cases of intestinal metaplasia and 27% cases of gastric adenoma. Promotor hypermethylation of retinoic acid receptor beta (RARβ) is demonstrated in 64% cases of gastric carcinoma [60]. Thrombospondin-1 (TSP-1) is a potent peptide linked with angiogenesis in many tumors. Promoter hypermethylation of TSP1 gene was found in 33% cases of gastric carcinoma [61]. DLC-1 (deleted in liver cancer) gene showed hypermethylation in 30% cases of primary gastric cancer (Kim et al. 2003) [62]. Aberrant methylation of COX-2 has also been described in gastric carcinoma [53, 63].
Poplawski et al. (2008) [64] observed aberrant methylation in promoter regions of multiple genes (CASP8, hMLH1, CDH1 and MDR1) involved in gastric cancer. Interestingly, the hypermethylation of hMLH1 occurred more frequently in females than in men [64]. Lee et al. (2002) [5] reported promoter methylation of DAPK, E-cadherin, GSTP1, p15 and p16 in 70%, 76%, 18%, 69% and 67% of gastric carcinoma respectively. Kang et al. (2003) [65] investigated methylation of multiple genes in gastric cancer tissue, gastric adenoma, intestinal metaplasia and chronic gastritis [65]. Five different classes of methylation behaviors were found: (a) genes methylated in gastric cancer only (GSTP1 and RASSF1A) (b) genes showing significantly higher methylation frequency in gastric cancer in other lesion (COX-2, hMLH1, p16) (c) genes with high and similar methylation frequency in all four lesions (APC, E-cadherin) (d) a gene with low and similar methylation frequency in four step lesion (MGMT), and (e) genes showing an increasing methylation frequency in the progression of the disease (DAP- kinase, p14, THBS1 and TIMP-3). They concluded that tumor related genes show a gene type specific methylation profile along the multistep carcinogenesis. Oue et al. (2002) [66] showed CpG island hypermethylation of the p16 (INK4a), CDH1 and RARβ promoters in 27%, 58% and 53% cases of gastric carcinoma respectively. In this study, hypermethylation of p16 (INK4a) promoter was more common in intestinal type than in diffuse type gastric carcinoma. CDH1 and RAR-beta promoter hypermethylation was more frequently in diffuse scattered type of gastric carcinoma [66]. Recently, Sepulveda et al. (2016) [67] identified 13 genes (BRINP1, CDH11, CHFR, EPHA5, EPHA7, FGF2, FLI1, GALR1, HS3ST2, PDGFRA, SEZ6L, SGCE, and SNRPN) which are hypermethylated in gastric carcinoma as compared with normal mucosa, which in turn may be useful in developing diagnostic and prognostic tool for this lethal malignancy.
Hypomethylation of specific genes also contribute to gastric carcinogenesis. Demethylation of MAGE, synuclein-γ (SNCG) and cyclin D2 has been described in gastric carcinoma [10]. Melanoma antigen (MAGE) expression is known to be activated by promethylation. Demethylation of both MAGE-A 1 and A 3 promoters is more frequently observed (29% and 66% respectively) in advanced clinical stages of gastric carcinoma and also associated with poor prognosis [68]. SNCG demethylation is common in cases with LN metastasis [69]. Hypomethylation of cyclin D2 promoter was found in 71% cases of gastric carcinoma. This event is more common in stage-III and IV tumor than in stage-I and II tumor [70]. Modification of histone by methylation which occurs at lysine or arginine residues is generally associated with gene inactivation or silencing [71,72,73,74,75]. Histone modifications also regulate genes that participate in cell cycle. It has also been reported that methylation in histone H3 plays an important role in carcinogenesis by silencing tumor suppressor genes [76, 77]. Park et al. (2008) [75] found global histone modification pattern through immunohistochemistry and reported that trimethylation of H3K9 correlate positively with the tumor stage and lymphovascular invasion in gastric carcinoma. On the other hand, acetylation of histone, which mostly occurs at lysine residues of N- terminal domains, is known to be associated with transcriptional activation. The acetylation of histone H3 at K9 was associated with a poorly differentiated or diffuse type of histology [75]. Histone H4 acetylation is reduced in gastric carcinoma compared to normal mucosa. Reduction of histone H4 acetylation correlates with a more advanced stage, deeper invasion and greater extent of lymph node metastasis [78]. In gastric cancer cell, p21WAF1 is associated with extensive histone acetylation. It has also been reported that reduced histone H3 acetylation is associated with reduced tumor suppressor gene p21WAF1/CIP1 expression in gastric carcinoma [79]. Histone H4 acetylation at lysine 16 directs the tumor towards a better prognosis, possibly by activating tumor suppressor genes [75]. Xia et al. (2008) [80] investigated the modulation of cell cycle control protein p21 (WAF1) by H. pylori in gastric carcinoma cell line and primary gastric cells derived from healthy tissue. Study revealed that increase expression of p21 (WAF1) induced by H. pylori is associated with the release of HDAC-1 from p21 (WAF1) promoter and hyperacetylation of histone 4 [80].
Non Coding RNAs
Protein coding genes accounts to only 1.5% to 2% of total genome. A large portion of human genome is noncoding, was once thought to be non functional. However, recently there is increase in knowledge regarding functional aspects of these RNAs particularly in normal development and physiology [81]. They mainly consists of small nucleolar RNAs (snoRNAs), promoter-associated non coding RNAs (paRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs), repeat-associated siRNAs (rasiRNAs), small nuclear RNAs (snRNAs), piwi interacting RNA (piRNA), long non coding RNAs (lncRNAs), mitochondrial non coding RNAs, satellite non coding RNAs, repeat associated non coding RNAs, long intergenic non coding RNAs (lincRNAs) and ultraconserved regions (UCRs) [82]. The functional relevance of the non coding portion of genome is evident for miRNAs and lncRNAs [81] particularly, and hence only these non coding RNAs are detailed here. Figure 1 summarizes of major hypermethylated genes, hypomethylated genes, dysregulated miRNAs and long noncoding RNAs in gastric cancer.
MicroRNAs (miRNAs)
MicroRNAs (miRNAs) are recently discovered evolutionary conserved, single stranded, non-coding RNAs of about 22 nucleotides. They play vital role in post-transcriptional regulation of gene expression by binding to 3′-untranslated regions (UTRs) region of target mRNA. A single miRNA regulates several mRNAs and hence their dysregulation affects multiple signaling pathways leading to cancer progression [83]. The miRNA signature is different for various cancers and hence they can use as a diagnostic tool. Table 2 summarizes major dysregulated miRNAs in gastric cancer known till date. Downregulated miRNAs in cancers suggests that these molecules may act as tumor suppressors; however the exact mechanism is not clear yet. Studies have shown that regulation of miRNA expression is modulated by promoter CpG island hypermethylation during tumorigenesis [84]. Yin et al. (2016) [85] identified miR-33b to be tumor suppressor for gastric cancer and its downregulation is tightly regulated by aberrant DNA methylation in the upstream CpG island. Li et al. (2012) [86] identified significant association between DNA methylation and downregulation of miR-155. MiR-155, an important multifunctional microRNA regulates gastric cancer cell metastasis by targeting SMAD2. Similarly, CpG island methylation profile of miR-338-3p may be utilized as a potential diagnostic marker for gastric cancer [87]. Tsai et al. (2011) [88] found significant downregulation of miR-9 and its aberrant hypermethylation during gastric cancer progression. Similarly, miR-137, miR-378, miR-449 and miR-375 are found to be downregulated and hypermethylated in gastric carcinoma [89,90,91,92]. In contract to hypermethylation, demethylation of miR-196 promoter region leading to its upregulation has also been observed in GC [93]. Also, higher expression of miR-34c-5p significantly increases chemoresistance in gastric cancer by downregulating microtubule associated protein tau protein expression [94]. Zhu et al. (2012) [95] observed significant role of miR-200bc/429 cluster in the development of multidrug resistance by targeting BCL2 and XIAP. These findings may suggest significant role of miRNAs promoter methylation in gastric cancer pathology and hence targeting such genes will help in developing new strategies for gastric cancer treatment.
Epigenetic alteration, especially methylation plays a vital role in changes in miRNA expression. It has been well demonstrated that various tumour suppressor miRNA are silenced by epigenetic events in cancer. Expression of miRNAs was found to be upregulated after 5-aza 2′ deoxycytidine treatment, a potent inhibitor of DNA methylation. These finding leads to the conclusion that miRNAs expression are under tight regulation of epigenetic events. Moreover, it is hypothesized that miRNAs may have their own CpG islands containing promoters within introns of protein coding genes which are regulated by DNA methylation. However, exact mechanism is not clear and further studies are much needed to understand complete mechanism of epigenetic regulation of miRNAs. PIWI-interacting RNAs (piRNA) are normally 24–32 nucleotides newly discovered noncoding RNAs which is supposed to regulate gene silencing. It is hypothesized that piRNAs may induce changes in DNA methylation in the CpG islands near to its binding sites. However, the mechanisms of its biogenesis and functions remain largely unknown and needs to be explored.
Long Non Coding RNAs (lncRNAs)
Long non coding RNAs are about 200nucleotides long, non protein coding transcripts constituting around 75% of human genome [96, 97]. A large number of human lncRNAs have been discovered and their numbers are increasing with time. They are like mRNAs specifically in respects of 5’capping, splicing, and poly-adenylation, but have little or no open reading frame [98]. The functions of lncRNAs are poorly understood, but there are evidence of their role in pre splicing editing, transcription, post transcriptional modifications like splicing and translation. It may also play important role in chromatin remodeling through epigenetic modification of DNA [82]. Dysregulation of lncRNAs has been identified in much human cancers including gastric carcinoma [99] (Table 3).
Arita et al. (2013) [100] detected circulating lncRNAs (H19, HOX antisense intergenic RNA (HOTAIR), and metastasis associated lung adenocarcinoma transcript-1 (MALAT1) in plasma of gastric cancer patients. Later on based on these findings, Zhou et al. (2015) [101] identified higher expression of H19 in plasma of GC patients with respect to controls and can be used as a diagnostic biomarker. Moreover, expression of HOTAIR is associated with GC pathology and worse prognosis [102]. Also, overexpression of HULC is associated with proliferation, tumor metastasis and apoptosis for gastric cancer [103]. Maternally expressed gene 3 regulates cell proliferation and apoptosis and its downregulation is associated with decrease survival of GC patients [104]. Cao et al. (2013) [105] identified a set of differentially expressed lncRNAs in gastric cancer through publicly available data set. Similarly, Li et al. (2016) [106] identified diagnostic and prognostic utility of two lncRNAs (HOTAIR and UCA1) selected from large sample sequencing database. In spite of limited knowledge regarding lncRNAs in gastric carcinoma, they have tremendous potential to be used as tumor markers in clinical settings and hence this field needs to be explored.
Clinical Implication of Epigenetics
The knowledge of epigenetic alterations could be potentially useful for cancer diagnosis and treatment. The aberrant promoter methylation occurs very early during carcinogenesis. The CpG island hypermethylation may become one of the most promising biomarkers for the early detection of tumor. It will be more beneficial than genetic counterpart as promoter methylation occur more frequently in tumors than genetic alterations and also because several methylated loci can be analyzed simultaneously. Since promoter methylation occurs within a well defined region of a gene, epigenetic study will be more efficient and cost effective. The overall survival of gastric carcinoma is poor (10–20%) [12]. The early detection of lesion and/or reliable biomarker for monitoring locoregional recurrence may increase the survival in gastric carcinoma patients.
Epigenetic alterations in tumor may also be utilized in predicting the tumor behaviour or prognosis of the gastric carcinoma. The promoter hypermethylation of E-cadherin is associated with poor prognosis [35]. CDH 1 promoter hypermethylation is more frequent in diffuse histological type and significantly higher with lymph node (LN) metastasis and serosal invasion [36, 107]. DAPK is correlated with the presence of LN metastasis, advanced stage and poor survival [40]. Methylation of SOCS-1 present with reduced expression of SOCS gene, lymph node metastases and advanced tumor stage [56]. Demethylation of both MAGE-A1 and A3 promoters is observed in advanced clinical stages and associated with poor prognosis [68]. SNCG demethylation is common in cases with LN metastasis. The trimethylation of H3K9 in gastric carcinoma is associated with advanced tumor stage and lymphovascular invasion and acetylation of histone H3 at K9 is present in poorly differentiated or diffuse type of histology [71]. Reduction of histone H4 acetylation correlates with a more advanced stage, deeper invasion and greater extent of lymph node metastasis [78].
The recent development in the understanding of the relevant gene silencing by epigenetic mechanisms in cancer development is closely linked to epigenetic drug design and development. These compounds basically act via three different processes: DNA cytosine methylation, histone modification and nucleosomal remodeling. The two main classes of drugs are DNA methylation inhibitors and HDAC inhibitors [11]. The various drugs which are widely used in clinical practice for treating other conditions like hydralazine (hypertension), procainamide (cardiac arrhythmia), valproic acid (epilepsy) may be used now for cancer treatment [107,108,109].
DNA methylation inhibitors is an attractive approach for anticancer treatment as toxicity of these drugs to normal cells is potentially lower than in conventional anticancer chemotherapeutic agents [14]. DNA methylation inhibitors are divided into nucleoside and non-nucleoside analogues. The former are compounds that form a covalent intermediate complex with DNMT, preventing the cell from being methylated correctly. DNMT inhibitors such as 5-azacytidine (Vidaza), 5-aza- deoxycytidine (decitabine) are the only two cytidine analogues that have been approved by US Food and Drug Administration (FDA) for haematological malignancies [110, 111]. Non-nucleoside analogues have an advantage over the analogues since they bind to the catalytic site of the enzyme DNMT and are not integrated into the DNA. Thus, it avoids the nonspecific effects of the nucleoside analogues (Mulero-Navarro & Esteller 2008). Hydralazine a potent peripheral vasodilator is now also used as demethylating agent for treating cervical cancer [112].
The HDAC inhibitors are divided into four groups: short chain fatty acids, hyroxamic acids, cyclic tetra peptides and benzamides [11]. The hydroxamic acid trichostatin A (TSA) is found to be a chemosensitizer which increases the efficacy of chemotherapeutic drug in gastric carcinoma [113]. TSA is a promising chemotherapeutic agent in combination with 5-fluorouracil, paclitaxel and irinotecan in gastric cancer cell lines [113]. Recently, (in October 2006) the US Food and Drug Administration have approved the first HDAC inhibitor, Vorinostat 1 (SAHA) for the treatment of cutaneous T cell lymphoma [114]. The various other HDAC inhibitors are under clinical trial.
The other novel method of using epigenetics in treatment of cancer is the reactivation of key enzymes controlling the cellular response to anticancer drug. Satoh et al. (2003) [115] demonstrated that microtubule inhibitors such as docetaxel and paclitaxel induce apoptosis in gastric cell lines with CHFR (Checkpoint with fork head associated and ring finger) methylation. They found that gastric cancer cell lines not expressing CHFR lack a mitotic checkpoint and are highly susceptible to microtubule inhibitors. Thus, CHFR methylation may be a useful molecular marker to predict the responsiveness of gastric cancer to a treatment with microtubule inhibitors. Koga et al. (2006) [116] also reported CHFR methylation in predicting the response of microtubule inhibitor in the treatment of gastric cancer. Also, more detailed studies regarding long coding RNAs, particularly miRNAs and long noncoding RNAs will be very useful for early diagnosis and for prediction of this common gastrointestinal problem.
Conclusion
There are a number of publications regarding epigenetic regulations in gastric cancer; however its complete picture is not clear. In future, these events may be used as a potential screening marker for the early detection of gastric carcinoma. It may also be used as risk assessment tool for the person who might develop cancer later on. Epigenetic markers may be utilized in clinical practice in predicting the tumor behavior and prognosis of the gastric carcinoma patient’s. It may be used as biomarker in monitoring the response to therapeutic agents. Also, DNA methylation inhibitors and HDAC inhibitors may be used as monotherapy or in combination with other anticancer drug for treatment of this lethal malignancy.
References
Ushijima T, Sasako M (2004) Focus on gastric cancer. Cancer Cell 5(2):121–125
Tahara E (2004) Genetic pathways of two types of gastric cancer. IARC Sci Publ 157:327–349
Smith MG, Hold GL, Tahara E, El-Omar EM (2006) Cellular and molecular aspects of gastric cancer. World J Gastroenterol 12(19):2979–2990
Park WS, Oh RR, Park JY, Lee SH, Shin MS, Kim YS, Kim SY, Lee HK, Kim PJ, Oh ST, Yoo NJ, Lee JY (1999) Frequent somatic mutations of the beta-catenin gene in intestinal type gastric cancer. Cancer Res 59(17):4257–4260
Lee TL, Leung WK, Chan MW, Ng EK, Tong JH, Lo KW, Chung SC, Sung JJ, To KF (2002) Detection of gene promoter hypermethylation in the tumor and serum of patients with gastric carcinoma. Clin Cancer Res 8(6):1761–1766
Maesawa C, Tamura G, Suzuki Y, Ogasawara S, Sakata K, Kashiwaba M, Satodate R (1995) The sequential accumulation of genetic alterations characteristic of the colorectal adenoma carcinoma sequence does not occur between gastric adenoma and adenocarcinoma. J Pathol 176(3):249–258
Becker KF, Atkinson MJ, Reich U, Becker I, Nekarda H, Siewert JR, Höfler H (1994) E-cadherin gene mutations provide clues to diffuse type gastric carcinomas. Cancer Res 54(14):3845–3852
Li QL, Ito K, Sakakura C, Fukamachi H, Ki I, Chi XZ et al (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109(1):113–124
Hirst M, Marra MA (2009) Epigenetics and human disease. Int J of Biochem Cell Bio 41(1):136–146
Panani AD (2008) Cytogenetic and molecular aspects of gastric cancer: clinical implications. Cancer Letter 266(2):99–115
Mulero-Navarro S, Esteller M (2008) Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol 68(1):1–11
Khan FA, Shukla AN (2006) Pathogenesis and treatment of gastric carcinoma: “an up-date with brief review”. J Cancer Res Ther 2(4):196–199
Nitti D, Mocellin S, Marchet A, Pilati P, Lise M (2008) Recent advances in conventional and molecular prognostic factors for gastric carcinoma. Surg Oncol Clin N Am 17(3):467–483
Fan CY (2004) Epigenetic alterations in head and neck cancer: prevalence, clinical significance, and implications. Curr Oncol Rep 6(2):152–161
Tischoff I, Wittekind C, Tannapfel A (2006) Role of epigenetic alterations in cholangiocarcinoma. J Hepato-Biliary-Pancreat Surg 13(4):274–279
Robertson KD (2005) DNA methylation and human disease. Nat Rev Genet 6(8):597–610
Bird AP (1986) CpG-rich islands and the function of DNA methylation. Nature 321(6067):209–213
Nakajima T, Enomoto S, Ushijima T (2008) DNA methylation: a marker for carcinogen exposure and cancer risk. Environ Health Prev Med 13(1):8–15
Chen CZ (2005) Micro-RNAs as oncogenes and tumour suppressors. N Engl J Med 353(17):1768–1771
Calin GA, Croce CM (2006) Micro-RNA signatures in human cancers. Nat Rev Cancer 6(11):857–866
Hinshelwood RA, Clark SJ (2008) Breast cancer epigenetics: normal human mammary epithelial cells as a model system. J Mol Med 86(12):1315–1328
Jones PA, Baylin SB (2002) The fundamental role of epigenetic events in cancer. Nat Rev Genet 3(6):415–428
Karpf AR, Jones DA (2002) Reactivating the expression of methylation silenced genes in human cancer. Oncogene 21(35):5450–5461
Lund AH, van Lohuizen M (2004) Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol 16(3):239–246
Sparmann A, van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6(11):846–856
An C, Choi IS, Yao JC, Worah S, Xie K, Mansfield PF, Ajani JA, Rashid A, Hamilton SR, Wu TT (2005) Prognostic significance of CpG island methylator phenotype and microsatellite instability in gastric carcinoma. Clin Cancer Res 11(2 Pt 1):656–663
Song SH, Jong HS, Choi HH, Kang SH, Ryu MH, Kim NK, Kim WH, Bang YJ (2000) Methylation of specific CpG sites in the promoter region could significantly down-regulate p16 (INK4a) expression in gastric adenocarcinoma. Int J Cancer 87(2):236–240
Guimarães AC, Lima EM, Khayat AS, Girão Faria MH, Barem Rabenhorst SH, Pitombeira MV, Assumpção PP, de Oliveira Bahia M, Lima de Lima PD, de Arruda Cardoso Smith M, Burbano RR (2007) Interrelationships among chromosome aneuploidy, promoter hypermethylation, and protein expression of the CDKN2A gene in individuals from northern Brazil with gastric adenocarcinoma. Cancer Genet Cytogenet 179(1):45–51
Lee JH, Park SJ, Abraham SC, Seo JS, Nam JH, Choi C, Juhng SW, Rashid A, Hamilton SR, Wu TT (2004) Frequent CpG island methylation in precursor lesions and early gastric adenocarcinomas. Oncogene 23(26):4646–4654
Fleisher AS, Esteller M, Wang S, Tamura G, Suzuki H, Yin J, Zou TT, Abraham JM, Kong D, Smolinski KN, Shi YQ, Rhyu MG, Powell SM, James SP, Wilson KT, Herman JG, Meltzer SJ (1999) Hypermethylation of the hMLH1 gene promoter in human gastric cancers with microsatellite instability. Cancer Res 59(5):1090–1095
Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC (1999) hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res 59(1):159–164
Jung HY, Jung KC, Shim YH, Ro JY, Kang GH (2001) Methylation of the hMLH1 promoter in multiple gastric carcinomas with microsatellite instability. Pathol Int 51(6):445–451
Oue N, Shigeishi H, Kuniyasu H, Yokozaki H, Kuraoka K, Ito R, Yasui W (2001) Promoter hypermethylation of MGMT is associated with protein loss in gastric carcinoma. Int J Cancer 93(6):805–809
Zazula M, Ferreira AM, Czopek JP, Kolodziejczyk P, Sinczak-Kuta A, Klimkowska A, Wojcik P, Okon K, Bialas M, Kulig J, Stachura J (2006) CDH1 gene promoter hypermethylation in gastric cancer: relationship to Goseki grading, microsatellite instability status, and EBV invasion. Diagn Mol Pathol 15(1):24–29
Graziano F, Arduini F, Ruzzo A, Bearzi I, Humar B, More H, Silva R, Muretto P, Guilford P, Testa E, Mari D, Magnani M, Cascinu S (2004) Prognostic analysis of E-cadherin gene promoter hypermethylation in patients with surgically resected, node-positive, diffuse gastric cancer. Clin Cancer Res 10(8):2784–2789
Liu WT, Jiao HL, Yang YL, Wang D, Zhang WM (2007) Correlation of E-cadherin hypermethylation to tumorigenesis and development of gastric cancer. Ai Zheng 26(11):1199–1203
Terrés AM, Pajares JM, O'Toole D, Ahern S, Kelleher D (1998) Helicobacter pylori infection is associated with downregulation of E-cadherin, a molecule involved in epithelial cell adhesion and proliferation control. J Clin Pathol 51(5):410–412
Chan AO, Lam SK, Wong BC, Wong WM, Yuen MF, Yeung YH, Hui WM, Rashid A, Kwong YL (2003) Promoter methylation of E-cadherin gene in gastric mucosa associated with helicobacter pylori infection and in gastric cancer. Gut 52(4):502–506
Chan AO (2006) E-cadherin in gastric cancer. World J Gastroenterol 12(2):199–203
Chan AW, Chan MW, Lee TL, Ng EK, Leung WK, Lau JY, Tong JH, Chan FK, To KF (2005) Promoter hypermethylation of death associated protein-kinase gene associated with advance stage gastric cancer. Oncol Rep 13(5):937–941
Byun DS, Lee MG, Chae KS, Ryu BG, Chi SG (2001) Frequent epigenetic inactivation of RASSF1A by aberrant promoter hypermethylation in human gastric adenocarcinoma. Cancer Res 61(19):7034–7038
Balgkouranidou I, Matthaios D, Karayiannakis A, Bolanaki H, Michailidis P, Xenidis N, Amarantidis K, Chelis L, Trypsianis G, Chatzaki E, Lianidou ES, Kakolyris S (2015) Prognostic role of APC and RASSF1A promoter methylation status in cell free circulating DNA of operable gastric cancer patients. Mutat Res 778:46–51
Dote H, Toyooka S, Tsukuda K, Yano M, Ota T, Murakami M, Naito M, Toyota M, Gazdar AF, Shimizu N (2005) Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in gastrointestinal tumour. Br J Cancer 92(6):1117–1125
Qu Y, Dang S, Hou P (2013) Gene methylation in gastric cancer. Clin Chim Acta 424:53–65. https://doi.org/10.1016/j.cca.2013.05.002
Byun DS, Cho K, Ryu BK, Lee MG, Kang MJ, Kim HR, Chi SG (2003) Hypermethylation of XIAP associated factor 1, a putative tumor suppressor gene from the 17p13.2 locus, in human gastric adenocarcinomas. Cancer Res 63(21):7068–7075
Jee CD, Lee HS, Bae SI, Yang HK, Lee YM, Rho MS, Kim WH (2005) Loss of caspase-1 gene expression in human gastric carcinomas and cell lines. Int J Oncol 26(5):1265–1271
Jung Y, Park J, Bang YJ, Kim TY (2008) Gene silencing of TSPYL5 mediated by aberrant promoter methylation in gastric cancers. Lab Investig 88(2):153–160
Lee JH, Byun DS, Lee MG, Ryu BK, Kang MJ, Chae KS, Lee KY, Kim HJ, Park H, Chi SG (2008) Frequent epigenetic inactivation of hSRBC in gastric cancer and its implication in attenuated p53 response to stresses. Int J Cancer 122(7):1573–1584
Cheng YY, Yu J, Wong YP, Man EP, To KF, Jin VX, Li J, Tao Q, Sung JJ, Chan FK, Leung WK (2007) Frequent epigenetic inactivation of secreted frizzled-related protein 2 (SFRP2) by promoter methylation in human gastric cancer. Br J Cancer 97(7):895–901
Sato H, Suzuki H, Toyota M, Nojima M, Maruyama R, Sasaki S, Takagi H, Sogabe Y, Sasaki Y, Idogawa M, Sonoda T, Mori M, Imai K, Tokino T, Shinomura Y (2007) Frequent epigenetic inactivation of DICKKOPF family genes in human gastrointestinal tumours. Carcinogenesis 28(12):2459–2466
Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, Hirata K, Itoh F, Tokino T, Mori M, Imai K, Shinomura Y (2007) Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene 26(32):4699–4713
Ebert MP, Yu J, Hoffmann J, Rocco A, Röcken C, Kahmann S, Müller O, Korc M, Sung JJ, Malfertheiner P (2003) Loss of beta-catenin expression in metastatic gastric cancer. J Clin Oncol 21(9):1708–1714
Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN, Geng H, Tian LW, Wong YP, Tong JH, Ying JM, Jin H, To KF, Chan FK, Sung JJ (2009) Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology 136(2):640–651
Takada H, Imoto I, Tsuda H, Nakanishi Y, Sakakura C, Mitsufuji S, Hirohashi S, Inazawa J (2006) Genomic loss and epigenetic silencing of very-low-density lipoprotein receptor involved in gastric carcinogenesis. Oncogene 25(49):6554–6562
Choi MC, Jong HS, Kim TY, Song SH, Lee DS, Lee JW, Kim TY, Kim NK, Bang YJ (2004) ACAP12/Gravin is inactivated by epigenetic mechanism in human gastric carcinoma and shows growth suppressor activity. Oncogene 23(42):7095–7103
Oshimo Y, Oue N, Mitani Y, Nakayama H, Kitadai Y, Yoshida K, Chayama K, Yasui W (2004) Frequent epigenetic inactivation of RIZ1 by promoter hypermethylation in human gastric carcinoma. Int J Cancer 110(2):212–218
Yamamichi N, Inada K, Ichinose M, Yamamichi-Nishina M, Mizutani T, Watanabe H, Shiogama K, Fujishiro M, Okazaki T, Yahagi N, Haraguchi T, Fujita S, Tsutsumi Y, Omata M, Iba H (2007) Frequent loss of Brm expression in gastric cancer correlates with histologic features and differentiation state. Cancer Res 67(22):10727–10735
Hamai Y, Oue N, Mitani Y, Nakayama H, Ito R, Matsusaki K, Yoshida K, Toge T, Yasui W (2003) DNA hypermethylation and histone hypoacetylation of the HLTF gene are associated with reduced expression in gastric carcinoma. Cancer Sci 94(8):692–698
Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW, Bang YJ, Kang GH (2004) Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Investig 84(4):479–484
Hayashi K, Yokozaki H, Goodison S, Oue N, Suzuki T, Lotan R, Yasui W, Tahara E (2001) Inactivation of retinoic acid receptor beta by promoter CpG hypermethylation in gastric cancer. Differentiation 68(1):13–21
Oue N, Matsumura S, Nakayama H, Kitadai Y, Taniyama K, Matsusaki K, Yasui W (2003) Reduced expression of the TSP1 gene and its association with promoter hypermethylation in gastric carcinoma. Oncology 64(4):423–439
Kim TY, Jong HS, Song SH, dimtchev A, Jeong SJ, Lee JW, Kim TY, Kim NK, Jung M, Bang YJ (2003) Transcriptional silencing of the DLC-1 tumor suppressor gene by epigenetic mechanism in gastric cancer cells. Oncogene 22(25):3943–3951
de Maat MF, van de Velde CJ, Umetani N, de Heer P, Putter H, van Hoesel AQ, Meijer GA, van Grieken NC, Kuppen PJ, Bilchik AJ, Tollenaar RA, Hoon DS (2007) Epigenetic silencing of cyclooxygenase-2 affects clinical outcome in gastric cancer. J Clin Oncol 25(31):4887–4894
Poplawski T, tomaszewska K, Galicki M, Morawiec Z, Blasiak J (2008) Promoter methylation of cancer related genes in gastric carcinoma. Exp Oncol 30(2):112–116
Kang GH, Lee S, Kim JS, Jung HY (2003) Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Investig 83(4):519–526
Oue N, Motoshita J, Yokozaki H, Hayashi K, Tahara E, Taniyama K, Matsusaki K, Yasui W (2002) Distinct promoter hypermethylation of p16INK4a, CDH1, and RAR-beta in intestinal, diffuse-adherent, and diffuse-scattered type gastric carcinomas. J Pathol 198(1):55–59
Sepulveda JL, Gutierrez-Pajares JL, Luna A, Yao Y, Tobias JW, Thomas S, Woo Y, Giorgi F, Komissarova EV, Califano A, Wang TC, Sepulveda AR (2016) High-definition CpG methylation of novel genes in gastric carcinogenesis identified by next-generation sequencing. Mod Pathol 29(2):182–193
Honda T, Tamura G, Waki T, Kawata S, Terashima M, Nishizuka S, Motoyama T (2004) Demethylation of MAGE promoters during gastric cancer progression. Br J Cancer 90(4):838–843
Yanagawa N, Tamura G, Honda T, Endoh M, Nishizuka S, Motoyama T (2004) Demethylation of the synuclein gamma gene CpG island in primary gastric cancers and gastric cancer cell lines. Clin Cancer Res 10(7):2447–2451
Lima EM, Leal MF, Burbano RR, Khayat AS, Assumpcao PP, Bello MJ, Rey JA, Smith MA, Casartelli C (2008) Methylation status of ANAPC1, CDKN2A and TP53 promoter genes in individuals with gastric cancer. Braz J Med Biol Res 41(6):539–543
Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL (2001) Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107(6):727–738
Mermoud JE, Popova B, Peters AH, Jenuwein T, Brockdorff N (2002) Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol 12(3):247–251
Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA (2002) Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res 62(22):6456–6461
Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18(11):1251–1262
Park YS, Jin MY, Kim YJ, Yook JH, Kim BS, Jang SJ (2008) The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol 15(7):1968–1976
Kondo Y, Shen L, Issa JP (2003) Critical role of histone methylation in tumour suppressor gene silencing in colorectal cancer. Mol Cell Biol 23(1):206–215
Watanabe Y, Toyota M, Kondo Y, Suzuki H, Imai T, Ohe-Toyota M, Maruyama R, Nojima M, Sasaki Y, Sekido Y, Hiratsuka H, Shinomura Y, Imai K, Itoh F, Tokino T (2007) PRDM5 identified as a target of epigenetic silencing in colorectal and gastric cancer. Clin Cancer Res 13(16):4786–4794
Ono S, Oue N, Kuniyasu H, Suzuki T, Ito R, Matsusaki K, Ishikawa T, Tahara E, Yasui W (2002) Acetylated histone H4 is reduced in human gastric adenomas and carcinomas. J Exp Clin Cancer Res 21(3):377–382
Mitani Y, Oue N, Hamai Y, Aung PP, Matsumura S, Nakayama H, Kamata N, Yasui W (2005) Histone H3 acetylation is associated with reduced p21(WAF1/CIP1) expression by gastric carcinoma. J Pathol 205(1):65–73
Xia G, Schneider-Stock R, Diestel A, Habold C, Krueger S, Roessner A, Naumann M, Lendeckel U (2008) Helicobacter pylori regulates p21(WAF1) by histone H4 acetylation. Biochem Biophys Res Commun 369(2):526–531
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874
Tekcham DS, Tiwari PK (2016) Non-coding RNAs as emerging molecular targets in gallbladder cancer. Gene 588(1):79–85
Kang C, Song JJ, Lee J, Kim MY (2014) Epigenetics: an emerging player in gastric cancer. World J Gastroenterol 20(21):6433–6447
Suzuki H, Maruyama R, Yamamoto E, Kai M (2012) DNA methylation and microRNA dysregulation in cancer. Mol Oncol 6(6):567–578
Yin H, Song P, Su R, Yang G, Dong L, Luo M, Wang B, Gong B, Liu C, Song W, Wang F, Ma Y, Zhang J, Wang W, Yu J (2016) DNA methylation mediated down-regulating of MicroRNA-33b and its role in gastric cancer. Sci Rep 6:18824. https://doi.org/10.1038/srep18824
Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY, Yan M, Qu QL, Zhu ZG, Liu BY (2012) microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep 27(6):1960–1966
Li P, Chen X, Su L, Li C, Zhi Q, Yu B, Sheng H, Wang J, Feng R, Cai Q, Li J, Yu Y, Yan M, Liu B, Zhu Z (2013) Epigenetic silencing of miR-338-3p contributes to tumorigenicity in gastric cancer by targeting SSX2IP. PLoS One 8(6):e66782. https://doi.org/10.1371/journal.pone.0066782
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC, Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, Huang KH, Lin WC (2011) Aberrant hypermethylation of miR-9 genes in gastric cancer. Epigenetics 6(10):1189–1197
Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X (2011) miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci 56(7):2009–2016
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu Z, Yu X, Xia T, Cui L, Guo J (2013) MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 518(2):351–359
Wei B, Song Y, Zhang Y, Hu M (2013) microRNA-449a functions as a tumor-suppressor in gastric adenocarcinoma by targeting Bcl-2. Oncol Lett 6(6):1713–1718
Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T (2010) MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res 20(7):784–793
Niinuma T, Suzuki H, Nojima M, Nosho K, Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki Y, Nishida T, Bamba T, Kanda T, Ajioka Y, Taguchi T, Okahara S, Takahashi H, Nishida Y, Hosokawa M, Hasegawa T, Tokino T, Hirata K, Imai K, Toyota M, Shinomura Y (2012) Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 72(5):1126–1136
Wu H, Huang M, Lu M, Zhu W, Shu Y, Cao P, Liu P (2013) Regulation of microtubule-associated protein tau (MAPT) by miR-34c-5p determines the chemosensitivity of gastric cancer to paclitaxel. Cancer Chemother Pharmacol 71(5):1159–1171
Zhu W, Xu H, Zhu D, Zhi H, Wang T, Wang J, Jiang B, Shu Y, Liu P (2012) miR-200bc/429 cluster modulates multidrug resistance of human cancer cell lines by targeting BCL2 and XIAP. Cancer Chemother Pharmacol 69(3):723–731
Eddy SR (2001) Non–coding RNA genes and the modern RNA world. Nat Rev Genet 2(12):919–929
S D, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A et al (2012) Landscape of transcription in human cells. Nature 489(7414):101–108
Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato A, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SP, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Madan Babu M, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schönbach C, Sekiguchi K, Semple CA, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, FANTOM Consortium, RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) (2005) The transcriptional landscape of the mammalian genome. Science 309(5740):1559–1563
Yang Q, Zhang RW, Sui PC, He HT, Ding L (2015) Dysregulation of non-coding RNAs in gastric cancer. World J Gastroenterol 21(39):10956–10981
Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E (2013) Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res 33(8):3185–3193
Zhou X, Yin C, Dang Y, Ye F, Zhang G (2015) Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep 5:11516. https://doi.org/10.1038/srep11516
Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, Iijima K, Shimosegawa T, Sugamura K, Satoh K (2013) Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One 8(10):e77070. https://doi.org/10.1371/journal.pone.0077070.eCollection2013
Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y (2014) Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep 31(1):358–364
Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X (2014) Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol 35(2):1065–1073
Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY (2013) Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol 19(23):3658–3664
Li CY, Liang GY, Yao WZ, Sui J, Shen X, Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, Zhang WH, Yin LH, Pu YP (2016) Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol 48(5):1965–1976
Rich S, Ganz R, Levy PS (1983) Comparative actions of hydralazine, nifedipine and amrinone in primary pulmonary hypertension. Am J Cardiol 52(8):1104–1107
Strolin Benedetti M, Rumigny JF, Dostert P (1984) Mechanisms of action and biochemical toxicology of valproic acid. Encéphale 10(4):177–188
Windle J, Prystowsky EN, Miles WM, Heger JJ (1987) Pharmacokinetic and electrophysiologic interactions of amiodarone and procainamide. Clin Pharmacol Ther 41(6):603–610
Issa JP, Gharibyan V, Cortes J, Jelinek J, Morris G, Verstovsek S, Talpaz M, Garcia-Manero G, Kantarjian HM (2005) Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol 23(17):3948–3956
Kaminskas Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, Wang YC, Pazdur R (2005) Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res 11(10):3604–3608
Zambrano P, Segura-Pacheco B, Perez-Cardenas E, Cetina L, Revilla-Vazquez A, Taja-Chayeb L, Chavez-Blanco A, Angeles E, Cabrera G, Sandoval K, Trejo-Becerril C, Chanona-Vilchis J, Duenas-González A (2005) A phase I study of hydralazine to demethylate and reactivate the expression of tumor suppressor genes. BMC Cancer 5:44
Zhang X, Yashiro M, Ren J, Hirakawa K (2006) Histone deacetylase inhibitor, trichostatin A, increases the chemosensitivity of anticancer drugs in gastric cancer cell lines. Oncol Rep 16(3):563–568
Riester D, Hildmann C, Schwienhorst A (2007) Histone deacetylase inhibitors--turning epigenic mechanisms of gene regulation into tools of therapeutic intervention in malignant and other diseases. Appl Microbio Biotechnol 75(3):499–514
Satoh A, Toyota M, Itoh F, Sasaki Y, Suzuki H, Ogi K, Kikuchi T, Mita H, Yamashita T, Kojima T, Kusano M, Fujita M, Hosokawa M, Endo T, Tokino T, Imai K (2003) Epigenetic inactivation of CHFR and sensitivity to microtubule inhibitors in gastric cancer. Cancer Res 63(24):8606–8613
Koga Y, Kitajima Y, Miyoshi A, Sato K, Sato S, Miyazaki K (2006) The significance of aberrant CHFR methylation for clinical response to microtubule inhibitors in gastric cancer. J Gastroenterol 41(2):133–139
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Puneet, Kazmi, H.R., Kumari, S. et al. Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol. Oncol. Res. 24, 757–770 (2018). https://doi.org/10.1007/s12253-018-0410-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-018-0410-z