Abstract
Mycobacterium avium subsp. paratuberculosis (MAP) causes major problem in a wide range of animal species. In ruminant livestock including cattle, it causes a chronic disease called Johne’s disease, or paratuberculosis (pTB) which is currently considered as potential zoonosis, causing Crohn’s disease in humans. MAP infection susceptibility is suspected to be controlled by host genetics. Thus, selecting individuals according to their genetic structure could help to obtain bovine populations that are increasingly resistant to MAP infection. The aim of the present work was to investigate the association between toll-like receptor (TLR) \({ 1}\) (+1380 G/A), TLR1 (+1446 C/A), TLR4 (+10 C/T), TLR9 (+1310 G/A) and solute carrier family 11 member 1 (SLC11A1) (+1066 C/G) mutations and MAP infection status in 813 cattle comprising East Anatolian Red crossbred, Anatolian Black crossbred and Holstein breed. TLR1 (+1380 G/A) mutation showed an association with bovine MAP (\(P\!<\!0.05\)). For the TLR1 (+1380 G/A) locus, the odds ratio for AG and AA genotypes versus GG genotypes were 2.31 (1.24–4.30; 95% confidence interval (CI)) and 0<0.001 (<0.001 to >999.999; 95% CI) which indicated that a proportion of AG homozygote was significantly higher in pTB-affected animals as compared with the control. General linear model analysis demonstrated higher MAP antibody response in TLR1 (+1380 AG) genotype as compared with TLR1 (+1380 GG) (\(P\!<\! 0.0001\)). Present findings suggest that selection against TLR1 (+1380 G/A) may reduce the risk of pTB in bovine herds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Paratuberculosis (pTB), or Johne’s disease is an intestinal infection caused by Mycobacterium avium subsp. paratuberculosis (MAP) (Chaubey et al. 2016). Animals are chronically infected, usually at a young age, and are generally believed to undergo an extended latent period (Marcé et al. 2010). The clinical signs of pTB in cattle are easily recognized by pipestream diarrhoea, weight loss and ultimately death (Fecteau 2018). Infections with MAP are difficult to control because of the long incubation periods (1–7 years), absence of clinical signs until advanced stages of the disease and lack of completely reliable diagnostic methods in the preclinical stages of the disease (Fecteau 2018). MAP infection is known to result in lower milk production, decreased reproductive performance, early culling or latter fatal diarrhoea (Smith et al. 2017). Livestock disease costs have been estimated to be up to 20% and 35–50% of turnover within the livestock sector in developed and developing countries, respectively. In the US, MAP disease causes estimated losses of $200 million every year (Cho et al. 2012). Eradication has been considered as the efficient way for controlling MAP. However, diagnosis of the disease could be problematic due to the lack of efficient diagnostic tools (Collins 2011). Therefore, the high costs involved in testing and culling approaches have shifted the focus to control strategies providing better benefit/cost ratios of more conservative strategies like vaccination (Juste 2012). Vaccination using killed inactivated vaccines has also been attempted for MAP control. The vaccines are said to afford protection by delaying the onset of clinical disease, but they does not reduce the number of infected animals (Wentink et al. 1994; Tewari et al. 2014).
Susceptibility to MAP infection can also be genetically influenced. Previous studies indicated that certain cattle breeds (Guernsey, Jersey and Bos indicus breeds) and family lines might have a genetic predisposition for MAP infections (Roussel et al. 2005; Elzo et al. 2006). Breeding for disease susceptibility is a central focus of livestock breeding programmes, as any successful variety must have the complete package of high yield, disease resistance, agronomic performance and end-use quality (Bishop and Woolliams 2014). Therefore, selection may be used as an additional tool for control of pTB (Bishop and Woolliams 2014; Bishop 2015). The use of genetic markers with the phenotypes in a process called marker-assisted selection (MAS) is mainly important and useful for phenotypes with low heritability (Meuwissen et al. 2013) such as trait-like antibody response to MAP which was estimated to be about 0.1 (Mortensen et al. 2004; Hinger et al. 2008). Recently, five single-nucleotide polymorphisms (SNPs) namely toll-like receptor (TLR) \({ 1}\) (+1380 G/A), TLR1 (+1446 C/A), TLR4 (+10 C/T), TLR9 (+1310 G/A) and solute carrier family 11 member 1 (SLC11A1) (+1066 C/G) have been found to be associated with bovine brucellosis (Prakash et al. 2014). TLRs are cell-surface receptors mediating activation of innate immunity that controls host defence against pathogens (Takeda and Akira 2005). TLRs target a range of microbial ligands, including lipopolysaccharide (TLR4), lipoproteins (TLR2) and unmethylated CpG motifs in DNA (TLR9) (Akira et al. 2001). Expression of TLRs on innate immune cells link microbial recognition to induction of antimicrobial mechanisms, such as production of reactive oxygen and nitrogen species and expression of antimicrobial peptides (AMPs) (Arpaia et al. 2011). Associations of different TLRs with various bovine diseases (Werling et al. 2009) including pTB infection (Fisher et al. 2011) were previously presented in the literature. SLC11A1 (formerly NRAMP1) has many pleiotropic effects on immune cells including regulation of chemokines and interleukins, major-histocompatibility complex (MHC) class II molecules, tumour necrosis factor \(\upalpha \) (TNF\(\upalpha )\), as well as antimicrobial activity (Blackwell et al. 2001). The SLC11A1 protein prevents intracellular bacterial growth through transportation of divalent metal ions, for the cellular recycling of the latter (Ruiz-Larrañaga et al. 2010).
Microorganisms are mainly destroyed by the host tissues through processes that usually involve phagocytosis and lysosomal disruption (Trangoni et al. 2015). However, some microorganisms establish themselves within the cell and spend all or part of their life intracellularly such as genus Mycobacterium and Brucella bacterium (Trangoni et al. 2015) and genetics of host protection could be common for different genera of intracellular pathogens (Yap et al. 2006). It is hypothesized in this study that SNPs associated with bovine brucellosis caused by intracellular pathogen, may also show association with another intracellular pathogen (MAP)-borne bovine pTB. Therefore, the aim of this study was to investigate associations between TLR4 (+10 C/T), TLR9 (+1310 G/A), TLR1 (+1380 G/A), TLR1 (+1446 C/A) and SLC11A1 (+1066 C/G) polymorphisms with bovine pTB in the Holstein population reared in Turkey and Turkish indigenous cattle crossbreds.
Materials and methods
Sample collection
All biological materials were collected from cattle slaughtered (showing clinical signs of pTB as cachexia and diarrhoea) at a local abattoir. Following the slaughter, blood samples were collected into vacutainer tubes (B.D. Bioscience, Germany) with ethylenediaminetetraacetic acid (EDTA) as an anticoagulant for genetic analysis and into vacutainer tubes (B.D. Bioscience, Germany) without an anticoagulant for serological analysis. All samples were delivered back to the laboratory in an ice box. Blood samples were collected from 2–3-year-old East Anatolian Red crossbred (\(n=288\)), Anatolian Black crossbred (\(n=112\)) and Holstein (\(n=418\)) breeds from Kayseri province and vicinity, Turkey. Animals used in the present study were housed under similar environmental conditions and not vaccinated for pTB. Blood samples were used for the isolation of genomic DNA for restriction fragment length polymorphism (RFLP) genotyping of the cattle under study, whereas serum samples were used for detection of MAP antibodies by an enzyme-linked immunosorbent assay (ELISA). All experimental procedures were performed in accordance with the guidelines of the Local Ethics Committee for Animal Experiments at Erciyes University.
Genomic DNA isolation and genotyping
Genomic DNA was extracted from whole blood using the standard phenol–chloroform extraction procedure. DNA concentration of the samples was quantified by using a Nano Drop (NanoDrop, Thermo Fisher Scientific, Waltham, USA) and stored at −20\(^{\circ }\)C until use. Primer pairs and restriction enzymes were obtained from Prakash et al. (2014) (see table 1). Polymerase chain reaction (PCR) was carried out using 50 ng of genomic DNA in a total reaction volume of 25 \(\mu \)L containing 10\(\times \) PCR buffer, 1.5 mM MgCl\(_{2}\), 200 \(\mu \)M dNTPs, 10 pM of forward and reverse primers and 0.5 U of Taq DNA polymerase (Sigma-Aldrich, Darmstadt, Germany). Amplification reactions were performed with an initial denaturation step of 5 min at 94\(^{\circ }\)C, followed by 35 cycles of 30 s at 94\(^{\circ }\)C, 40 s of x \(^{\circ }\)C (provided in table 1) and 30 s at 72\(^{\circ }\)C with a final extension step of 7 min at 72\(^{\circ }\)C. PCR products were screened by 1.5% agarose gel (Sigma-Aldrich) electrophoresis stained with ethidium bromide and visualized in a gel documentation system (Bio-Rad, Hercules, USA). These PCR fragments obtained with the primers were analysed by means of RFLP. Results from the PCR amplification were scored on agarose gel according to the band pattern of 100 base-pair ladder (Thermo Fisher Scientific). Suitable restriction enzymes (Thermo Fisher Scientific) were selected to identify RFLP patterns. All enzymes were allowed to react in a PCR machine at a temperature of 37\(^{\circ }\)C for 6 h. Enzymes used for the PCR-RFLP are indicated in table 1. The digestion products were separated by horizontal electrophoresis in 3% agarose gel in 0.5\(\times \) tris/borate/EDTA (135 V for 20 min) stained with ethidium bromide solution prior to visualization under ultraviolet light.
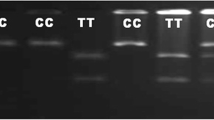
(a) Restriction digestion with the BstUI enzyme of bovine TLR4 (+10 C/T) SNP-amplified PCR products showing different genotypes in 3% agarose gel (a, 246 bp; b, 405 bp; c, 159 bp). (b) Restriction digestion with the BseRI enzyme of bovine TLR9 (+1310 G/A) SNP-amplified PCR products showing only a genotype in 3% agarose gel (only 392 bp). (c) Restriction digestion with the BslI enzyme of bovine TLR1 (+1380 G/A) SNP-amplified PCR products showing different genotypes in 3% agarose gel (a, 179 bp; b, 93 bp; c, 86 bp). (d) Restriction digestion with the BstXI enzyme of bovine TLR1 (+1446 C/A) SNP-amplified PCR products showing different genotypes in 3% agarose gel (a, 354 bp; b, 187 bp; c, 167 bp). (e) Restriction digestion with the PstI enzyme of bovine SLC11A1 (+1066 C/G) SNP-amplified PCR products showing different genotypes in 3% agarose gel (a, 374 bp; b, 293 bp; c, 81 bp). A 100-bp ladder was used in all figures.
Detection of MAP-specific antibodies by ELISA
Serum samples (\(n=818\)) were screened for detection of MAP-specific antibodies by using an ELISA kit (Idexx Paratuberculosis Screening, Maine, USA). Optical density (OD) values were measured at 450 nm using a microplate reader Elx800 (Bio-Tek Instruments, Winooski, USA). Positive and negative sera were included as controls. As per the manufacturer’s instruction, serum samples with corrected sample/positive control ratio above 55% were considered as positive for pTB.
Statistical analysis
Data were analysed using SAS 9.2 software (SAS Institute, Cary, USA). Allele and genotype frequencies were calculated using the ALLELE procedure of SAS v9.2. The univariable analysis for logistic regression considered the infection status as categorical response variable (yes/no), and SNPs (TLR4 and TLR9 have two genotypes, TLR1 and SLC11A1 have three genotypes, therefore respective loci have two or three levels), breed (three groups, i.e. two indigenous crossbred and Holstein) and sex (male and female) were included as possible explanatory variables. Genotypes were considered as ordinal variables and as class variables with the major homozygous genotype deemed as baseline. Data were analysed using the PROC LOGISTIC procedure and odds ratio (OR) with 95% confidence interval (CI) were calculated. ELISA OD data were checked for normality before analyses with the PROC UNIVARIATE procedure of SAS v9.2 (SAS Institute). Associations of different genotypes and OD values were tested using PROC GLM. Genotypic comparisons were reported following Tukey–Kramer adjustment; \(P\!\le \! 0.05\) was considered significant.
Results
Five SNP loci detected by PCR-RFLP are shown in figure 1. Initially, a total of 818 individuals were subjected to RFLP analysis, however, 813 cattle forTLR4 (+10 C/T), TLR9 (+1310 G/A), TLR1 (+1380 G/A), 817 animals for TLR1 (+1446 C/A) and 815 animals for SLC11A1 (+1066 C/G) were genotyped due to PCR or RFLP artefacts. As presented in table 1 in electronic supplementary material at http://www.ias.ac.in/jgenet/, the genotypic and allelic frequencies of all SNPs, except for TLR9 (+1310 G/A) in Holstein cattle and for SLC11A1 (+1066 C/G) in Anatolian Black crossbred cattle, exceeded threshold as present in both the case and control groups, suggesting that these SNPs were significant polymorphic loci.
The presence of pTB in East Anatolian Red crossbred, Anatolian Black crossbred and Holstein cattle was tested by ELISA. The number of positive results were reported in table 2. The resulting ratio of cases to controls was 1:14.4 (53 cases versus 765 controls) in the cattle aged \(\ge \)2 years both for cases and controls. The seropositivity of pTB was 2.5, 3.7 and 11.1% for East Anatolian Red crossbred, Anatolian Black crossbred and Holstein cattle, respectively. The lowest was noticed in the East Anatolian Red crossbred and the highest in Holstein cattle. Among the animals with pTB, mean OD values were 1.96, 2.18 and 2.61, respectively, for East Anatolian Red crossbred, Anatolian Black crossbred and Holstein cattle (table 2).
Two analyses were applied to investigate association between SNPs and pTB status of cattle. Firstly, association analysis was applied in accordence to case (pTB) and control (disease free) status of animals. Secondly, 53 animals which have pTB according to ELISA OD values (\(\ge \)1.4) were subjected to association analysis. The results from the univariate logistic regression for case–control and generalized linear model (GLM) for OD values indicated a non-significant effect for sex and a significant effect for breed except for TLR1 (+1380 G/A) SNP (table 3). Significant differences in genotype frequencies between two groups of individuals tested positive or negative for pTB infection were found only for TLR1 (+1380 G/A) polymorphism (\(P=0.0042\), table 3). The estimated ORs for the univariable logistic regression models are presented in table 3. At the TLR1 (+1380 G/A) locus, the OR of AG genotype versus GG genotype was 0.31 (0.16–0.62; 95% CI) in case (pTB) which revealed that relative proportion GG genotypes was significantly higher in case population than in control population, whereas AA genotype was exclusively not present in pTB positive animal and was present in three positive animals in the control population and its OR was towards infinity. Results of GLM are given in table 4. TLR1 (+1380 G/A) was found to be associated with ELISA OD values (\(P\!<\!0.0001\), table 4). For the TLR1 (+1380 G/A), locus genotype AG yielded higher antibody response as compared with genotype GG and none of the pTB animals showed genotype AA.
No significant differences in genotype frequencies between the cases and controls were found for the TLR4 (+10 C/T), TLR9 (+1310 G/A), TLR1 (+1446 C/A) and SLC11A1 (+1066 C/G) genotypes (table 3). Similarly, the GLM analysis did not show association between any of the polymorphisms analysed and ELISA OD values (table 4).
Discussion
pTB creates significant problem to the world dairy and beef industries and it is considered as a potential zoonotic disease (Pinedo et al. 2009). MAP has been incriminated as a triggering agent for Crohn’s disease in genetically susceptible individuals in human (Sweeney et al. 2012). The role of genetic factors in association with susceptibility and resistance to mycobacterial infection including bovine pTB had been discussed in studies which are focussed on a candidate gene level (Mucha et al. 2009; Koets et al. 2010; Ruiz-Larrañaga et al. 2011; Pinedo et al. 2013) and estimations of heritability to MAP infection ranging from 0.01 to 0.183 have been reported in Holstein cattle (Kirkpatrick and Shook 2011). Currently, a few studies are involved in exploring association of host-candidate genes with susceptibility to MAP infection in cattle (Kirkpatrick and Shook 2011). Some of previous works reported positive association with MAP for the selected candidate genes, such as interferon gamma (BoIFNG), TLR4 and SLC11A1 (Mucha et al. 2009; Pinedo et al. 2009) which were also associated with bovine brucellosis (Prakash et al. 2014). However, in the present study, no associations were identified between genes including TLR4 (+10 C/T), TLR9 (+1310 G/A), TLR1 (+1380 G/A) and SLC11A1 (+1066 C/G) and MAP infections. This may have occurred due to two reasons. Firstly, none of the markers investigated in the current study may not be in linkage disequilibrium with genes affecting disease susceptibility. Secondly, genotype frequency did not show any difference between pTB-infected and noninfected animals. Unlike association mapping techniques that are attempting to detect linkage disequilibrium indirectly between marker alleles and phenotype, in case–control tests, this is done by comparing marker allele or genotype frequencies among affected individuals with their frequencies among unaffected individuals (Nielsen and Weir 2001). On the other hand, coinciding with our results, some experiments did not identify any association between investigated gene polymorphisms such as caspase recruitment domain 15 (CARD15) and TLR4 and pTB (White et al. 2003; Taylor et al. 2006).
The results of this study, performed with the Turkish indigenous cattle crossbreds for the first time, suggest that TLR1 (+1380 G/A) gene mutation might have a role in pTB susceptibility, but none of these have been previously highlighted by the other authors. Breed effect was reported as one of the factors that influence variation in susceptibility to pTB (Cetinkaya et al. 1997; Roussel et al. 2005; Elzo et al. 2006). Similarly, it was in the present study that breed had effects on pTB. The MAP-specific serum antibody level was significantly different between the investigated breeds of the present study (table 2). Holstein cattle breed showed the highest ELISA OD values, East Anatolian Red crossbred showed the lowest and Anatolian Black crossbred remained in the middle (table 2) which shows that Turkish native cattle breeds exhibit better resistance against MAP infections compared with Holstein cattle breed. The present findings are consistent with the findings of a previous experiment that investigated susceptibility to bovine tuberculosis in which native Zebu cattle were found more resistant to bovine tuberculosis than the exotic Holstein–Friesian cattle (Vordermeier et al. 2012). Roussel et al. (2005) also stated that cattle from B. indicus-based herds were more than 17 times as likely to be seropositive as were cattle from B. taurus-based herds, and cattle from interspecies-based herds were 3.6 times as likely to be seropositive as were cattle from B. taurus-based herds. However, no study was devoted to compare MAP infection status of exotic cattle breeds with the native or native crossbred cattle breeds in Turkey.
TLR1, when dimerized with TLR2, is a cell-surface receptor that, upon recognition of bacterial lipoproteins, activates the innate immune system (Misch et al. 2008). In the present experiment, TLR1 (+1380 G/A) SNP was found to be associated with MAP infections in cattle which are reared in Turkey (tables 3&4). TLR1 has been implicated as a coreceptor together with TLR2 in the recognition of bacterial triacyl lipopeptides which is an extracellular mycobacterial ligand (Wiersinga et al. 2007) and TLR1-deficient mice were defective in their response to bacterial lipopeptides that are triacylated (Takeda et al. 2002). Different TLR1 variants were found to be associated with mycobacterial signalling including cattle MAP (Mucha et al. 2009; Ruiz-Larrañaga et al. 2011) and M. bovis in Holstein cattle (Sun et al. 2012). Additionally, two quantitative trait loci associated with susceptibility to MAP in Holstein cattle were identified in the same region where TLR1 located on BTA6 (Kirkpatrick et al. 2011).
For the TLR1 (+1380 G/A) locus, the OR for AG genotypes versus GG genotypes was 2.31 (1.24–4.30; 95% CI) which revealed that relative proportion AG genotypes was significantly higher in the case population than in the control population, whereas the AA genotype was not present in any pTB positive animals and its OR was low and near to zero in pTB-affected animals as compared with the control. It indicated that the AG genotype at the TLR1 (+1380 G/A) locus was responsible for increasing the relative risk of bovine pTB and hence selection against the AG genotype may reduce seropositivity of pTB in cattle (table 3). Prakash et al. (2014) also investigated association between TLR1 (+1380 G/A) SNP and brucella susceptibility in indigenous Shri Mataji Goshala and Barsana cattle and found controversial to present results; proportion of GG homozygote was significantly higher in brucellosis-affected animals as compared with the control. Although, both pTB and brucella are intracellular bacterial pathogen caused diseases, susceptibility in terms of TLR1 (+1380 G/A) SNP maintained with different genotypes among the cattle reared in Turkey and in India. This may be due to genetic origins of cattle breeds in Turkey and India. While cattle breeds in Turkey belong to humpless, the B. taurus lineage, humped Indian cattle breeds belong to B. indicus and clear phylogenetic distinction among these cattle breeds were shown genetically (Lin et al. 2010; Randhawa et al. 2016).
In conclusion, in the present study, genetic polymorphism was found in Holstein and two Turkish indigenous cattle crossbreds, and the association between genetic polymorphism of the synonymous TLR1 (+1380 G/A) and risk of pTB infection. An ELISA antibody response showed that East Anatolian Red crossbred and Anatolian Black crossbred were more resistant to pTB as compared with Holsteins reared in Turkey. Additionally, selecting animals against TLR1 +1380 AG genotype may decrease the risk of pTB in cattle which belongs to B. taurus species. Further analyses through expanded sampling in some breeds and detailed studies under various molecular levels are recommended to provide more consistent explanations for the molecular mechanisms of mutation-related risk of susceptibility.
References
Akira S., Takeda K. and Kaisho T. 2001 Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680.
Arpaia N, Godec J, Lau L, Sivick K. E., McLaughlin L. M., Jones M. B. et al. 2011 TLR signaling is required for Salmonella typhimurium virulence. Cell 144, 675–688.
Bishop S. C. 2015 Genetic resistance to infections in sheep. Vet. Microbiol. 181, 2–7.
Bishop S. C. and Woolliams J. A. 2014 Genomics and disease resistance studies in livestock. Livest. Sci. 166, 190–198.
Blackwell J. M., Goswami T., Evans, C. A. W., Sibthorpe D., Papo N., White J. K. et al. 2001 SLC11A1 (formerly NRAMP1) and disease resistance. Cell. Microbiol. 3, 773–784.
Cetinkaya B., Erdogan H. M. and Morgan K. L. 1997 Relationships between the presence of Johne’s disease and farm and management factors in dairy cattle in England. Prev. Vet. Med. 32, 253–266.
Chaubey K. K., Gupta R. D., Gupta S., Singh S. V., Bhatia A. K., Jayaraman S. et al. 2016 Trends and advances in the diagnosis and control of paratuberculosis in domestic livestock. Vet. Q. 36, 203–227.
Cho J., Tauer L. W., Schukken Y. H., Gómez M. I. Smith R. L., Lu Z. et al. 2012 Economic analysis of Mycobacterium avium subspecies paratuberculosis vaccines in dairy herds. J. Dairy Sci. 95, 1855–1872.
Collins M. T. 2011 Diagnosis of paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 27, 581–591.
Elzo M. A., Rae D. O., Lanhart S. E., Wasdin J. G., Dixon W. P. and Jones J. L. 2006 Factors associated with ELISA scores for paratuberculosis in an Angus-Brahman multibreed herd of beef cattle. J. Anim. Sci. 84, 41–48.
Fecteau M. E. 2018 Paratuberculosis in cattle. Vet. Clin. North Am. Food Anim. Pract. 34, 209–222.
Fisher C. A., Bhattarai E. K., Osterstock J. B., Dowd S. E., Seabury P. M., Vikram M. et al. 2011 Evolution of the bovine TLR gene family and member associations with Mycobacterium avium subspecies paratuberculosis infection. PLoS One 6, e27744.
Hinger M., Brandt H. and Erhardt G. 2008 Heritability estimates for antibody response to Mycobacterium avium subspecies paratuberculosis in German Holstein cattle. J. Dairy Sci. 91, 3237–3244.
Juste R. A. 2012 Current strategies for eradication of paratuberculosis and issues in public health. Vet. Immunol. Immunopathol. 148, 16–22.
Kirkpatrick B. W. and Shook G. E. 2011 Genetic susceptibility to paratuberculosis. Vet. Clin. North Am. Food Anim. Pract. 27, 559–571.
Kirkpatrick B. W., Shi X., Shook G. E. and Collins M. T. 2011 Whole-genome association analysis of susceptibility to paratuberculosis in Holstein cattle. Anim. Genet. 42, 149–160.
Koets A., Santema W., Mertens H., Oostenrijk D., Keestra M., Overdijk M. et al. 2010 Susceptibility to paratuberculosis infection in cattle is associated with single nucleotide polymorphisms in Toll-like receptor 2 which modulate immune responses against Mycobacterium avium subspecies paratuberculosis. Prev. Vet. Med. 93, 305–315.
Lin B. Z., Sasazaki S. and Mannen H. 2010 Genetic diversity and structure in Bos taurus and Bos indicus populations analyzed by SNP markers. Anim. Sci. J. 81, 281–289.
Marcé C., Ezanno P., Weber M. F., Seegers H., Pfeiffer D. U. and Fourichon C. 2010 Invited review: modeling within-herd transmission of Mycobacterium avium subspecies paratuberculosis in dairy cattle: a review. J. Dairy Sci. 93, 4455–4470.
Meuwissen T., Hayes B. and Goddard M. 2013 Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 1, 221–237.
Misch E. A., Macdonald M., Ranjit C., Sapkota B. R., Wells R. D., Siddiqui M. R. et al. 2008 Human TLR1 deficiency is associated with impaired mycobacterial signaling and protection from leprosy reversal reaction. PLoS Neglected Trop. Dis. 2, e231.
Mortensen H., Nielsen S. S. and Berg P. 2004 Genetic variation and heritability of the antibody response to Mycobacterium avium subspecies paratuberculosis in Danish Holstein cows. J. Dairy Sci. 87, 2108–2113.
Mucha R., Bhide M. R., Chakurkar E. B., Novak M. and Mikula I. 2009 Toll-like receptors TLR1, TLR2 and TLR4 gene mutations and natural resistance to Mycobacterium avium subsp. paratuberculosis infection in cattle. Vet. Immunol. Immunopathol. 128, 381–388.
Nielsen D. M. and Weir B. S. 2001 Association studies under general disease models. Theor. Popul. Biol. 60, 253–263.
Pinedo P. J., Buergelt C. D., Donovan G. A., Melendez P., Morel L., Wu R. et al. 2009 Candidate gene polymorphisms (BoIFNG, TLR4, SLC11A1) as risk factors for paratuberculosis infection in cattle. Prev. Vet. Med. 91, 189–196.
Pinedo P. J., Galvão K. N. and Seabury C. M. 2013 Innate immune gene variation and differential susceptibility to uterine diseases in Holstein cows. Theriogenology 80, 384–390.
Prakash O., Kumar A., Sonwane A., Rathore R., Singh R. V., Chauhan A. et al. 2014 Polymorphism of cytokine and innate immunity genes associated with bovine brucellosis in cattle. Mol. Biol. Rep. 41, 2815–2825.
Randhawa I. A. S., Khatkar M. S., Thomson P. C. and Raadsma H. W. 2016 A meta-assembly of selection signatures in cattle. PLoS One 11, e0153013.
Roussel A. J., Libal M. C., Whitlock R. L., Hairgrove T. B., Barling K. S. and Thompson J. A. 2005 Prevalence of and risk factors for paratuberculosis in purebred beef cattle. J. Am. Vet. Med. Assoc. 226, 773–778.
Ruiz-Larrañaga O., Garrido J. M., Manzano C., Iriondo M., Molina E., Gil A. et al. 2010 Identification of single nucleotide polymorphisms in the bovine solute carrier family 11 member 1 (SLC11A1) gene and their association with infection by Mycobacterium avium subspecies paratuberculosis. J. Dairy Sci. 93, 1713–1721.
Ruiz-Larrañaga O., Manzano C., Iriondo M., Garrido J. M., Molina E., Vazquez P. et al. 2011 Genetic variation of toll-like receptor genes and infection by Mycobacterium avium ssp. paratuberculosis in Holstein-Friesian cattle. J. Dairy Sci. 94, 3635–3641.
Smith R. L., Al-Mamun M. A. and Gröhn, Y. T. 2017 Economic consequences of paratuberculosis control in dairy cattle: a stochastic modeling study. Prev. Vet. Med. 138, 17–27.
Sun L., Song Y., Riaz H., Yang H., Hua G., Guo A. et al. 2012 Polymorphisms in toll-like receptor 1 and 9 genes and their association with tuberculosis susceptibility in Chinese Holstein cattle. Vet. Immunol. Immunopathol. 147, 195–201.
Sweeney R. W., Collins M. T., Koets A. P., McGuirk S. M. and Roussel A. J. 2012 Paratuberculosis (Johne’s disease) in cattle and other susceptible species. J. Vet. Intern. Med. 26, 1239–1250.
Takeda K. and Akira S. 2005 Toll-like receptors in innate immunity. Int. Immunol. 17, 1–14.
Takeda K., Takeuchi O. and Akira S. 2002 Recognition of lipopeptides by toll-like receptors. J. Endotoxin Res. 8, 459–463.
Taylor K. H., Taylor J. F., White S. N. and Womack J. E. 2006 Identification of genetic variation and putative regulatory regions in bovine CARD15. Mamm. Genome 17, 892–901.
Tewari D., Hovingh E., Linscott R., Martel E., Lawrence J., Wolfgang D. et al. 2014 Mycobacterium avium subsp. paratuberculosis antibody response, fecal shedding, and antibody cross-reactivity to Mycobacterium bovis in M. avium subsp. paratuberculosis-infected cattle herds vaccinated against Johne’s disease. Clin. Vaccine Immunol. 21, 698–703.
Trangoni M. D., Gioffré A. K., Cerón Cucchi M. E., Caimi K. C., Ruybal P., Zumárraga M. J. et al. 2015 LAMP technology: rapid identification of Brucella and Mycobacterium avium subsp. paratuberculosis. Braz. J. Microbiol. 46, 619–626.
Vordermeier M., Ameni G., Berg S., Bishop R., Robertson B. D., Aseffa A. et al. 2012 The influence of cattle breed on susceptibility to bovine tuberculosis in Ethiopia. Comp. Immunol. Microbiol. Infect. Dis. 35, 227–232.
Wentink G. H., Bongers J. H., Zeeuwen A. A. and Jaartsveld F. H. 1994 Incidence of paratuberculosis after vaccination against M. paratuberculosis in two infected dairy herds. Zentralbl. Vet. B 41, 517–522.
Werling D., Jann O. C., Offord V., Glass E. J. and Coffey T. J. 2009 Variation matters: TLR structure and species-specific pathogen recognition. Trends Immunol. 30, 124–130.
White S. N., Taylor K. H., Abbey C. A., Gill C. A. and Womack J. E. 2003 Haplotype variation in bovine Toll-like receptor 4 and computational prediction of a positively selected ligand-binding domain. Proc. Natl. Acad. Sci. U.S.A. 100, 10364–10369.
Wiersinga W. J., Wieland C. W., Dessing M. C., Chantratita N., Cheng A. C., Limmathurotsakul D. et al. 2007 Toll-like receptor 2 impairs host defense in gram-negative sepsis caused by Burkholderia pseudomallei (Melioidosis). PLoS Med. 4, e248.
Yap G. S., Shaw M. H., Ling Y. and Sher A. 2006 Genetic analysis of host resistance to intracellular pathogens: lessons from studies of Toxoplasma gondii infection. Microbes Infect. 8, 1174–1188.
Acknowledgements
This research was financially supported by the Turkish Research Council (TUBITAK) grant number 115O478.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Silvia Garagna
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cinar, M.U., Hizlisoy, H., Akyüz, B. et al. Polymorphisms in toll-like receptor (TLR) 1, 4, 9 and SLC11A1 genes and their association with paratuberculosis susceptibility in Holstein and indigenous crossbred cattle in Turkey. J Genet 97, 1147–1154 (2018). https://doi.org/10.1007/s12041-018-1008-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-1008-7