Abstract
Rice blast caused by fungal pathogen Pyricularia oryzae has a major impact on reducing yield potential of rice. In this study, homozygous plants were selected using microsatellite markers from the \(\hbox {BC}_{3}\hbox {F}_{2}\) population pyramided with four major genes in elite rice variety ADT 43. Background and selected lines with various blast resistance gene combinations were screened under natural conditions to study the effects of various gene combinations. Upon inspection of lines with different gene combinations, the three-gene pyramided line Pi54+Pi33+Pi1 was found to be highly resistant with the score of 3.3 followed by other three-gene pyramided lines Pi54+Pi2+Pi1 and Pi33+Pi2+Pi1, with the scores of 3.9 and 3.8, respectively. Two-gene pyramided lines Pi54+Pi1, Pi33+Pi1 and Pi2+Pi1 were found to be moderately resistant with a mean score of 4.0 each. In the case of monogenic lines, positive plants for Pi54 performed almost equal to three-gene pyramided lines with a mean score of 3.6. Lines with Pi2 and Pi1 were found to be moderately resistant and moderately susceptible with the mean scores of 4.1 and 4.5, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is the staple food of more than half of the world’s population—more than 3500 million people depend on rice for more than 20% of their daily calories (Maclean et al. 2013). Globally, 472.09 million tonne of rice is produced from 163.46 million hectares of land, of which India accounts for 106.05 million tonnes produced from 42.25 million hectares (FAOSTAT 2015). More than 90% of rice is consumed in Asia where it is a staple food for the majority of the population. India being one of the largest and fast growing Asian countries requires around 350 million tonnes of rice in future (Mohanty 2013). Following the green revolution in 1960s, numerous high yielding varieties were released to increase the rice production; however, the full potentiality of these varieties is limited by various biotic and abiotic stresses. Numerous pests and diseases threaten rice cultivation in which blast, the major disease caused by the fungus Magnaporthe oryzae, can infect all parts of shoot region throughout the crop period and can cause severe yield loss up to an extent of 85% at the global level (Sirithunya et al. 2002).
Rice blast is caused by heterothallic fungi Pyricularia oryzae (anamorph; P. grisea, synonym; Magnaporthe grisea). The fungi infect parts of rice, namely, collar, leaf, leaf sheath, neck and node. Leaves show typical spindle-shaped lesions with dark-brown borders having a grey coloured centre. In neck blast, brown-coloured lesions are found in the collar region of the panicle that is also called as panicle blast and the rotten panicle will drop down making the grains chaffy. In nodal blast, the lower nodes of the stem turn black; brown to black spots can be seen on the rachis. Pathogen races of this fungus evolve continuously and develop new races that are diverse in virulence (Tharreau et al. 2009). Centre for Disease Control and Prevention has recognized and listed rice blast as a potential biological threat as no part of the world is now safe from this disease and each year it causes destruction of rice grains that can be fed up to 60 million people (Zeigler 1994). This disease can be controlled by chemicals but spraying more chemicals is hazardous to environmental degradation and in turn increases the production cost. Apart from fungicides, cultural practices such as reduction of nitrogen fertilizers and water management can limit the infection; however, these practices cannot control the infection completely. Utilizing the diversity of rice for its resistance to blast, identification and development of rice varieties with highly effective and durable resistance serves as the most economically feasible and environmentally sound management approach to overcome the disease.
Genes which show resistance to rice blast are named with a prefix ‘Pi’. To date, 100 blast resistance genes have been mapped to different genotypes of rice; 19 genes have been cloned and characterized at the molecular level. To date, 350 quantitative trait loci have been identified in the rice genome (Sharma et al. 2012). Host–plant resistance against rice blast was found even before 1960s. Nakamori and Kosato (1949) suggested a backcrossing method in rice breeding to introduce resistance genes into superior varieties. Introduction of marker-assisted selection paved a way for effective identification and introgression of blast resistance genes from various landraces and wild relatives to high yielding commercial varieties susceptible to rice blast. As of today, hundreds of closely related molecular markers for various genes are identified and few gene-specific markers are reported for nine cloned blast resistance genes (Koide et al. 2009). The rapid evolutionary changes that occur in the virulence characteristics of population raise a continuous threat to the effectiveness of the existing blast resistant varieties (Hittalmani et al. 2000).
Gene pyramiding is one of the strategies that hold greater prospects to attain durable resistance against rice blast. In this method, more than one broad-spectrum gene is introgressed into a single-genetic background to confer resistance against different races (Joshi and Nayak 2010). The gene-pyramiding strategy is deployed for various rice varieties against different pests and diseases to create broad-spectrum horizontal resistance. For rice blast, three major genes Pi1+Pi2+Pi33 were pyramided and proved their durable resistance against blast for more than 10 years in various states of Colombia (Correa-Victoria et al. 2002). Pyramiding for blast (Pi5 and Pi54)+bacterial leaf blight (xa13+xa21) and brown plant hopper (Bph3, Bph17 and Bph18) were performed in Pusa basmathi 1121 and Pusa basmathi 6 (Singh et al. 2011). Pi-d(t)1, Pi-b and Pi-ta2 were pyramided in susceptible G46B, a promising line used for a three-line breeding strategy in hybrid rice (Chen et al. 2004).
Studying the reaction of blast resistance genes individually and in combinations against rice blast in a specific region provides valuable information about the efficiency and compatibility of various combinations that aids in designing future gene pyramiding schemes. In this study, four major resistance genes Pi1, Pi2, Pi33 and Pi54 pyramided into the elite rice variety ADT 43 were used to study the level of resistance as individual and in combinations against rice blast in the disease hot spot region.
Materials and methods
Plant materials
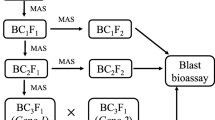
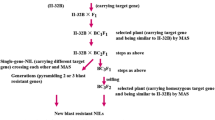
Experimental materials consist of advanced backcross \(\hbox {BC}_{3}\hbox {F}_{2}\) population pyramided with a combination of four blast resistance genes Pi1+Pi2+Pi33+Pi54. This population was obtained by a cross between ADT 43 \(\times \) CT 13432-3R. \(\hbox {F}_{1}\)s obtained from the cross were backcrossed with the recurrent parent ADT 43. The population was forwarded till \(\hbox {BC}_{3}\) to recover the recurrent parent genome (ADT 43) (Divya 2012). The donor parent used in this study (CT 13432-3R) is a near isogenic line of CO39 pyramided four blast resistance genes Pi1 + Pi2 + Pi33 + Pi54. The recurrent parent ADT 43 is a popular variety in Tamil Nadu, known for its high yielding ability and medium slender grain. It is a cross derivative of IR50\(\times \)I.W. Ponni, released in 1998 by Tamil Nadu Rice Research Institute, Aduthurai. The major disadvantage of this variety is its susceptibility to rice blast disease which often causes substantial yield reduction.
Foreground selection
Homozygous individuals from the \(\hbox {BC}_{3}\hbox {F}_{2}\) population with single genes and gene combinations were selected using four microsatellite markers linked to the four resistance genes Pi1, Pi2, Pi33 and Pi54. The markers used in this study are given in table 1.
The abovementioned microsatellite markers are reported by Correa-Victoria et al. (2002) for screening various CO 39 NILs for the same genes and the same set of markers was used to select the previous generations of this population by Divya (2012).
Fresh leaf samples were collected from the parents and \(\hbox {BC}_{3}\hbox {F}_{2}\) plants at tillering stage (45 days after sowing (DAS)). DNA was extracted following the cetyltrimethylammonium bromide (CTAB) method (Doyle 1987). DNA concentration was quantified and diluted to \(20\hbox { ng}/ \mu \hbox {L}\). Polymerase chain reaction (PCR) was carried out with 10 \(\mu \)L reaction mixture containing 1 \(\mu \)L of template DNA, 0.5 \(\mu \)L forward primer, 0.5 \(\mu \)L reverse primer, 5 \(\mu \)L of Go-prime red dye master mix and 3 \(\mu \)L of sterile water. PCR products were resolved on 1.5–3% agarose or 6% polyacrylamide gels. Agarose gels were stained using ethidium bromide and polyacrylamide gels were stained with silver nitrate as per the protocol suggested by Benbouza et al. (2006).
Phenotypic screening under natural epiphytotic conditions
Screening for natural infection was carried out at Hybrid Rice Evaluation Centre, Gudalur located at the latitude of \(11{^{\circ }}30^\prime \hbox {N}\), longitude of \(76{^{\circ }}30^\prime \hbox {E}\) and an elevation of 1317.00 MASL, which is the most favourable condition for blast disease development. Gudalur is considered as a ‘hot spot’ for leaf blast disease where disease occurrence is observed throughout the year and is maximum in winter and rainy seasons (Selvaraj et al. 2011). Raised beds were prepared with a length of 1.0 m, 70 cm width and 10 cm height. Furrows of 2-cm depth were formed on the bed parallel to its width with a 10 cm gap. Selected plants were sown in two replications and susceptible varieties such as ADT 43 and CO 39 were sown in the side furrows and after every three rows of entries as a spreader source or infector rows for pathogen. These infector rows are sown one week ahead of sowing of the entries. Each test accessions were sown in a single row with 100 seeds for each entry. Observation of disease occurrence was recorded when susceptible check was severely infected by leaf blast. Disease severity was assessed on 10 plants of each entry for leaf blast, lesion number, lesion type and infested leaf area adopting the Standard Evaluation System on a 0–9 scale (SES, IRRI, 2002). The potential disease incidence was calculated using the formula:
Results
Firstly, all the 572 plants were screened for the gene Pi1 using the SSR marker RM 1233 from which 138 homozygous positive plants, 202 heterozygous plants and 187 negative plants were obtained. All the plants were again screened for Pi33 using RM 72 which resulted in 109 positive plants, 137 heterozygous plants and 326 negative plants. From the above analysis, 21 homozygous plants with both genes Pi1 and Pi33 have been selected. The selected two gene pyramided lines were screened using RM 206 for Pi54 from which eight homozygous, eight heterozygous and five negative plants were obtained. Twenty-one triple gene pyramided individuals were again screened for Pi2 using RM 527 and six homozygous, seven heterozygous and eight negative plants were obtained (figure 1). Overall, foreground analysis (figure 2) resulted in the identification of 28 plants with different gene combinations for blast resistance screening (table 2). The selected progeny families were categorized based on gene combinations and three progeny families per combination were randomly selected for phenotypic evaluation.
Phenotypic screening of selected plants
The selected \(\hbox {BC}_{3}\hbox {F}_{3}\) plants with different gene combinations and single-gene introgression were screened under epiphytotic natural conditions at Hybrid Rice Evaluation Centre, Gudalur. The infected plants scored 15 DAS to evaluate the performance of selected plants. The susceptible check CO 39 and susceptible recipient parent ADT 43 were highly affected with the mean scores of 9.0 and 8.7, respectively, which confirms the level of blast infection as severe. In this condition, the three gene pyramided plants Pi54+Pi33+Pi1 were found to be highly resistant under Gudalur conditions with the mean score of 3.3 followed by other three gene pyramided lines Pi54+Pi2+Pi1 and Pi33+Pi2+Pi1, with the mean scores of 3.9 and 3.8, respectively. Two-gene pyramided lines Pi54+Pi1, Pi33+Pi1 and Pi2+Pi1 were found to be moderately resistant with the mean score of 4.0 each. In case of monogenic lines, plants positive for the Pi54 gene performed almost equal to three gene pyramided lines with the mean score of 3.6. But, the other lines with the Pi2 and Pi1 genes were found to be moderately resistant and moderately susceptible with the mean scores of 4.1 and 4.5, respectively. Per cent disease index is calculated for all the lines to appraise the severity of incidence which is given in table 3 along with the lesion number found in the plants.
Durability of resistance
In order to ascertain the durability of resistance rendered by different gene combinations, all the plants are again scored on 45 DAS. In three-gene pyramided lines, Pi54+Pi33+Pi1 rendered durable resistance with the score of 3.4 on 15 DAS and 3.3 on 45 DAS, the donor parent (CT 13432-3R) with four resistance genes Pi54+Pi33+Pi2+Pi1 recorded score 3 on 15 DAS and score 3.6 on 15 DAS, three gene combinations Pi54+Pi2+Pi1 and Pi33+Pi2+Pi1scored 3.4 and 3.0 on 15 DAS and 3.7 and 4.0 on 45 DAS (table 4). Gene combination Pi54+Pi1 found with least variation within two gene pyramided lines has maintained the mean score of 4.0 from beginning to 45 DAS, the other two gene pyramided lines Pi33+Pi1 and Pi2+Pi1 failed to render durable resistance with higher variation of mean scores 3.8 and 3.5 on 15 DAS and 4.5 each on 45 DAS.
Between the monogenic lines, Pi54 was found with smaller variation 3.4 on 15 DAS and 3.8 on 45 DAS, the other two monogenic lines Pi2 and Pi1 had higher variation with the mean score of 3.8 each on 15 DAS and 4.5 and 5.2, respectively, on 45 DAS. The difference in mean scores is depicted in figure 3.
Discussion
Use of resistant varieties against rice blast is an effective and eco-friendly strategy for disease management but resistance rendered by single gene doesn’t provide long term resistance due to the highly evolving nature of rice blast pathogen (Chen et al. 1996). A combination of the major genes can provide durable resistance against the various pathogen races (Chen et al. 1995; Zeigler et al. 1994, 1995). Liu et al. (2016) reported gene pyramiding as an effective strategy to develop broad-spectrum resistance varieties. Four blast resistance genes Pi1, Pi2, Pi33 and Pi54 were pyramided into an elite variety ADT 43 and the effects of these genes and combinations were studied. Three- and two-gene pyramided lines with any of these four genes are identified but no plant pyramided with all four genes was found in this study. The lines used in this study are primarily selected for three genes Pi1, Pi2 and Pi33 up to \(\hbox {BC}_{3}\); the introgression and presence of Pi54 from the parent CT 13432-3R is identified later. The four genes used in this study are the few of the major blast resistance genes reported to be rendering broad-spectrum resistance against rice blast and used in various studies: Vasudevan et al. (2015) reported that the rice-blast resistance gene Pi54 confers broad-spectrum resistance across India; Pi33 confers high degree of resistance to the rice blast fungus and corresponds to the ACE1 avirulence gene (Berruyer et al. 2003; Bohnert et al. 2004), the gene was identified in different rice varieties including IR64 (Berruyer et al. 2003); Pi2 is an effective gene that can be used in combination with other genes (Chen et al. 1996) and the Pi1R gene, derived from the durably resistant West African cv. LAC23, has displayed a high level of durable resistance against blast (Hittalmani et al. 2000; Fuentes et al. 2008). The above-mentioned four genes have been introgressed into ADT 43 which is a cross derivative of IR50 \(\times \) I.W. Ponni, released in 1998 by Tamil Nadu Rice Research Institute, Aduthurai. It is a short duration variety maturing in 110 days with medium slender grains. It is a high-yielding variety with an average yield of 6.0 t/ha (RKMP 2011, http://www.rkmp.co.in/search/node/ADT%204). This variety is highly preferred by Tamil Nadu farmers because of its high yielding ability with medium slender grains. The major drawback of this variety is its susceptibility to rice-blast disease. The donor parent CT 13432-3R is a near isogenic line pyramided with four genes pyramided with four resistant genes Pi1, Pi2, Pi33 and Pi54 in blast susceptible variety CO 39 and reported to be having broad spectrum resistance across Colombia (Correa-Victoria et al. 2002).
Gene pyramiding in elite susceptible varieties is a broadly used strategy to improve variety, blast resistance genes Pi54+Piz5 and bacterial blight resistance genes xa13+Xa21 were pyramided into the background of the elite variety ‘Pusa basmathi 1’ through marker-assisted backcross breeding (MABB) and successfully released as a variety ‘Improved Pusa basmathi’ (Pusa 1460), it was the first variety developed in India by molecular breeding (Singh et al. 2011). Three major bacterial blight resistance genes (Xa21, xa13 and xa5) in Samba Mahsuri, a high preference variety from a donor line (SS1113); BB infection in the three-gene pyramid lines, exhibited a significant yield advantage over the susceptible parent Samba Mahsuri (Sundaram et al. 2008). The MABB approach was employed to incorporate blast resistance genes namely, Piz-5 and Pi54, from the donor lines C101A51 and Tetep into the genetic background of PRR78 an elite restorer line (Singh et al. 2013).
Studying the effects of various genes as individuals and combinations is important to ascertain the compatibility and efficiency of the genes, the results of the study provide valuable inputs for selection genes for future pyramiding programmes and the effect of gene combinations was studied in the pyramided populations of various crops. Differential reactions of soybean aphid resistance genes Rag1, Rag2 and Rag3 as iso lines and combinations were studied by Ajayi-Oyetunde et al. (2016); the effects of Pi1+Pi2+Pi33 as combinations and isogenic lines were studied to ascertain the resistance of these genes across Colombia (Correa-Victoria 2004). In this study, different gene combinations were screened under natural conditions at Gudalur, the donor parent CT 13432-3R with four genes showed higher level of resistance with a mean score of 3.3 and PDI of 32%, before the genotype was reported to be highly resistance under Gudalur conditions (Divya et al. 2014) and found to be resistant to nine lineages of rice blast pathogen in Colombia (Correa-Victoria et al. 2002). Another three gene combination Pi54+Pi33+Pi1 was also found to be resistant with a score of 3.4 and PDI of 32.95%; the performance of this genotype is on par with the donor parent. In plants with a single gene, Pi54 was found be highly resistant to rice blast with a mean score of 3.4, plants with Pi1+Pi2 was found to be moderately resistant to rice blast with a score of 3.8 and in combination (Pi1+Pi2), it showed a score of 3.5. The gene Pi54 performs well (3.4) when it was single but the performance reduced when it was combined with Pi1 (3.8), this may be due to interaction effect of the gene. Interaction effects in gene pyramiding have been reported using several gene combinations (Yoshimura et al. 1995; Fukuta et al. 1998; Fujita et al. 2010).
Compared with the combination plants having Pi54 combined with other genes and as monogenic line shown high level of resistance with the mean score between 3.0 and 3.8, plants without the gene Pi54 showed slight resistance with the mean score ranging from 4.1 to 4.5. The Pi54 gene is reported as a major blast resistance gene which confers a high degree of resistance to diverse strains of the fungus P. oryzae. The single gene Pi54 activates a complex defence mechanism involving numerous genes and enzymes (Gupta et al. 2011), NLR145 a variety introgressed with Pi54 showed high degree of resistance against blast with a mean disease score of 3.0 for two consecutive years 2012 and 2013 and in two locations (Arunakumari et al. 2016). The function of Pi54 was isolated and transformed through an agrobacterium into the susceptible variety Taipei 309 to evaluate the efficiency of the gene, transgenic lines carrying stable Pi54 gene were highly resistant to all the four isolates of P. oryzae (Rai et al. 2011). As reported in the above studies, Pi54 showed high degree of resistance in this study against rice blast in Gudalur. Though Pi54 as a single gene endowed higher degree of resistance on par with plants with multiple genes, the purpose of pyramiding is to develop lines which provide long-term and broad-spectrum resistance from different races of P. oryzae. Pathogens can overcome resistance within a short span in varieties with single-resistance gene cultivated on a large scale, this can be tackled by pyramiding multiple genes because the probability of mutation which enables virulence to multiple genes is lower than a single gene (Singh et al. 2001). The principal objective of gene pyramiding is to prevent resistance breakdown to study the durability of pyramided lines disease reaction was evaluated on 45 DAS. No previous study is available which compares different time periods in the same season. This study is conducted to confirm the long-term resistance without breakdown until the plant reaches the grand growth stage. There is no major variation in scores noted in three-gene lines, Pi54+Pi33+Pi1 rendered durable resistance with a score of 3.4 on 15 DAS and 3.3 on 45 DAS, donor parent (CT 13432-3R) with four resistance genes Pi54+Pi33+Pi2+Pi1 recorded a score of 3 on 15 DAS and a score of 3.6 on 15 DAS, three gene combinations Pi54+Pi2+Pi1 and Pi33+Pi2+Pi1 scored 3.4 and 3.0 on 15 DAS and 3.7 and 4.0 on 45 DAS but the breakdown of resistance found in monogenic and two gene pyramided lines Pi33+Pi1 and Pi2+Pi1 with higher variation of mean scores 3.8 and 3.5 on 15 DAS and both scored 4.5 on 45 DAS, two monogenic lines Pi2 and Pi1 had higher variation with a mean score of 3.8 both on 15 DAS and 4.5 and 5.2, respectively, on 45 DAS. But, the two gene combination with Pi54 and monogenic Pi54 found to render durable resistance with smaller variation 3.4 on 15 DAS and 3.8 on 45 DAS and the combination Pi54+Pi1 found with least variation within two-gene pyramided lines, it maintained the mean score of 4.0 from the beginning to 45 DAS. This again proves ability of Pi54 to provide durable resistance against rice blast. The breakdown of resistance of monogenic lines may be due to infection by different races of blast pathogen in later period through air, wind and water from other rice fields of Gudalur or through alternate hosts such as finger millet or Digitaria sanguinalis. P. oryzae can rapidly produce thousands of spores readily spread through air, wind or rain, onto neighbouring plants (Srivastava et al. 2014). P. oryzae from nonrice hosts such as D. sanguinalis, Eleusine indica and Lolium boucheanum could also serve as sources of inoculum for rice crops (Narayanasamy 2011).
References
Ajayi-Oyetunde O. O., Diers B. W., Lagos-Kutz D., Hill C. B., Hartman G. L., Reuter-Carlson U. et al. 2016 Differential reactions of soybean isolines with combinations of aphid resistance genes Rag1, Rag2, and Rag3 to four soybean aphid biotypes. J. Econ. Entomol. 109, 1431–1437.
Arunakumari K., Durgarani C. V., Satturu V., Sarikonda K. R., Chittoor P., Vutukuri B. et al. 2016 Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into Indian rice variety MTU1010. Rice Sci. 23, 306–316.
Benbouza H., Jacquemin J., Baudoin J. and Mergeai G. 2006 Optimization of a reliable, fast, cheap and sensitive silver staining method to detect SSR markers in polyacrylamide gels. Biotechnol. Agron. Biotechnol. Agron. Soc. Environ. 10, 77–81.
Berruyer R., Adreit H., Milazzo J., Gaillard S., Berger A., Dioh W. et al. 2003 Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Theor. Appl. Genet. 107, 1139–1147.
Bohnert H. U., Fudal I., Dioh W., Tharreau D., Notteghem J. and Lebrun M. 2004 A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513.
Chen D., Zeigler R. S., Leung H. and Nelson R. J. 1995 Population structure of Pyricularia grisea at two screening sites in the Philippines. Phytopathology 85, 1011–1020.
Chen D. H., Zeigler R. S., Ahn S. W. and Nelson R. J. 1996 Phenotypic characterization of the rice blast resistance gene Pi-2 (t). Plant Disease (USA) 80, 52–56.
Chen X. W., Li S. G., Ma Y. Q., Li H. Y., Zhou K. D. and Zhu L. H. 2004 Marker-assisted selection and pyramiding for three blast resistance genes, Pi-d (t) 1, Pi-b, Pi-ta2, in rice. Chin. J. Biotechnol. 20, 708–714.
Correa-Victoria F. J. 2004 Interactions of gene combinations against rice blast in blast: interaction with rice and control. Kluwer Academic Publishers, Dordrecht.
Correa-Victoria F. J., Martinez C., Tharreau F. J., Vales C., Escobar M. and Aricada G. 2002 Gene combination for durable blast resistance in Colombia. Fitopatol. Colomb. 26, 47–54.
Divya B. 2012 Marker assisted backcross breeding to introgress blast (Pyricularia oryzae) resistance genes into the susceptible rice (Oryza sativa L.) variety ADT 43. Ph.D. thesis, TNAU, Coimbatore.
Divya B., Robin S., Rabindran R., Manjunath H., Valarmathi P. and Joel A. J. 2014 Resistance reaction of gene introgressed lines against rice blast (Pyricularia oryzae) disease. Australas. Plant Pathol. 43, 177–191.
Doyle J. J. 1987 A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Eizenga G. C., Agrama H. A., Lee F. N., Yan W. and Jia Y., 2006. Identifying novel resistance genes in newly introduced blast resistant rice germplasm. Crop Sci. 46, 1870–1878.
FAOSTAT 2015 Statistical database of food and agriculture. FAO, Rome.
Fjellstrom R., Conaway-Bormans C. A., McClung A. M., Marchetti M. A., Shank A. R. and Park W. D. 2004 Development of DNA markers suitable for marker assisted selection of three Pi genes conferring resistance to multiple Pyricularia grisea pathotypes. Crop Sci. 44, 1790–1798.
Fuentes J. L., Correa-Victoria F. J., Escobar F., Prado G., Aricapa G., Duque M. C. et al. 2008 Identification of microsatellite markers linked to the blast resistance gene Pi-1 (t) in rice. Euphytica 160, 295–304.
Fujita D., Yoshimura A. and Yasui H. 2010 Development of near-isogenic lines and pyramided lines carrying resistance genes to green rice leafhopper (Nephotettix cincticeps Uhler) with the Taichung 65 genetic background in rice (Oryza sativa L.). Breed. Sci. 60, 18–27.
Fukuta Y., Tamura K., Hirae M. and Oya S. 1998 Genetic analysis of resistance to green rice leafhopper (Nephotettix cincticeps Uhler) in rice parental line, Norin-PL6, using RFLP markers. Jpn. J. Breed. 48, 243–249.
Gupta S. K., Rai A. K., Kanwar S. S., Chand D., Singh N. K. and Sharma T. R. 2011 The single functional blast resistance gene Pi54 activates a complex defence mechanism in rice. J. Exp. Bot. 63, 757–772.
Hittalmani S., Parco A., Mew T. V., Zeigler R. S. and Huang N. 2000 Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 100, 1121–1128.
Joshi R. K. and Nayak S. 2010 Gene pyramiding – a broad spectrum technique for developing durable stress resistance in crops. Biotechnol. Mol. Biol. Rev. 5, 51–60.
Koide Y., Kobayashi N., Xu D. and Fukuta Y. 2009 Resistance genes and selection DNA markers for blast disease in rice (Oryza sativa L.). Jpn. Agric. Res. Q.: JARQ 43, 255–280.
Liu Y., Jia Y., Gealy D., Goad D. M., Caicedo A. L. and Olsen K. M. 2016 Marker development for rice blast resistance gene and application in the USDA rice mini-core collection. Crop Sci. 56, 1001–1008.
Maclean J., Hardy B. and Hettel G. 2013 Rice Almanac: source book for one of the most important economic activities on earth (ed. J. L. Macclean, D. C. Dawe, B. Hardy and G. P. Hettel), \(3{\text{rd}}\) edition, pp. 88. IRRI, Los Banos.
Mohanty S. 2013 Trends in global rice consumption. Rice Today 12, 44–45.
Nakamori E. and Kosato U. 1949 Theoretical consideration on the backcross method for breeding of rice varieties. Jpn. J. Breed. 3, 10–18.
Narayanasamy P. 2011 Detection of fungal pathogens in plants. In Microbial plant pathogens-detection and disease diagnosis (ed. Anonymous), pp 5–199. Springer.
Rai A. K., Kumar S. P., Gupta S. K., Gautam N., Singh N. K. and Sharma T. R. 2011 Functional complementation of rice blast resistance gene Pi-kh (Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J. Plant Biochem. Biotechnol. 20, 55–65.
Selvaraj I., Nagarajan P. and Thiyagarajan K. 2011 Identification of microsatellite (SSR) and RAPD markers linked to rice blast disease resistance gene in rice (Oryza sativa L.). Afr. J. Biotechnol. 10, 3301.
Sharma T. R., Rai A. K., Gupta S. K., Vijayan J., Devanna B. N. and Ray S. 2012 Rice blast management through host-plant resistance: retrospect and prospects. Agric. Res. 1, 37–52.
Sharma T. R., Madhav M. S., Singh B. K., Shanker P., Jana T. K., Dalal V. et al. 2005 High-resolution mapping, cloning and molecular characterization of the Pi-k h gene of rice, which confers resistance to Magnaporthe grisea. Mol. Genet. Genom. 274, 569–578.
Singh S., Sidhu J. S., Huang N., Vikal Y., Li Z., Brar D. S. et al. 2001 Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 102, 1011–1015.
Singh A. K., Gopalakrishnan S., Singh V. P., Prabhu K. V., Mohapatra T., Singh N. K. et al. 2011 Marker assisted selection: a paradigm shift in Basmati breeding. Indian J. Genet. Plant Breed. 71, 120.
Singh V. K., Singh A., Singh S. P., Ellur R. K., Singh D., Gopala Krishnan S. et al. 2013 Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR78’. Plant Breed. 132, 486–495.
Sirithunya P., Tragoonrung S., Vanavichit A., Pa-In N., Vongsaprom C. and Toojinda T. 2002 Quantitative trait loci associated with leaf and neck blast resistance in recombinant inbred line population of rice (Oryza sativa). DNA Res. 9, 79–88.
Srivastava D., Shamim M., Kumar D., Pandey P., Khan N. A. and Singh K. N. 2014 Morphological and molecular characterization of Pyricularia oryzae causing blast disease in rice (Oryza sativa) from north India. Int. J. Sci. Res. Publ. 4, 1–9.
Sundaram R. M., Vishnupriya M. R., Biradar S. K., Laha G. S., Reddy G. A., Rani N. S. et al. 2008 Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160, 411–422.
Tharreau D., Fudal I., Andriantsimialona D., Utami D., Fournier E., Lebrun M. H. and Nottéghem J. L. 2009 World population structure and migration of the rice blast fungus, Magnaporthe oryzae. In Advances in genetics, genomics and control of rice blast disease, pp. 209–215. Springer, Dordrecht.
Vasudevan K., Gruissem W. and Bhullar N. K. 2015 Identification of novel alleles of the rice blast resistance gene Pi54. Sci. Rep. 5, 15678.
Yoshimura S., Yoshimura A., Iwata N., McCouch S. R., Abenes M. L., Baraoidan M. R. et al. 1995 Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol. Breed. 1, 375–387.
Zeigler R. S. 1994 Linking blast population analysis to resistance breeding: a proposed strategy for durable resistance. In Rice blast disease (ed. R. S. Zeigler, S. A. Leong and P. S. Teng) pp. 2. International Rice Research Institute, Los Banos.
Zeigler R. S., Thome J., Nelson R., Levy M. and Correa-Victoria F. J. 1994 Lineage exclusion: a proposal for linking blast population analysis to rice breeding. In The rice blast disease (ed. R. S. Zeigler, S. A. Leong and P. S. Teng) pp. 269–291. International Rice Research Institute, Los Banos.
Zeigler R. S., Cuoc L. X., Scott R. P., Bernardo M. A., Chen D. H., Valent B. et al. 1995 The relationship between lineage and virulence in Pyricularia grisea in the Philippines. Phytopathology 85, 443–451.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Manoj Prasad
Rights and permissions
About this article
Cite this article
Pandian, B.A., Joel, J., Nachimuthu, V.V. et al. Marker-aided selection and validation of various \({ Pi}\) gene combinations for rice blast resistance in elite rice variety ADT 43. J Genet 97, 945–952 (2018). https://doi.org/10.1007/s12041-018-0988-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-018-0988-7