Abstract
Rhizobacteria are root-associated bacteria that influence plant growth by various direct and indirect mechanisms. In quest of efficient plant growth–promoting rhizobacteria (PGPR) with multiple plant growth–promoting traits, a total of 52 rhizobacterial isolates were isolated from the rhizospheric soil collected at Pohang beach, Republic of Korea. The bacterial isolates were evaluated in vitro for their plant growth–promoting traits like production of 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, indole-3-acetic acid (IAA), siderophore, and phosphate solubilization activities. More than 28% of the isolates revealed all of the multi-trait plant growth–promoting activities, whereas 11.54% exhibited robust results for producing IAA, ACC deaminase, siderophore, and phosphate solubilization activities. Similarly, 36% isolates were capable for the production of IAA, siderophore, and ACC deaminase, while 32% revealed phosphate solubilization and siderophore production. The isolates with prominent multi-trait plant growth–promoting activities were identified based on 16S rRNA gene sequences and matched to Pseudomonas koreensis-(S4T10), Pseudomonas fluorescens-(S3B1), Serratia fonticola-(S1T1), Sphingobacterium multivorum-(S1B1), Brevundimonas vesicularis-(S1T13), and Arthrobacter sp.-(S2T9) with 99–100% similarity. Our results confirm that further evaluation of these PGPR (exhibiting multi-traits for plant growth promotion) is required on crop plants to reveal their pragmatic role under normal and abiotic stress conditions and add into the consortium of biofertilizers for sustainable agriculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In 1904, the German agronomist Lorenz Hiltner, a pioneer in rhizosphere microbial ecology research, first coined the term “rhizosphere” to describe the plant-root interface (Hartmann et al. 2008; Hiltner 1904). The organic material of growing plant’s roots provides a driving force for the development of active microbial population in the near vicinity of the roots which is known as rhizospheric effect (Whipps 2001). Thus, plant nutrition is greatly influenced by microbial composition of the rhizosphere, although rhizosphere support a large number of active microbial population proficient of exerting beneficial, neutral, or detrimental rhizospheric effects on the plants. In other words, plant-microbe interactions in the rhizospheric region may be beneficial to the plant, the microbe, or to neither of them (Ali et al. 2017; Kumar et al. 2012; Singh et al. 2004). The heterogeneous group of bacteria colonizing in the rhizospheric region are known as rhizobacteria that support the growth and development of the host plant directly or indirectly are termed as plant growth–promoting rhizobacteria (PGPR), which enhance plant health and soil fertility in the rhizospheric region (Ahmad et al. 2008; Kumar et al. 2012).
Plant growth–promoting bacteria use one or more direct or indirect mechanisms to facilitate the growth and development of the plants (Grover et al. 2021; Kumar et al. 2021; Penrose and Glick 2003; Upadhyay et al. 2021). The direct promotion of plant growth by PGPR involves either providing the host plant with plant growth–promoting substances which are synthesized by PGPR or assisting the uptake of plant nutrients available in the near vicinity of the plant. The direct promotion of plant growth by PGP bacteria involves different mechanisms such as the synthesis of plant growth hormones, i.e., auxin and cytokinins (Khan et al. 2016), solubilization of minerals (El-Tarabily et al. 2008; Umar et al. 2020), stimulation of ion uptake or transport systems in plants (Mantelin and Touraine 2004), and fixation of atmospheric nitrogen (Hirota et al. 1978), whereas in indirect mechanisms of plant growth promotion, these bacteria mitigate or halt the deleterious effects of abiotic stress conditions or a plant pathogen by producing antibiotics, enzymes, and siderophores (Grover et al. 2021; Kumar et al. 2021). Indirect mechanism requires the involvement of plant defensive metabolic processes that react to the signals directed by PGPR and influence the plant; the examples are the induction of systemic resistance to plant pathogens and protection against abiotic stress conditions (Bal et al. 2013; Glick 2014; Goswami et al. 2016; Ijaz et al. 2021; Kumar et al. 2012; Nelson 2004).
In plant growth promotion, the promotion of root growth is one of the prominent effects of PGPR because root exudates contain a greater amount of sugar, amino acids, and organic acids. These root exudates are used by PGPR as their food source and numbers of bacteria around the root of the plant “microbial niches in the rhizosphere” are 10–1000 times higher compared to bulk soil (Glick 2014). The study of Li et al. (2019) reported ACC as strongest chemoattractant for ACC deaminase–producing bacteria and revealed that the colonization of the PGPR is based on chemotaxis of PGPR and ACC exuded from host plant roots (Li et al. 2019). Similarly, the plant’s roots exuding tryptophan are utilized by associated PGPR which synthesize and secrete indole acetic acid (IAA), some of which are provided to the host plant. The availability of exogenous IAA to the host plant not only stimulates plant cell proliferation and elongation but also induces the production of ACC synthase that catalyzes the synthesis of ACC, which is a non-protein amino acid and precursor of hormone ethylene. PGPR with ACC deaminase activity has the potential to enhance plant growth and development under abiotic stress conditions. Additionally, several PGPR such as Bacillus sp., Pseudomonas putida, Pseudomonas fluorescens, Pseudomonas veronii, Aneurinibacillus sp., and Paenibacillus sp. revealed ACC deaminase activity that enhance plant growth by reducing stress ethylene (Ali et al. 2018; Ali and Kim 2018; Glick 2014; Gupta and Pandey 2019; Penrose and Glick 2003; Penrose et al. 2001; Vanderstraeten and Van Der Straeten 2017). Under stressful conditions, ACC is exuded from plant tissues taken up by associated bacteria and cleaved into ammonia and α-ketobutyrate by bacterial ACC deaminase enzymes (Ali and Kim 2018; Glick 2014).
Additionally, the prolific effect of PGPR application is not only limited to their capability to promote the growth of the plant but also to their ability to metabolize different substrates and provide energy to the host plant. The use of strains with multiple plant growth–promoting traits could be a more effective approach for environmentally friendly crop production. The inoculation of multi-trait PGPR such as Brevundimonas sp., Bacillus sp., Pseudomonas sp., and Providencia sp. revealed consistent results in the growth and development of their host plant under normal and stressful conditions. Therefore, PGPRs with multiple traits are a good candidate for the formulation of biofertilizers and microbial biostimulants (Rana et al. 2011; Ruzzi and Aroca 2015; Tahir et al. 2006). Recently, significant results in crop growth and productivity have been reported by using multi-trait PGPR (Batista et al. 2018; Rana et al. 2011).
The coastal sand dune is considered a specialized ecosystem characterized by conditions which are hostile for life forms such as fluctuating temperature, high salt, and low organic matter content. Rhizospheric bacteria in such conditions have been found to produce plant growth–promoting biochemicals (i.e., IAA, siderophore, etc.) and are of great importance for agriculture. The rhizosphere of plants in sand dunes regions are rich in microbial diversity; however, the composition of microbial communities varies with salinity and fluctuating temperature (Nayak et al. 2019). PGPR isolation from the plants in coastal sand dunes (such as Pohang beach) has generally been neglected and attracted little attention from the researchers. Thus, the present study was aimed to isolate and identify multi-trait plant growth–promoting rhizospheric bacteria from the rhizospheric soils of Pohang beach.
Materials and methods
The study site
The soil samples were collected from closely related sites at Pohang beach (latitude: 36° 51′ 32.20″ N; longitude: 129° 45′ 34.90″ E), Republic of Korea. In Pohang, the summers are warm, wet, and mostly cloudy while the winters are very cold, windy, and mostly clear. Temperature typically varies from −2 to 30 °C throughout the year and both hot and cold seasons’ lasts for 3.0 to 3.5 months each. At the time of sampling in the spring season (April 2021), weather conditions at the sampling site were moderate with average temperature fluctuating between 20 °C (68°F) and 10 °C (50 °F), and average relative humidity about 58%. Plants growing on the coastal sand dunes at Pohang beach vary in structure and composition. The major plants growing at Pohang beach includes Calystegia soldanella, Ammophila breviligulata, Carex arenaria, Elymus mollis, Artiplex (orache or saltbush), Artemisia, Honckenya peploides, Spergularia, and Ipomoea pes-caprae (beach morning glory). The soil used in the study was collected from different sites which were located in the same region and each site was inhabiting the same types of plants. Three replicates of rhizospheric soil samples were collected from closely related sites for four different plants (Ammophila breviligulata, Artiplex, Calystegia soldanella, and Ipomoea pes-caprae).
Sampling of rhizospheric soil and isolation of rhizobacteria
A total of four soil samples (three replicates) were collected for the isolation of rhizosphere bacterial isolates. The plant’s roots were excised with rhizospheric soil by digging out intact root systems and were carefully taken in plastic zip bags, stored at 4 °C, and brought to Laboratory of Vegetables and Molecular Physiology, Yeungnam University for the isolation of bacteria. Rhizospheric bacteria were isolated from 1 g of soil sample tightly adhered to the root by serial dilution plating on Tryptic Soy Agar (TSA) and Luria-Bertani (LB) agar plates as described by Somasegaran and Hoben (2012). The plates of each sample were incubated at 28 °C ± 2 °C till the appearance of bacterial colonies. After the appearance of robust colonies, individual colonies were picked up and streaked on fresh LB plates for further purification.
Maintenance of the isolates
All selected isolates were maintained at 4 °C in equal volumes of nutrient broth and 40% glycerol for long-term use.
In vitro screening of bacterial isolates for their multi-trait PGP activities
Screening for indole acetic acid production
To screen the rhizobacterial isolates for IAA production, all the isolates obtained were grown in nutrient broth (50 mL) supplemented with and without L-tryptophan (0.15% w/v) and incubated at 28 °C ± 2 °C in shaking incubator at 200 rpm for 72 h in dark conditions. The bacterial culture was centrifuged at 6000 g for 10 min and screening of IAA in the supernatant was evaluated according to the detail method of Khan et al. (2016, 2020), with some modifications. One milliliter of supernatant was mixed with 1 mL of Salkowski reagents (2 mL of 0.5M FeCl3, 49 mL of 70% HClO4, and 49 mL of distilled water mixed in a glassware on ice in the fume hood) and subjected to dark condition for 30 min. The resultant change in color of the mixture was observed. The test was replicated two times.
Evaluation of phosphate solubilization capability of rhizobacterial isolates
The pure culture of selected isolates was screened for phosphate solubilization according to the detail method described by Khan et al. (2016) with slight modifications. The slight modifications were the addition of MgCl2 (5 g) and 16 g of agar in the media. A modified Pikovskaya’s agar media contains (per liter) dextrose (10 g), Ca3 (PO4)2 (5 g), yeast extract (0.500 g), MgCl2 (5 g), MgSO4 (0.100 g), KCl (0.200 g), (NH4)2SO4 (0.50 g), MnSO4 (0.10 g), FeSO4 (0.0001 g), and 16 g of agar. The reaction of the medium was then adjusted to pH 7.0 with sterile 1 M NaOH. The media was autoclaved and plated under sterile conditions. Each bacterial isolate was spot inoculated in the center of the plate and incubated at 28 °C ± 2 °C for 04 days until the formation of transparent “Halos” around each phosphate solubilizing bacterial colony. The isolates that formed a clear zone around the colonies were considered positive. A clear zone around a growing colony revealed phosphate solubilization and was measured as phosphate solubilization index (PSI), and was measured using the following formula (Pande et al. 2017). All the observations were recorded in triplicate.
Siderophores production
The detailed methods of Louden et al. (2011) and Khan et al. (2019) were followed for the assessment of siderophores production. Bacterial isolates were spot inoculated on chromeazurol “S” agar plates and incubated at 28 °C ± 2 °C for five consecutive days. The cultures were analyzed for the appearance of orange halos in contrast to blue backgrounds. The isolates that induced the formation of halos in contrast to blue background were considered positive for siderophores production.
ACC deaminase activity
The rhizobacterial isolates were screened for their ability to utilize ACC as a sole source of nitrogen in the Dworkin and Foster (DF) minimal medium according to the detail method of Penrose and Glick (2003) with some modifications. Two sets of DF minimal medium plates were prepared for the identification of ACC deaminase–producing bacteria. One set of plates was supplemented with ammonium sulfate ((NH4)2 SO4, 2 g/L), while the other set was supplemented with substrate ACC (3 mM) comprising a lawn of the substrate. The rhizobacterial culture was inoculated onto the medium plates and incubated at 28 °C for 3 days and the growth on the plates was checked on a daily basis.
Ninhydrin-ACC assay for selected rhizobacterial isolates
The detail method of Li et al. (2011) was used to determine the consumption of ACC by selected bacterial isolates through microtiter plate ninhydrin-ACC assay (Li et al. 2011). The consumption capability of ACC was tested for 06 rhizobacterial isolates grown in the DF-ACC medium and evaluated through a modified colorimetric ninhydrin-ACC assay with chimney-top 96-well microtiter plates with different concentrations of ACC (0.01, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 mmol/L). The reaction mix contained 60 µL of each tenfold diluted supernatant from bacterial culture and 160 µL of the ninhydrin reagents in a chimney-top 96-well microtiter plate and heated on boiling water for 30 min. The absorbance of microplate wells was measured at 570 nm with a spectrophotometer. The rhizobacterial isolates revealed visibly reduced color depth or lower absorbance of supernatant was compared with DF-ACC medium without inoculation and were considered ACC-utilizing bacteria.
Molecular identification
The bacterial isolates were identified on the basis of 16S ribosomal RNA (16S rRNA) gene sequence. The 16S rRNA gene was amplified by using the primers 27-F (5′–AGAGTTTGATC(AC)TGGCTAG–3′) and 1492-R (5′–CGG(CT)TACCTTGTTACGACTT–3′). The primers were complementary to the 5′ end and 3′ end of the prokaryotic 16S rRNA, respectively. The BLAST search program (http://www.ncbi.nlm.nih.gov/BLASTn) was used to compare the sequence homology of nucleotides, and closely related sequences obtained were aligned through CLUSTALW by using MEGA (version 6.0) software for phylogenetic analysis (Tamura et al. 2013). The maximum parsimony tree was constructed by using the same software, while the bootstrap replications (1000) were used as a statistical support for the nodes in the phylogenetic tree of the isolates.
Results
Isolation of plant growth–promoting rhizobacteria
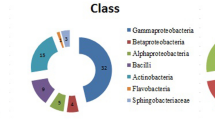
Rhizospheric soil samples from closely related sites at Pohang beach resulted in different numbers of rhizospheric bacteria; a total of 52 bacterial isolates were selected based on their cultural and morphological characteristics and processed for their plant growth–promoting traits. An equal number (13) of isolates were selected from each sample (Fig. 1) for the evaluation of their plant growth–promoting traits. The selected isolates from each sample (S1–S4) were labelled as S1 (S1T1, S1T2, S1T3, S1T4, S1T5, S1T6, S1T7, S1B1, S1T9, S1T10, S1T11, S1T12, S1T13), S2 (S2T1, S2T2, S2T3, S2T4, S2T5, S2T6, S2T7, S2B1, S2T9, S2T10, S2T11, S2T12, S2T13), S3 (S3T1, S3T2, S3T3, S3T4, S3T5, S3T6, S3T7, S3B1, S3T9, S3T10, S3T11, S3T12, S3T13), and S4 (S4T1, S4T2, S4T3, S4T4, S4T5, S4T6, S4T7, S4B1, S4T9, S4T10, S4T11, S4T12, S4T13).
Evaluation of multi-trait plant growth–promoting rhizobacteria
All isolates revealed varied plant growth–promoting traits. The evaluation of direct and indirect mechanisms for facilitating plant growth revealed the capabilities such as IAA and siderophore production, solubilization of phosphate, and production of ACC deaminase. All these activities were performed for a total 52 isolates. Our study revealed a large variation among different isolates with respect to plant growth–promoting traits and several of them possessed more than one PGP trait. A total of thirty isolates were positive for IAA production and thirty-one isolates were positive for phosphate solubilization, while twenty-six were positive for siderophore and twenty-nine revealed ACC deaminase production. Out of 52 isolates, 15 were positive for all four activities such as the production of ACC deaminase, siderophore, IAA, and phosphate solubilization (Fig. 2). More intestinally, six isolates revealed more robust results for all of the four plant growth–promoting traits. Based on initial screening, the isolates with multiple traits were selected for further processing and molecular identification.
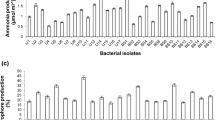
In individual traits assessment, most of the isolates were phosphate solubilizers (59.62%) and IAA producers (57.69%), whereas a low level of IAA production was observed in the absence of L. tryptophan while higher in the presence of L. tryptophan in the growth media. In the case of IAA, siderophore, and ACC deaminase, about 36% isolates showed positive results, whereas 40% were IAA and siderophore producers, and 44% were phosphate solubilizers along with IAA-producing capabilities. Similarly, 32% were phosphate solubilizers and siderophore producers, while 30% revealed production of siderophores, IAA, and phosphate solubilization (Table 1). Phosphate solubilizing potential was estimated by observing the halo zones around the colonies of selected rhizobacterial isolates on Pikovskaya agar media and phosphate solubilization index (PSI) was calculated. In the selected multi-trait PGPR isolates, five were found to have ≥ 3.0 phosphate solubilization index. Maximum PSI was observed by S3B1 (4.53 ± 0.22) followed by S2T9 (4.36 ± 0.13), S4T10 (4.17 ± 0.29), S1T1 (3.43 ± 0.26), and S1T13 (3.11 ± 0.08). In the present study, phosphate solubilizing multi-trait PGPR formed clear zones by solubilizing the suspended tricalcium phosphate in the media. On the other hand, 28% of the isolates were positive for all of the four activities in which 11.54% were showing robust results for all activities and were selected for further verification and screening to determine the consumption of ACC by selected bacterial isolates through microtiter plate ninhydrin-ACC assay. The consumption capability of ACC was tested for six rhizobacterial isolates because of their robust results on the DF media supplemented with ACC, and all other activities (Table 1). The selected rhizobacterial isolates were grown in the DF-ACC medium and evaluated through a modified colorimetric ninhydrin-ACC assay with chimney-top 96-well microtiter plates with different concentrations of ACC.
Ninhydrin-ACC assay for selected rhizobacteria
ACC determination through colorimetric ninhydrin assays authenticated our results of ACC utilization by selected rhizobacterial isolates and the bacterial isolates that revealed robust growth on DF-ACC media were further verified through 96-well microtiter plate assay and the concentration of ACC in the bacterial cultures was checked at 570 nm. The absorbance value at 570 nm of the 10-fold diluted supernatant of six ACC-utilizing rhizobacterial isolates revealed significant decrease as compared with diluted non-inoculated DF-ACC medium. Our results revealed that the concentration of ACC in the supernatant was reduced by 60–70% after 16 h and all isolates revealed differences in ACC utilization (Fig. 3).
Demonstrates the utilization of ACC by different bacterial isolates measured through 96-well plate ninhydrin-ACC assay. ACC concentration left behind (after 8, 16, and 24 h) in the DF-ACC medium which was supplemented with 3.0 mmol/L of ACC. Three replicates of each isolate were used for the utilization of ACC
Identification of multi-trait plant growth–promoting rhizobacteria
The rhizobacterial isolates with multi-trait plant growth–promoting attributes were subjected for molecular identification. For molecular identification and phylogenetic analysis of the isolates (06), the 16S rRNA genes were amplified and sequenced and compared against 16S rRNA sequences in the database. The sequences were submitted to NCBI to get the accession numbers (Fig. 4). Our detail analysis revealed that the rhizospheric bacteria with multiple traits S1T1, S1B1, S1T13, S2T9, S3B1, and S4T10 showed sequence identity with Serratia fonticola, Sphingobacterium multivorum, Brevundimonas vesicularis, Arthrobacter sp., Pseudomonas fluorescens, and Pseudomonas koreensis respectively. Moreover, the neighbor joining (NJ) methodology was used to construct the phylogenetic tree for 16S with MEGA 6.0 after sequence alignment by using CLUSTAL W. Our results revealed a high level of 16S rRNA sequence identity (99–100%) with their respective clades (Fig. 4) in which S1T1 shows a high level of 16S rRNA sequence identity (100%) with Serratia fonticola, while S3B1 and S4T10 exhibited a high level of sequence identity with P. fluorescens and P. koreensis. Similarly, S1T13 and S2T9 formed clades with Brevundimonas vesicularis and Arthrobacter sp., respectively, while S1B1 revealed a high level of 16S rRNA sequence identity (99%) with Sphingobacterium multivorum (Fig. 4).
Phylogenetic analysis of multi-trait plant growth–promoting rhizobacteria isolated from root rhizosphere of different plants at Pohang beach. The BLAST search program was used to compare nucleotide sequence similarity. The tree was constructed by using MEGA 6.0 package from 16S rRNA regions of the most similar rhizobacteria type strains related to those used in the present study retrieved from NCBI. The bootstrap replications (1K) were used as a statistical support for the nodes in the phylogenetic tree
Discussion
All plants host distinct microbial niches in their near vicinity (phyllosphere and rhizosphere) and inside their tissues (endosphere) designated as plant microbiota. The pragmatic analysis of these niches and plant-microbiome interactions develop our understanding in the profiling of microbial communities and type of relationship (mutualistic, commensalistic, and parasitic) of microbiota and host plant (Rodriguez et al. 2019; Schlaeppi and Bulgarelli 2015). Due to rich nutrients’ availability in the rhizospheric region, plant rhizosphere is known to be an ideal ecological niche for different types of soil microorganisms (Hakim et al. 2021; Khan et al. 2021; Prashar et al. 2014). Rhizobacteria with plant growth–promoting capabilities enhance plant growth and development under normal and stressful conditions by direct and indirect mechanisms including production of phytohormones, activation of phosphate solubilization, and production of siderophores and ACC deaminase (Moon and Ali 2022). There are various papers on the screening of PGPR from different regions but a few on the multi-trait PGPR isolated from the plant’s rhizosphere in the coastal sand dunes. Similarly, in the available literature, very little information is reported on the screening and using PGPRs isolated from the plants available at Pohang beach. In the present study, multi-trait PGPRs were isolated from rhizospheric soil samples of Pohang beach.
The isolated rhizobacterial isolates were screened for both direct and indirect plant growth–promoting traits and were identified on the basis of 16S rRNA gene sequences. Fifteen rhizobacterial isolates revealed multi-traits such as production of IAA, siderophore, ACC deaminase, and phosphate solubilization. On the other hand, six strains revealed prominent results for all activities. The rhizobacterial strains Pseudomonas fluorescence-S3B1 revealed maximum phosphate solubilization index (4.53 ± 0.22) followed by Arthrobacter sp.-S2T9 (4.36 ± 0.13), Pseudomonas koreensis-S4T10 (4.17 ± 0.29), and Serratia fonticola-S1T1 (3.43 ± 0.26), while Brevundimonas vesicularis-S1T13 (3.11 ± 0.08) and Sphingobacterium multivorum-S1B1 (2.61 ± 0.11) also revealed halo zones for phosphate solubilization in modified Pikovskaya’s agar media. Previous studies have also reported that some Pseudomonas sp., Serratia sp., and Arthrobacter sp. are efficient phosphate solubilizers (Banerjee et al. 2010; Zahir et al. 2011). The study of Reyes et al. (2007) and Kumar et al. (2012) also reported that phosphate solubilizing bacteria are more commonly found in the rhizospheric soil as compared to bulk soil and directly support plant growth (Kumar et al. 2012; Reyes et al. 2007). Our PSI results are consistent with the study of Pande et al. (2017) where they concluded that the application of PGPR that has the capability to solubilize phosphate could enhance the quantity of effective phosphate and enhance the growth and development of the plant (Pande et al. 2017; Yazdani et al. 2009). The PGPR strains reported in the present study have the ability to solubilize tricalcium phosphate and could be effective for enhancing crop growth and productivity under normal and abiotic stress conditions.
Similarly, another direct effect of PGPR on plants is the production of IAA which is an important phytohormone and basically functions as an essential signal molecule in the regulation of plant developmental processes. A number of different PGPRs are reported for the production of IAA while IAA-producing capability of PGPR is influenced by various factors such as cultural conditions, substrate availability, and growth stage (Lwin et al. 2012; Yousef 2018). In the present study, more than 57% bacterial isolates were positive for IAA production without adding L-tryptophan in the growth medium while upon the addition of L-tryptophan in the growth media, 73% PGPR revealed IAA-producing capabilities. The increased IAA production of some PGPR after addition of tryptophan suggests that the route of IAA biosynthesis is that these bacteria are dependent on tryptophan. Similarly, the stimulation of IAA synthesis by the availability of tryptophan was previously reported from gram-negative and gram-positive plant-associated bacteria. The capability of PGPR to colonize plant roots may depend on the ability of the bacteria to synthesize IAA because the synthesis of IAA contributes to rhizosphere competency (Idris et al. 2007; Koga et al. 1991). However, further experiments are needed to confirm the role of these PGPR in the mitigation of abiotic stress conditions and promotion of plant growth.
In different plants, the indirect effects of PGPR also alleviate or halt the deleterious effects of the stressful conditions by producing ACC deaminase and siderophores. Bacterial ACC deaminase lowers the levels of ethylene under stressful conditions and prevents the synthesis of stress ethylene by hydrolyzing ACC, which is an immediate precursor for ethylene biosynthesis. ACC deaminase–producing bacteria hydrolyze ACC into ammonia and α-ketobutyrate, limiting the availability of ACC for ethylene production and indirectly stimulating the growth of the plant (Ali and Kim 2018; Glick 2014). Initially, it was believed that the production of ACC deaminase is mostly related to free-living soil bacteria; however, recent studies revealed that various rhizospheric and endophytic strains are capable of producing ACC deaminase (Glick 2014; Khan et al. 2016). In the present study, the assessment of bacterial ACC deaminase revealed more than 55% rhizobacterial isolates positive for the production of ACC deaminase, whereas 21% showed prominent results for the production of ACC deaminase. The studies of Ali et al. (2018) and Bal et al. (2013) revealed that crop plants treated with ACC deaminase–producing bacteria (Pseudomonas and Bacillus) greatly alleviated the adverse effects of stressful conditions and enhanced the growth and development of the plants.
Similarly, siderophore-producing trait of PGPR is considered one of the important characteristics because siderophores are involved both in growth promotion and in the biocontrol of phytopathogens. In our results, about 50% PGPRs were siderophore producing while 19.23% showed prominent results on chromeazurol “S” agar plates and the appearance of orange halos in contrast to blue background. Multi-trait PGPRs have been reported by different researchers (Kumar and Gera 2014; Rana et al. 2011; Saeed et al. 2021) while such findings on indigenous stains of South Korea are less commonly examined. Siderophore-producing PGPRs play a pivotal role and are beneficial to the plants in mobilizing different metal ions and have also biocontrol properties (Matthijs et al. 2007). Siderophore and ACC deaminase–producing PGPR can mitigate the adverse effects of stressful conditions and help the plants to overcome stressful conditions.
In plant-microbe interactions, the potential and specificity of candidate microbes provide tolerance and mitigate the adverse effects of abiotic stressor in the plants. In the present study, rhizobacterial strains Pseudomonas koreensis-(S4T10), Serratia fonticola-(S1T1), Pseudomonas fluorescens-(S3B1), Brevundimonas vesicularis-(S1T13), Sphingobacterium multivorum-(S1B1), and Arthrobacter sp.-(S2T9) were found to most efficient multi-trait PGPR that produce phytohormone (IAA), solubilize phosphate, and produce siderophore and ACC deaminase. These types of studies are pivotal to substitute chemical fertilizers and pragmatise multi-trait PGPR as microbial biostimulants or biofertilizers.
Conclusion
The present study illustrates the significance of screening of multi-trait plant growth–promoting rhizobacteria under in vitro conditions and their anticipated role in plant growth promotion under normal and stressful conditions. This directed to the selection of effective plant growth–promoting rhizobacterial isolates—S4T10, S1T1, S3B1, S1T13, S1B1, and S2T9—which, as a result of their multiple PGPR traits, could prove effective in augmenting the growth and productivity of plants, even under stressful conditions. In the future, further studies are required to evaluate the roles of these PGPRs on crop plants under controlled conditions in a pot or field experiments and their role in root colonization under abiotic stress conditions.
References
Ahmad F, Ahmad I, Khan MS (2008) Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol Res 163:173–181. https://doi.org/10.1016/j.micres.2006.04.001
Ali MA, Naveed M, Mustafa A, Abbas A (2017) The good, the bad, and the ugly of rhizosphere microbiome. In: Kumar V, Kumar M, Sharma S, Prasad R (eds) Probiotics and plant health. Springer Singapore, Singapore, pp 253–290. https://doi.org/10.1007/978-981-10-3473-2_11
Ali S, Khan MA, Kim W-C (2018) Pseudomonas veronii KJ mitigates flood stress-associated damage in Sesamum indicum L. Appl Biol Chem 61:575–585
Ali S, Kim W-C (2018) Plant growth promotion under water: decrease of waterlogging-induced ACC and ethylene levels by ACC deaminase-producing bacteria. Front microbiol 9:1096
Bal HB, Das S, Dangar TK, Adhya TK (2013) ACC deaminase and IAA producing growth promoting bacteria from the rhizosphere soil of tropical rice plants. J Basic Microbiol 53:972–984
Banerjee S, Palit R, Sengupta C, Standing D (2010) Stress induced phosphate solubilization by ‘Arthrobacter’ sp. And ‘Bacillus’ sp. isolated from tomato rhizosphere. Aust J Crop Sci 4:378–383
Batista BD et al (2018) Screening of tropically derived, multi-trait plant growth-promoting rhizobacteria and evaluation of corn and soybean colonization ability. Microbiol Res 206:33–42
El-Tarabily KA, Nassar AH, Sivasithamparam K (2008) Promotion of growth of bean (Phaseolus vulgaris L.) in a calcareous soil by a phosphate-solubilizing, rhizosphere-competent isolate of Micromonospora endolithica. Appl Soil Ecol 39:161–171
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39. https://doi.org/10.1016/j.micres.2013.09.009
Goswami D, Thakker JN, Dhandhukia PC (2016) Portraying mechanics of plant growth promoting rhizobacteria (PGPR): a review. Cogent food agric 2(1)
Grover M, Bodhankar S, Sharma A, Sharma P, Singh J, Nain L (2021) PGPR mediated alterations in root traits: way toward sustainable crop production. Front Sustain Food Syst 287
Gupta S, Pandey S (2019) ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front Microbiol 10:1506
Hakim S et al (2021) Rhizosphere engineering with plant growth-promoting microorganisms for agriculture and ecological sustainability. Front Sustain Food Syst 5:16
Hartmann A, Rothballer M, Schmid M (2008) Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312:7–14. https://doi.org/10.1007/s11104-007-9514-z
Hiltner L (1904) Uber nevere Erfahrungen und Probleme auf dem Gebiet der Boden Bakteriologie und unter besonderer Beurchsichtigung der Grundungung und Broche. Arbeit Deut Landw Ges Berlin 98:59–78
Hirota Y, Fujii T, Sano Y, Iyama S (1978) Nitrogen fixation in the rhizosphere of rice. Nature 276:416–417
Idris EE, Iglesias DJ, Talon M, Borriss R (2007) Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant-Microbe Interact 20:619–626
Ijaz A et al (2021) Insights into manganese solubilizing Bacillus sp. for improving plant growth and manganese uptake in maize. Front in Plant Sci 12
Khan AL et al (2016) Indole acetic acid and ACC deaminase from endophytic bacteria improves the growth of Solanum lycopersicum. Elect J of Biotech 21:58–64
Khan M et al (2020) Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol 22:850–862
Khan MA et al (2019) Halotolerant rhizobacterial strains mitigate the adverse effects of NaCl stress in soybean seedlings. BioMed Res Internat 2019
Khan N, Ali S, Shahid MA, Mustafa A, Sayyed R, Curá JA (2021) Insights into the interactions among roots, rhizosphere, and rhizobacteria for improving plant growth and tolerance to abiotic stresses: a review. Cells 10:1551
Koga J, Adachi T, Hidaka H (1991) Molecular cloning of the gene for indolepyruvate decarboxylase from Enterobacter cloacae. Mol General Genet MGG 226:10–16
Kumar A, Kumar A, Devi S, Patil S, Payal C, Negi S (2012) Isolation, screening and characterization of bacteria from Rhizospheric soils for different plant growth promotion (PGP) activities: an in vitro study. Recent Res Sci Technol 4:1–5
Kumar SS, Mahale AG, Faizullah M, Krishna JR, Channa TK (2021) A critical review on plant growth-promoting rhizobacteria (PGPR) as a drought sress management tool. Inter. J Plant Soil Sci. https://doi.org/10.9734/IJPSS/2021/v33i2130657
Kumar V, Gera R (2014) Isolation of a multi-trait plant growth promoting Brevundimonas sp. and its effect on the growth of Bt-cotton 3. Biotech 4:97–101
Li T, Zhang J, Shen C, Li H, Qiu L (2019) 1-Aminocyclopropane-1-carboxylate: a novel and strong chemoattractant for the plant beneficial rhizobacterium Pseudomonas putida UW4. Mol Plant-Microbe Interact 32:750–759
Li Z, Chang S, Lin L, Li Y, An Q (2011) A colorimetric assay of 1-aminocyclopropane-1-carboxylate (ACC) based on ninhydrin reaction for rapid screening of bacteria containing ACC deaminase. Lett Appl Microbiol 53:178–185
Louden BC, Haarmann D, Lynne AM (2011) Use of blue agar CAS assay for siderophore detection. J Microbiol Biol Educ 12:51–53
Lwin KM, Myint MM, Tar T, Aung WZM (2012) Isolation of plant hormone (indole-3-acetic acid-IAA) producing rhizobacteria and study on their effects on maize seedling. Eng J 16:137–144
Mantelin S, Touraine B (2004) Plant growth-promoting bacteria and nitrate availability: impacts on root development and nitrate uptake. J Exper Bot 55:27–34
Matthijs S, Tehrani KA, Laus G, Jackson RW, Cooper RM, Cornelis P (2007) Thioquinolobactin, a Pseudomonas siderophore with antifungal and anti-Pythium activity. Envi Microbiol 9:425–434
Moon YS, Ali S (2022) Possible mechanisms for the equilibrium of ACC and role of ACC deaminase-producing bacteria Appl Microbiol Biotech 1–11. https://doi.org/10.1007/s00253-022-11772-x
Nayak S, Behera S, Dash PK (2019) Potential of microbial diversity of coastal sand dunes: need for exploration in Odisha Coast of India. Sci World J
Nelson LM (2004) Plant growth promoting rhizobacteria (PGPR): prospects for new inoculants. Crop Manag 3:1–7. https://doi.org/10.1094/CM-2004-0301-05-RV
Pande A, Pandey P, Mehra S, Singh M, Kaushik S (2017) Phenotypic and genotypic characterization of phosphate solubilizing bacteria and their efficiency on the growth of maize. J Genet Eng Biotech 15:379–391
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plantar 118:10–15
Penrose DM, Moffatt BA, Glick BR (2001) Determination of 1-aminocycopropane-1-carboxylic acid (ACC) to assess the effects of ACC deaminase-containing bacteria on roots of canola seedlings. Canad J Microbiol 47:77–80
Prashar P, Kapoor N, Sachdeva S (2014) Rhizosphere: its structure, bacterial diversity and significance. Rev Envi Sci Biotech 13:63–77
Rana A, Saharan B, Joshi M, Prasanna R, Kumar K, Nain L (2011) Identification of multi-trait PGPR isolates and evaluating their potential as inoculants for wheat. Annal Microbiol 61:893–900
Reyes I, Valery A, Valduz Z (2007) Phosphate-solubilizing microorganisms isolated from rhizospheric and bulk soils of colonizer plants at an abandoned rock phosphate mine. In: First international meeting on microbial phosphate solubilization. Springer, pp 69–75
Rodriguez PA, Rothballer M, Chowdhury SP, Nussbaumer T, Gutjahr C, Falter-Braun P (2019) Systems biology of plant-microbiome interactions. Mole Plant 12:804–821
Ruzzi M, Aroca R (2015) Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Scientia Horti 196:124–134
Saeed Q et al (2021) Rhizosphere bacteria in plant growth promotion, biocontrol, and bioremediation of contaminated sites: a comprehensive review of effects and mechanisms. Internat J Mol Sci 22:10529
Schlaeppi K, Bulgarelli D (2015) The plant microbiome at work. Mol Plant-Microbe Interact 28:212–217
Singh BK, Millard P, Whiteley AS, Murrell JC (2004) Unravelling rhizosphere–microbial interactions: opportunities and limitations. Trend Microbiol 12:386–393. https://doi.org/10.1016/j.tim.2004.06.008
Somasegaran P, Hoben HJ (2012) Handbook for rhizobia: methods in legume-Rhizobium technology. Springer Science & Business Media
Tahir M, Arshad M, Naveed M, Zahir Z, Shaharoona B, Ahmad R (2006) Enrichment of recycled organic waste with N fertilizer and PGPR containing ACC-deaminase for improving growth and yield of tomato. Soil Environ 25:105–112
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mole Biol Evol 30:2725–2729
Umar W et al (2020) Nitrogen and phosphorus use efficiency in agroecosystems. Resources Use Efficiency in Agriculture. Springer, pp 213–257
Upadhyay H, Gangola S, Sharma A, Singh A, Maithani D, Joshi S (2021) Contribution of zinc solubilizing bacterial isolates on enhanced zinc uptake and growth promotion of maize (Zea mays L.). Folia Microbiol 1–11
Vanderstraeten L, Van Der Straeten D (2017) Accumulation and transport of 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: current status, considerations for future research and agronomic applications. Front Plant Sci 8:38
Whipps JM (2001) Microbial interactions and biocontrol in the rhizosphere. J Exper Bot 52:487–511. https://doi.org/10.1093/jexbot/52.suppl_1.487
Yazdani M, Bahmanyar MA, Pirdashti H, Esmaili MA (2009) Effect of phosphate solubilization microorganisms (PSM) and plant growth promoting rhizobacteria (PGPR) on yield and yield components of corn (Zea mays L.). World Acad Sci Eng Technol 49:90–92
Yousef NM (2018) Capability of plant growth-promoting rhizobacteria (PGPR) for producing indole acetic acid (IAA) under extreme conditions. Eur J Biol Res 8:174–182
Zahir ZA, Zafar-ul-Hye M, Sajjad S, Naveed M (2011) Comparative effectiveness of Pseudomonas and Serratia sp. containing ACC-deaminase for coinoculation with Rhizobium leguminosarum to improve growth, nodulation, and yield of lentil. Biol Fertil Soils 47:457–465
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yong Sun Moon and Sajid Ali are equally contributed.
Rights and permissions
About this article
Cite this article
Moon, YS., Ali, S. Isolation and identification of multi-trait plant growth–promoting rhizobacteria from coastal sand dune plant species of Pohang beach. Folia Microbiol 67, 523–533 (2022). https://doi.org/10.1007/s12223-022-00959-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-022-00959-4