Abstract
Swarna-Sub1, a ruling submergence tolerant variety of rice, showed a significant decrease in the yield in major blast disease prone areas, which is caused by fungus Magnaporthe oryzae (Pyricularia oryzae). To overcome this problem, three major blast resistance genes viz., Pi1, Pi2, and Pi54 were pyramided through marker-assisted backcross breeding using donors, Swarna-LT (having Pi1 + Pi54) and Swarna-A51 (having Pi2). Foreground selection was carried out using molecular markers tightly linked to three blast resistance genes and also submergence tolerance for retaining the Sub1 gene in the recurrent parent. Based on disease resistance and agro-morphological performance of backcross-derived lines, we selected the best 25 plants of two- and three-gene pyramided BC3F2 homozygous lines with a maximum of ~ 93.5% recurrent parent genome and were further advanced to the BC3F5 generation. The stringent recurrent parent genome recovery analysis using SSR markers limited the linkage drag from a minimum of 0.2 Mb to a maximum of 2 Mb in all three-gene pyramided lines. The two and three blast resistance genes pyramided lines, i.e., SS30-24-82, SS30-24-73, and SS30-24-46, displayed a high level of blast resistance and submergence tolerance. The successful use of marker-assisted backcrossing strategy coupled with phenotypic selection helped in the development of Swarna-Sub1 lines having multiple blast resistance genes with superior agro-morphological and grain quality traits, which could serve as valuable genetic stocks in the rice breeding program.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice is the staple food crop of India with an annual production of 110 MT. In order to meet the demands of the ever-increasing population, the annual production of rice needs to be increased to 125 MT by 2020, which is being affected by both biotic and abiotic stresses. Breeding for developing resistant lines along with yield enhancement is of priority for maintaining stable rice production. Among abiotic stresses, submergence adversely affects poor farmers living on 15 million ha of rice-growing areas in the rainfed lowlands in South and South-East Asia. In India, about 15% rice area is often prone to flash floods, which may occur at any stages of the crop wherein the seedling stage is the more vulnerable. Swarna is one of the popular varieties in South and South-East Asia, which is improved with submergence tolerance by incorporating the Sub1 gene and released as Swarna-Sub1 with good yield (Bailey-Serres et al. 2010; Septiningsih et al. 2009). The varieties having the Sub1 gene have been spreading fast in several countries over the last few years, and are currently grown by more than 4 million farmers in Asia including India, where over 12 million hectares of the total paddy cultivation is being grown. In flash-flood affected areas, due to the stagnation of water, which results in high humidity and lower canopy temperature, Swarna-Sub1 suffers from leaf blast disease, especially in the seedling stages, as they are very tender and more prone to the disease attack resulting in the complete seedling loss (Chaudhary and Sah 1998).
The rice blast caused by M. oryzae is an important limiting factor for rice cultivation worldwide, which causes not only an annual loss of more than 50% yield, but also affects the quality of harvested grain (Scardaci et al. 1997; Variar et al. 2009). Nearly 347 QTLs and 102 genes have been identified so far of which more than 27 genes were cloned and characterized (Khanna et al. 2015; Zheng et al. 2016). Among the identified blast resistance genes so far Pi1, Pi2, and Pi54 have been reported to provide broad-spectrum resistance and have been widely used in different combinations by breeders for improving blast resistance in the majority of the ruling varieties (Madhavi et al. 2016; Swathi et al. 2019; Khan et al. 2018; Balachiranjeevi et al. 2015). Since the resistance conferred by a single gene often breaks down in the varieties after a few years of their release due to the dynamic behavior of the blast pathogen (Hittalmani et al. 2000; Fukuoka et al. 2015). The blast genes Pi1 and Pi2 are derived from indica rice lines LAC23 and C101A51, respectively, which are highly effective against the pathogen population in Asia (Tacconi et al. 2010; Mackill and Bonman 1992). The Pi54 gene was identified and mapped on chromosome 11L from a rice variety Tetep and was reported to govern resistance against predominant races of the blast pathogens in India (Sharma et al. 2005, 2010). The exploitation of these genes in the marker-assisted breeding program is an effective and economic strategy for the development of blast-resistant lines. Marker-assisted backcross breeding (MABB) is a highly successful method for deployment of target traits for biotic and abiotic stress resistance in the rice-breeding program (Kumar et al. 2016; Balachiranjeevi et al. 2015; Hari et al. 2013; Bhatia et al. 2011). The MABB program is effective in retaining the gene(s) of interest and eliminating the unnecessary segments of donor genome during backcrossing, ultimately increasing the percentage of the recurrent genome in successive generations (Lewis and Kernodle 2009; Ribaut et al. 2002; Hospital et al. 1992). The selection of plant with desirable alleles is facilitated by foreground selection along with stringent phenotypic selection (Singh et al. 2013). The present study was aimed at pyramiding of three major blast resistance genes, Pi1, Pi2, and Pi54 in the genetic background of an elite cultivar Swarna-Sub1 through marker-assisted backcross breeding (MABB) and the improved versions of Swarna-Sub1 lines having different blast resistance gene combinations were evaluated for agronomic performance, grain quality traits, and resistance to blast disease.
Materials and methods
Plant material
Swarna-Sub1, a submergence-tolerant line of Swarna carrying Sub1 gene, was used as a recurrent parent for the incorporation of blast resistance genes. Two BC1F2 lines of Swarna namely, Swarna-LT (possessing Pi1 + Pi54) and Swarna-A51 (possessing Pi2 gene) were used as donors, and these lines were developed and screened at the Indian Institute of Rice Research, Hyderabad (Madhav et al. unpublished data). In addition to this, HR12 and TN1 were used as susceptible checks for the screening of blast disease.
Development of blast resistance lines
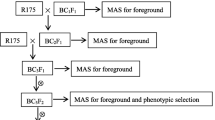
The recurrent parent Swarna-Sub1 was crossed with the donors, Swarna-LT and Swarna-A51, independently and the F1s were screened with the tightly linked markers for the target genes (Supplementary Table 1). The true F1s for the respective genes were inter-crossed and the ICF1s positive for all the three targeted genes were backcrossed with Swarna-Sub1 to produce the BC1F1 population. The plants harboring the target genes with maximum recurrent parent genome and phenotypically similar to recurrent parent were backcrossed to Swarna-Sub1 until BC3 generation and selfed to generate BC3F2 lines. The improved lines of Swarna-Sub1, which possess blast genes in homozygous condition and in different combinations, were screened for blast resistance, further forwarded to BC3F5 generation by following pedigree selection.
Marker-assisted selection for blast resistance
In the present study, the foreground and background analysis were carried out with the DNA extracted from parents and backcross progenies by following the procedure of Wen-Yue et al. (2006). The PCR and gel electrophoresis process was carried according to the protocol followed by Sundaram et al. 2008. For the foreground selection, RM224 and RM527 for Pi1 and Pi2 genes, respectively; and for Pi54, RM206 and PI54 MAS markers were used (Usatov et al. 2016; Ramkumar et al. 2011) whereas, for submergence tolerance Sub1AB1, Sub1BC2, and Sub1BC3 markers were used for the transfer of the entire Sub1 QTL (Septiningsih et al. 2009) (Supplementary Table 1). Although background selection is not necessary as both the donor parents used for the introgression of target genes have the same genetic background, for more precise selection of plants, their rapid advancement and to avoid undesirable linkages, the parental polymorphic survey was carried out using 700 SSR markers distributed among 12 chromosomes of rice genome in three parents, namely, Swarna-Sub1, Swarna-LT, and Swarna-A51. The recurrent parent genome recovery and the extent of linkage drag in backcross populations were estimated by polymorphic markers identified among the parents and a graphical representation of the genomic contribution of donors and the recurrent parent on target chromosomes were explained using the software program Graphical GenoTypes (GGTs) version 2.0 (Supplementary Fig. 1).
Screening for blast resistance and submergence tolerance
All the selected backcross-derived lines for three, two, and one blast resistance genes were artificially screened using IIRR blast isolate (NLR-1) in Uniform Blast Screening Nursery (UBN) at two locations viz., the Indian Institute of Rice Research (IIRR) and Agricultural Research Institute (ARI), PJTSAU, Rajendranagar, Hyderabad, India. The screening for blast disease was done following Prasad et al.’s (2011) procedure and the data on blast reaction was recorded three times following a 0–9 scale (SES 1996) at a 10-day interval started after 25 days of sowing unless and until the susceptible check gets totally infected by the disease (Fig. 1).
For screening of submergence, backcross-derived plants along with susceptible and resistant checks (IR42 and Swarna-Sub1) were sown in trays in the greenhouse, IIRR. After 14–16 days of sowings, the seedlings were subjected to complete submergence for 12–15 days. When the susceptible checks showed more than 50% damage, then the water was drained from the trays and the survival of plants was scored after 10–15 days of recovery.
Evaluation of agro-morphological traits
Twenty-five-day-old seedlings of backcross-derived lines of improved Swarna-Sub1 with three genes, two genes, and one gene were evaluated for the agronomical characters in a randomized complete block design (RCBD) in three replications, in a total plot size of 2500 m2 during Kharif 2014 at the research farm, IIRR. The agro-morphological yield-associated traits viz., days to maturity (DM), plant height (PH) (cm), number of productive tillers per plant (NPT), panicle length (PL) (cm), panicle weight (PW) (g), number of filled grains per panicle (NGP), spikelet fertility (SF) (%), grain yield per plant (GY) (g), 1000 seed weight (TGW) (g), and grain type (GT) observations were recorded in comparison with Swarna-Sub l. For the grain quality evaluation, amylose content (AC) of grain was determined according to the Juliano method (1981). The data were tabulated for each entry in three replications and statistical analysis was performed by Statistix 8.1 software for the determination of coefficient of variation (CV), standard error, and analysis of variance (ANOVA) to determine the significant variation among the lines (Table 1 and Fig. 1).
Results
Marker-assisted introgression of blast resistance genes
The development of blast resistance in Swarna-Sub1 was initiated with the development of F1s from the crosses, Swarna-Sub1 with Swarna-LT (possessing Pi1 and Pi54 genes) and Swarna-A51 (possessing Pi1 gene). Inter-crossing the true F1s confirmed by foreground analysis generated 20 ICF1 seeds. Four ICF1 plants were heterozygous for three genes, which on backcrossing with recurrent parent generated 300 BC1F1 plants with 48 plants heterozygous for three genes (Pi1 + Pi2 + Pi54). Among these, 13 plants showed the presence of the Sub1 gene when genotyped with submergence tolerance markers in addition to three blast genes. The background analysis revealed 120 SSR markers polymorphic between Swarna-Sub1and Swarna-LT, while 112 markers were polymorphic between Swarna-Sub1 and Swarna-A51. A total of 93 markers showed polymorphism in common between the recurrent parent and two donor parents. These 93 polymorphic SSR markers were used for background analysis covering an average marker distance of 5–7 Mb in the whole genome of rice. Out of 13 BC1F1 plants, one single best plant SS30-1, which had maximum recurrent parent genome (RPG) recovery of 75.8%, was selected and further backcrossed, which generated around 150 BC2F1 seeds. Among 150 BC2F1 plants, five elite plants having three blast resistance genes in heterozygous condition, and Sub1 with a maximum of 87.9% recurrent parent genome were selected and further backcrossed for producing BC3F1 population.
A total of 254 BC3F1 plants were genotyped and selected nine best plants belongs to the three combinations viz., Pi2 + Pi54, Pi1 + Pi54 and Pi1 + Pi2 + Pi54 having Sub1 gene in common with 92.3% of recurrent parent genome recovery (Supplementary Figure 2). Selfing of BC3F1 obtained 800 BC3F2 plants with three genes (Pi1 + Pi2 + Pi54), 600 two-gene (Pi2 + Pi54), and 700 two-gene (Pi1 + Pi54) combinations. Phenotypic and genotypic evaluation of BC3F2 plants revealed the best 100 plants with three genes, whereas there were 150 plants with two-gene combinations (Pi1 + Pi54 and Pi2 + Pi54). Based on agronomic performance and background analysis of BC3F2 plants, the eight best BC3F2 plants having three genes (Pi1 + Pi2 + Pi54) and eight plants having combinations of Pi1 + Pi54 and nine plants having combination of Pi2 + Pi54 with an average of 93.5% recurrent parent genome were selected.
The selected BC3F2 plants were further analyzed for the extent of linkage drag using the flanking markers of target genes on chromosome 11 and 6. At Pi54 locus, all eight of the three-gene harboring lines showed a gene segment of 2.0 Mb at the proximal end and 0.6 Mb at the distal end introgressed from the donor parent genome. At the Pi1 locus, all the lines showed a 0.2-Mb segment at the proximal end, while no segment was found at the distal end except in two lines, SS30-24-19 and SS30-24-76, which showed a 0.6-Mb segment from the donor parent genome. In lines SS30-24-19, SS30-24-107, and SS30-24-143, near the Pi2 locus 1.8 Mb of donor genome segment found at the proximal end and 0.5 Mb appeared at the distal end in S30-24-76, SS30-24-112, and SS30-24-131 (Supplementary Fig. 1). In eight two-gene harboring lines (Pi1 + Pi54), at Pi1 locus donor genome segment of 0.3 Mb at the proximal end and 0.6 Mb at the distal end were observed. At the Pi54 locus, 2.0 Mb and 1.1 Mb of the donor genome segments were detected at proximal and distal ends, respectively, in all the lines. In nine two-gene possessing plants (Pi2 + Pi54), at Pi2 locus, 0.5 Mb and 1.8 Mb while at Pi54 locus 2.0 Mb and 0.6 Mb of the donor genome segments were observed at the proximal and distal ends, respectively. These selected plants were initially subjected to evaluation of phenotypic and agro-morphological traits. The grain-quality traits selection was done in BC3F2 generation and later at BC3F5 generation after stabilization of targeted plant product.
Phenotypic screening for blast resistance
The blast resistance evaluation revealed that the donor parents, Swarna-LT and Swarna-A51, exhibited resistance with a score of 0–1 whereas the recurrent parent Swarna-Sub1 is highly susceptible with a score of 9. The blast reaction of five introgressed lines having three genes (Pi1 + Pi2 + Pi54) SS30-24-46, SS30-24-76, SS30-24-112, SS-30-24-131, and SS30-24-143 showed complete resistance with scores ranging from 0–1 to the IIRR isolate under artificial conditions in UBN test locations. Similarly, the lines with two genes, Pi2 + Pi54 (SS30-24-73, SS30-24-111, SS30-24-134) and Pi1 + Pi54 (SS30-24-82) showed resistance were also identified (Table 1). Six lines having only Pi1, 24 lines having only Pi2, and nine lines having Pi54 have shown the resistance with the score of 1–3 against IIRR isolate. Among those lines having single genes, the lines having Pi54 showed a high level of resistance compared to the lines having Pi1 or Pi2 (Supplementary Table 2).
Phenotypic screening for submergence tolerance
Evaluation of submergence tolerance showed that the survival percentage of the Swarna-Sub1 was 85 ± 5, whereas the susceptible check has shown 4 ± 6 percentage survivals. The improved Swarna-Sub1 lines with two and three blast resistance genes showed a survival percentage ranging from 81 ± 8 (SS30-24-111) to 84 ± 5 (SS30-24-46) (Table 1).
Agronomic performance of improved lines of Swarna-Sub1
The days to maturity of recurrent and donor parents ranged from 110 to 125 days. Among the three-gene harboring lines, SS30-24-46 displayed a significant difference for DM, NPT, PL, NGP, PY, TGW, and SF, whereas SS30-24-76 and SS30-24-107 showed the difference in PH and SS30-24-131 in NPT, PY, and SF compared to its parental line Swarna-Sub1 (Table 1 and Fig. 2). Among the lines carrying Pi1 and Pi54 genes, SS30-24-82 showed significant improvement for NPT, PW, PL, NGP, PY, TGW, and SF in comparison to the recurrent parent Swarna-Sub1 whereas among nine Pi2 and Pi54 genes carrying lines, SS30-24-73 had more PW, PL, NGP, PY, and SF (Table 1). The Amylose content of the recurrent parent was 25.8% and the backcross-derived plants showed almost similar content ranging from 25.1% (SS30-24-76 and SS30-24-131) to 26.6% (SS30-24-19) with similar grain type as that of Swarna-Sub1 (Table 1).
Phenotyping of selected BC3F5 plants for blast resistance. Swarna-Sub1, TN1 & HR12 showed a susceptible reaction, whereas the selected backcross-derived lines showed resistance reaction. RP Swarna-Sub1, P1 Swarna LT, P2 Swarna A51, S1 susceptible check TN1, S2 Susceptible check HR12, A, B, C three-gene pyramided lines SS30-24-46, SS30-24-131, and SS30-24-143
Discussion
The susceptibility of the elite rice variety, Swarna-Sub1, to blast disease is a major factor offsetting its overwhelming performance. The MABB approach coupled with phenotypic selection helped in improving the elite genotype, Swarna Sub-1, with blast resistance genes. The polymorphic markers between the parents play a crucial role in any MABB program. In the present study, the polymorphic markers between the donor and recurrent parents were only 13.2% since all the parents utilized in this study have similar genetic backgrounds, i.e., Swarna. The selection of markers that are evenly distributed on chromosomes was able to detect the percentage of recurrent parent genome recovered in any particular BC progeny and helps in reducing the donor genome and the number of generations essential to develop the lines with target genes (Visscher et al. 1996; Frisch et al. 1999). Khan et al. 2018 proposed the foreground selection using markers for transfer of target genes, which is more practical and economical but precise and accurate transfer of target genes relies mainly on the gene-based markers. In this study, both gene-based and tightly linked flanking markers were used for the foreground selection (Usatov et al. 2016; Ramkumar et al. 2011; Septiningsih et al. 2009). Similar studies were reported which employed these markers in rice improvement programs (Khan et al. 2018; Madhavi et al. 2016). The three-gene pyramided lines showed complete resistance to blast disease while two-gene combinations having Pi54 in common (Pi1 + Pi54, Pi2 + Pi54 and Pi2 + Pi54) also showed a high level of resistance. Even the lines having Pi54 showed resistance, which may be the due to the broad-spectrum resistance of the Pi54 gene (Ramkumar et al. 2011). Variation in the disease reaction among the two-gene harboring lines can be attributed to epistatic effects and background interactions with the QTL/genes (Singh et al. 2015). Single-gene harboring lines developed in the present study are also highly useful, which can be deployed in hotspot regions to understand the prevalence of predominant race, so as to decipher the effectiveness of the gene, thereby the deployment of gene combinations can be done effectively for durable resistance programs.
The extent of recurrent parent genome recovery in the developed lines was up to 93.5%. Such a high recovery can be attributed to recurrent genome recovery analysis employed from BC1 to BC3 generations. In the genetic background of landrace, Mushk Budji, Pi54, Pi1, and Pita genes were introgressed with a maximum RPG recovery up to 92% (Khan et al. 2018). An RPG recovery of 90.27% was observed in the Pi2 introgressed lines in the genetic background of BPT-5204 (Krishnamurthy et al. 2017). Both the foreground and background selection process reduces the transfer of undesirable donor genomic region besides recurrent parent genome recovery, thus minimizes the linkage drag. Such undesirable genes tightly linked to the target trait may have a negative effect on the agronomical performance of pyramided lines, likely low yield, or disease susceptibility. In the present study, two- and three-gene selected lines showed a less portion of donor genome segments of about 0.2–2.0 Mb on both sides of the target genes. The selected two- and three-gene lines have less linkage drag, as they have only target genomic regions. The foreground and background selection led to the development of improved PB 1, a basmati variety with xa13 and Xa21 with minimum linkage drag above 1.3 cM (Gopalakrishnan et al. 2008).
Stringent phenotypic selection is the key in any backcross breeding program, i.e., selection of plants in every generation which resembles the recurrent parent along with genotypic information was performed successfully in every generation of backcrossing and selfing in our present program. Agronomic performance of the three-gene pyramided line, SS-30-24-46, showed a high level of blast resistance as a well significant difference in plant yield (25.75%) compared to Swarna-Sub1. Similarly, the two-gene pyramided lines SS30-24-82 and SS30-24-73 also showed significant improvement in grain yield per plant but showed a delay in maturity and height compared to the recurrent parent. Earlier studies have reported that the yield mainly depends on the number of productive tillers and number of filled grains per panicle (Deshmukh et al. 2010). The increase in the productive tillers leads to an increase in panicle number and thereby higher grain yield per plant (Efisue et al. 2014). The results strongly support that, phenotypic selection practice was efficient which resulted in the identification of improved lines of Swarna-Sub1 lines with superior agronomic performance, grain quality compared to recurrent parent Swarna-Sub1 with an added advantage of blast resistance. In earlier studies, similar results were observed; the backcross-derived lines have shown a high level of resistance with best agro-morphological performance (Tanweer et al. 2015; Steele et al. 2006). The improved lines of Swarna-Sub1 having blast resistance are the better choice than the Swarna-Sub1 and these lines can also be used as donors for pyramiding blast resistance genes along with submergence in rice improvement programs.
References
Bailey-Serres J, Fukao T, Ronald P, Ismail A, Heuer S, Mackill D (2010) Submergence-tolerant rice: SUB1’s journey from landrace to modern cultivar. Rice 3:138–147. https://doi.org/10.1007/s12284-010-9048-5
Balachiranjeevi CH, Bhaskar NS, Kumar VA, Akanksha S, Viraktamath BC, Madhav MS, Hariprasad AS, Laha GS, Prasad MS, Balachandran SM, Neeraja CN, Kumar SM, Senguttuvel P, Kemparaju KB, Bhadana VP, Ram T, Harika G, Swamy HKM, Hajira SK, Yugander A, Pranathi K, Anila M, Rekha G, Kousik MBVN, Kumar TD, Swapnil A, Sundaram RM (2015) Marker-assisted introgression of bacterial blight and blast resistance into DRR17B, an elite, fine-grain type maintainer line of rice. Mol Breed 35:151. https://doi.org/10.1007/s11032-015-0348-8
Bhatia D, Sharma R, Vikal Y, Mangat GS, Mahajan R, Sharma N, Lore JS, Singh N, Bharaj TS, Singh K (2011) Marker-assisted development of bacterial blight resistant, dwarf, and high yielding versions of two traditional basmati rice cultivars. Crop Sci 51:759–770. https://doi.org/10.2135/cropsci2010.06.0358
Chaudhary B, Sah DN (1998) Efficacy of beam 75 WP in controlling leaf blast disease at the seedling stage of rice. Nepal Agirc Res J 2:42–47
Deshmukh R, Singh A, Jain N, Anand S, Gacche R, Singh A, Gaikwad K, Sharma T, Mohapatra T, Singh N (2010) Identification of candidate genes for grain number in rice (Oryza sativa L.). Funct Integr Genom 10:339–347. https://doi.org/10.1007/s10142-010-0167-2
Efisue AA, Umunna BC, Orluchukwu JA (2014) Effects of yield components on yield potential of some low land rice (Oryza sativa L.) in coastal region of south Nigeria. J Plant Breed Crop Sci 6:119–127. https://doi.org/10.5897/JPBCS2014.0449
Frisch M, Bohn M, Melchinger AE (1999) Minimum sample size and optimal positioning of flanking markers in marker-assisted backcrossing for transfer of a target gene. Crop Sci 39:967–975 (erratum: Crop Sci 39:1913)
Fukuoka S, Saka N, Mizukami Y, Koga H, Yamanouchi U, Yoshioka Y, Hayashi N, Ebana K, Mizobuchi R, Yano M (2015) Gene pyramiding enhances durable blast disease resistance in rice. Sci Rep 5:7773. https://doi.org/10.1038/srep07773
Gopalakrishnan S, Sharma RK, Rajkumar KA, Joseph M, Singh VP, Singh AK, Bhat KV, Singh NK, Mohapatra T (2008) Integrating marker-assisted background analysis with foreground selection for identification of superior bacterial blight resistant recombinants in basmati rice. Plant Breed 127:131–139. https://doi.org/10.1111/j.1439-0523.2007.01458.x
Hari Y, Srinivasa Rao K, Viraktamath BC, Hariprasad AS, Laha GS, Ahmed M, Nataraj Kumar P, Sujatha K, Srinivasprasad MS, Rani NS, Balachandran SM, Kemparaju S, Mohan KM, Sama VSAK, Shaik H, Balachiranjeevi CH, Pranathi K, Reddy GA, Madhav MS, Sundaram RM (2013) Marker-assisted introgression of bacterial blight and blast resistance into IR 58025B, an elite maintainer line of rice. J Plant Breed 132:586–594. https://doi.org/10.1111/pbr.12056
Hittalmani S, Parco A, Mew T, Zeigler R, Huang N (2000) Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet 100:1121–1128. https://doi.org/10.1007/s001220051395
Hospital F, Chevalet C, Mulsant P (1992) Using markers in gene introgression breeding programs. Genetics 132:1199–1210
Juliano BO, Perez CM, Blakeney AB, Castillo T, Kongseree N, Laignelet B (1981) International cooperative testing on the amylose content of milled rice. Starch 33:157–162. https://doi.org/10.1002/star.19810330504
Khan GH, Shikari AB, Vaishnavi R, Najeeb S, Padder BA, Bhat ZA, Parray GA, Bhat MA, Ramkumar Singh NK (2018) Marker-assisted introgression of three dominant blast resistance genes into an aromatic rice cultivar Mushk Budji. Sci Rep 8:4091. https://doi.org/10.1038/s41598-018-22246-4
Khanna A, Sharma V, Ellur RK, Shikari AB, Gopala Krishnan S, Singh UD, Prakash G, Sharma TR, Rathour R, Variar M, Prashanthi SK, Nagarajan M, Vinod KK, Bhowmick PK, Singh NK, Prabhu KV, Singh BD, Singh AK (2015) Development and evaluation of near-isogenic lines for major blast resistance gene(s) in basmati rice. Theor Appl Genet 128:1243–1259. https://doi.org/10.1007/s00122-015-2502-4
Krishnamurthy PS, Deshmukh DB, Kumar KJY, Patil S, Jakkeral S, Nemappa GH, Singh Variar M, Rathour R, Subbaiyan G, Singh AK, Sharma TR (2017) Introgression of Pi2 and Pi5 genes for blast (Magnaporthe oryzae) resistance in rice and field evaluation of introgression lines for resistance and yield traits. J Phytopathol 165:397–405. https://doi.org/10.1111/jph.12573
Kumar VA, Balachiranjeevi CH, Bhaskar NS, Rambabu R, Rekha G, Madhavi KR, Harika G, Vijay S, Pranathi K, Hajira SK, Srivastava A, Swamy HKM, Anila M, Yugander A, Aruna J, Hari Prasad AS, Madhav MS, Laha GS, Viraktamath BC, Balachandran SM, Senguttuvel P, Kemparaju KB, Ravindra Babu V, Sundaram Prasad MS (2016) Marker-assisted introgression of the major bacterial blight resistance gene, Xa21 and blast resistance gene, Pi54 into RPHR-1005, the restorer line of the popular rice hybrid, DRRH3. J Plant Biochem Biotechnol 25:400–409. https://doi.org/10.1007/s13562-016-0352-z
Lewis RS, Kernodle SP (2009) A method for accelerated trait conversion in plant breeding. Theor Appl Genet 118:1499–1508. https://doi.org/10.1007/s00122-009-0998-1
Mackill DJ, Bonman LM (1992) Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82:746–749
Madhavi KR, Rambabu R, Kumar AV, Kumar SV, Aruna J, Ramesh S, Sundaram RM, Laha GS, Madhav MS, Ravindrababu V, Prasad MS (2016) Marker-assisted introgression of blast (Pi-2 and Pi-54) genes in to the genetic background of elite, bacterial blight resistant indica rice variety, improved Samba Mahsuri. Euphytica 212:331. https://doi.org/10.1007/s10681-016-1784-1
Prasad MS, Madhav MS, Laha GS, Ladhalakshmi D, Krishnaveni D, Satendrakumar M, Balachandran SM, Sundaram RM, Arunakanthi B, Madhanmohan K, Ratnamadhavi K, Kumar V, Viraktamath BC (2011) Rice blast disease and its management. Indian Institute of Rice Research, Hyderabad
Ramkumar G, Srinivasarao K, Madhan Mohan K, Sudarshan I, Sivaranjani APK, Gopalakrishna K, Neeraja CN, Balachandran SM, Sundaram RM, Prasad MS, Shobha Rani N, Rama Prasad AM, Viraktamath BC, Madhav MS (2011) Development and validation of functional marker targeting an InDel in the major rice blast disease resistance gene Pi54 (Pik h). Mol Breed 27:129–135. https://doi.org/10.1007/s11032-010-9538-6
Ribaut J-M, Jiang C, Hoisington D (2002) Simulation experiments on efficiencies of gene introgression by backcrossing. Crop Sci 42:557–565
Scardaci SC, Webster RK, Greer CA, Hill JE, William JF, Mutters RG, Brandon DM, McKenzie KS, Oster JJ (1997) Rice blast: a new disease in California. Agronomy fact sheet series, Department of Agronomy and Range Science, University of California, Davis 1997–2
Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, Ismail AM, Mackill DJ (2009) Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot 103:151–160. https://doi.org/10.1093/aob/mcn206
SES-Standard Evaluation System for Rice (1996) International Rice Research Institute, Manila, Philippines
Sharma TR, Madhav MS, Singh BK, Shanker P, Jana TK, Dalal V, Pandit A, Singh A, Gaikwad K, Upreti HC, Singh NK (2005) High-resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to M. grisea. Mol Genet Genom 274:569–578. https://doi.org/10.1007/s00438-005-0035-2
Sharma TR, Rai AK, Gupta GK, Singh NK (2010) Broad-spectrum blast resistance gene Pi-kh cloned from the rice line Tetep designated as Pi54. J Plant Biochem Biotechnol 19:87–89. https://doi.org/10.1007/BF03323441
Singh VK, Singh A, Singh SP, Ellur RK, Singh D, Krishnan SG, Bhowmick PK, Nagarajan M, Vinod KK, Singh UD, Mohapatra T, Prabhu KV, Singh AK (2013) Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite basmati rice restorer line ‘PRR78’. Plant Breed 132:486–495. https://doi.org/10.1111/pbr.12077
Singh AK, Singh VK, Singh A, Ellur RK, Pandian RTP, Krishnan SG, Singh UD, Nagarajan M, Vinod KK, Prabhu KV (2015) Introgression of multiple disease resistance into a maintainer of basmati rice CMS line by marker-assisted backcross breeding. Euphytica 203:97–103. https://doi.org/10.1007/s10681-014-1267-1
Steele KA, Price AH, Shashidhar HE, Witcombe JR (2006) Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor Appl Genet 112:208–221. https://doi.org/10.1007/s00122-005-0110-4
Sundaram RM, Vishnupriya MR, Biradar SK, Laha GS, Reddy GA, ShobhaRani N, Sarma NP, Sonti RV (2008) Marker-assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160:411–422. https://doi.org/10.1007/s10681-007-9564-6
Swathi G, Rani CVD, Jamaloddin Md, Madhav MS, Vanisree S, Anuradha Ch, Kumar NR, Kumar NAP, Aruna Kumari K, Ramprasad E, Sravanthi P, Raju SK, Bhuvaneswari V, Rajan CPD, Jagadeeswar R (2019) Marker-assisted introgression of the major bacterial blight resistance genes, Xa21 and xa13, and blast resistance gene, Pi54, into the popular rice variety, JGL1798. Mol Breed 39:58–65. https://doi.org/10.1007/s11032-019-0950-2
Tacconi G, Baldassarre V, Lanzanova C, Faivre-Rampant O, Cavigiolo S, Urso S, Lupotto E, Vale G (2010) Polymorphism analysis of genomic regions associated with broad-spectrum effective blast resistance genes for marker development in rice. Mol Breed 26:595–617. https://doi.org/10.1007/s11032-010-9394-4
Tanweer FA, Rafii MY, Sijam K, Rahim HA, Ahmed F, Ashkani S, Latif MA (2015) Introgression of blast resistance genes (putative Pi-b and Pi-kh) into elite rice cultivar MR219 through marker-assisted selection. Front Plant Sci 6:1–11. https://doi.org/10.3389/fpls.2015.01002
Usatov AV, Kostylev PI, Azarin KV, Markin NV, Makarenko MS, Khachumov VA, Bibov MY (2016) Introgression of the rice blast resistance genes Pi1, Pi2 and Pi33 into Russian rice varieties by marker-assisted selection. Indian J Genet 76:18–23. https://doi.org/10.5958/0975-6906.2016.00003.1
Variar M, Cruz CV, Carrillo M, Bhatt J, Sangar R (2009) Rice blast in India and strategies to develop durably resistant cultivars. In: Wang GL, Valent B (eds) Advances in genetics, genomics and control of rice blast disease. Springer, Dordrecht, pp 359–373
Visscher PM, Haley CS, Thompson R (1996) Marker-assisted introgression in back cross breeding programs. Genetics 144:1923–1932
Wen-Yue C, Hai-Rui C, Jin-Song B, Xiang-Sheng Z, Qing-Yao S (2006) A simplified rice DNA extraction protocol for PCR analysis. Rice Sci 13:67–70
Zheng W, Wang Y, Wang L, Ma Z, Zhao J, Wang P, Zhang L, Liu Z, Lu X (2016) Genetic mapping and molecular marker development for Pi65(t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor Appl Genet 129:1035–1044. https://doi.org/10.1007/s00122-016-2681-7
Acknowledgements
The study is part of the Ph.D. research of the first author. We thank the University Grants Commission for the financial assistance under Rajiv Gandhi National Fellowship Scheme and Indian Institute of Rice Research, Hyderabad, for providing field and laboratory facilities.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patroti, P., Vishalakshi, B., Umakanth, B. et al. Marker-assisted pyramiding of major blast resistance genes in Swarna-Sub1, an elite rice variety (Oryza sativa L.). Euphytica 215, 179 (2019). https://doi.org/10.1007/s10681-019-2487-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2487-1