Abstract
Transposable elements usually comprise the most abundant nongenic fraction of eukaryotic genomes. Because of their capacity to selfreplicate and to induce a wide range of mutations, transposable elements have long been considered as ‘parasitic’ or ‘selfish’. Today, we recognize that the findings about genomic changes affected by transposable elements have considerably altered our view of the ways in which genomes evolve and work. Numerous studies have provided evidences that mobile elements have the potential to act as agents of evolution by increasing, rearranging and diversifying the genetic repertoire of their hosts. With large-scale sequencing becoming increasingly available, more and more scientists come across transposable element sequences in their data. I will provide examples that transposable elements, although having signatures of ‘selfish’ DNA, play a significant biological role in the maintainance of genome integrity and providing novel regulatoty networks. These features, along with the transpositional and mutagenic capacity to produce a raw genetic diversity, make the genome mobile fraction, a key player in species adaptation and microevolution. The last but not least, transposable elements stand as informative DNA markers that may complement other conventional DNA markers. Altogether, transposable elements represent a promising, but still largely unexplored research niche and deserve to be included into the agenda of molecular ecologists, evolutionary geneticists, conservation biologists and plant breeders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the late 1940s, Barbara McClintock challenged the existing concepts of genome organization and functioning when she discovered genes prone to mobility (McClintock 1950), which were later called ‘transposable elements’ (TEs). Although, the existence of TEs was accepted relatively soon after by the scientific community, the biology and applications of mobile genetic elements took decades to be widely recognized. With the discovery that many of these sequences are able to selfreproduce and to induce mutations, the selfish or parasitic DNA hypothesis was born. It said that these sequences served no function in the host organism, but were simply maintained by their ability to replicate and spread copies of themselves within and even between genomes (Doolittle and Sapienza 1980; Orgel and Crick 1980). In this ‘selfish’ way, TEs introduce genomic conflict trying to maximize their own fitness at the expense of the host’s genes (Burt and Trivers 2006; Werren 2011). Although, the TEs are primarily selfish and having deleterious effects, their activities may occasionally and stochastically confer a fitness advantage to their hosts. Nowadays, with the improvement of molecular tools for genome analysis including next generation sequencing technologies, the majority of scientists recognize that mobile elements, even behaving selfishly, play a significant biological role in the maintainance of genome integrity and diversification of the genetic repertoire of their hosts. Nevertheless, there is still an underestimation and/or lack of comprehension among scientists about the opportunity of studying TEs for resolving important research issues. The aim of this review was to highlight the significance of TEs as enhancers of genome dynamics and evolution, and to further disseminate this research issue to the biological community. First, I will provide a short overview of TEs, their distribution among eukaryotes and relation to genome size variation. Then, I will emphasize the evolutionary consequences of TEs for genome functioning and integrity through some examples in the plant and animal kingdoms. Finally, I will focus on the practical applications and perspectives of TEs for genome analysis and manipulation.
Structure and abundance of TEs
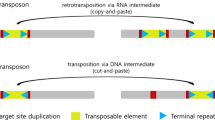
TEs are DNA sequences that can change their positions within the genome through the mechanism called transposition. There are two major types of TEs depending on their mechanism of transposition (Wicker et al. 2007). Class II elements (or DNA transposons) move through ‘cut and paste’ mechanism which comprises excision of a TE copy from one place in the genome followed by its reinsertion into another place. DNA transposons are present in low–moderate copy numbers in almost all eukaryotes. Class I retrotransposons transcribe RNA intermediates from genomic copies that are reverse-transcribed into a double-stranded DNA and integrated into a new position. Several daughter copies can be produced from a mother copy and insert throughout the genome, this proliferation has made retrotransposons a major fraction of large genomes. Eukaryote genomes are populated with large fractions of TEs. In plants, the repeat sequences range from 3% of the small genome of Utricularia gibba (Ibarra-Laclette et al. 2013) to more than 85% of the large genomes of cereals and maize (Li et al. 2004; Schnable et al. 2009). The human genome consists of about 65% of TEs (de Koning et al. 2011).
C-value paradox, transposable elements and the concept of ‘junk’ DNA
With the availability of many sequenced genomes (Michael and Jackson 2013) and the recently updated information of whole-genome sequences, it became evident that there is a striking variation in genome size in angiosperms which is often poorly correlated with the number of genes that code for proteins or the presumed evolutionary complexity of species (although we still do not understand how to meaningfully measure an organism’s complexity). This observation is the core of the C-value paradox raised more than 30 years ago where ‘C’ stands for the haploid genome size (Gall 1981). Genome sizes substantially vary between taxas: more than 7000-fold among animals (Gregory and DeSalle 2005) and 2400-fold across land plants as evidenced in the recent update of the plant genome size database containing data for 8510 species (Bennett and Leitch 2012; Garcia et al. 2014). A similar trend is observed between close species at a same ploidy level and apparently comparable complexity. A typical example is the plant genus, Eleocharis, which comprises more than 250 species, in which E. acicularis (2 n= 20, C = 0.25 pg) is 20 times smaller than E. palustris (2 n= 16, C = 5.5 pg) (Zedek et al. 2010). This genome size variation results from a substantial fraction of extra DNA other than for protein-coding genes and regulatory sequences, and is comprised of introns, pseudogenes, satellite sequences and TEs. This noncoding DNA has been considered a ‘junk DNA’ that has no biological function and conveys little or no selective advantage to the organisms (Orgel and Crick 1980).
The recent discovery that some regions of the noncoding DNA play important functions in genome and cell integrity has challenged the concept of useless ‘junk’ DNA. The noncoding DNA was found to be essential for maintaining the chromosome structure, the function of centromeres, binding of transcription factors and coding for RNA involved in gene silencing. The ENCODE project represents a significant breakthrough study into the understanding the proportion of human’s genome that is functional. A function was assigned for 80% (dominated by RNA transcription, which alone covered 62%) of the genome particularly outside of protein-coding genes (ENCODE Project Consortium 2012). ENCODE and other studies have indicated that 5–20% of the human genome is under detectable selective pressure. Interestingly, the genome fraction directly involved in gene regulation was found significantly higher (up to 8-fold) than that ascribed to protein-coding exons (1%). Soon after, the ‘function’ by any meaningful sense of the word of the ENCODE-defined functional elements has been criticized and questioned (Eddy 2012; Doolittle 2013; Graur et al. 2013; Niu and Jiang 2013). These authors argued that DNA is still ‘junk’ despite the fact that it may bind transcription factors and contain regions of modified chromatin. ENCODE project represents the first genomewide functional annotation of the human genome. However, it does not directly address the ‘junk’ DNA concept which still remains viable.
TEs, particularly retrotransposons, comprise a major fraction of the ‘junk’ DNA and to a larger extent contribute to genome size expansion and variation among species, even as gene numbers remain relatively constant (table 1). The Arabidopsis genome, e.g. contains about 27,000 genes and 20 to 25 Mb of retrotransposons, whereas the maize genome contains about 40,000 but more than 1800 Mb of retrotransposon sequences (Liu and Bennetzen 2008; Baucom et al. 2009; Schnable et al. 2009). The ‘selfish’ nature of TEs acting as molecular parasites and functioning for themselves rather than having an adaptive function for their host genome was postulated by Doolittle and Sapienza (1980) and Orgel and Crick (1980). Since then, TEs have been a subject of tremendous interest because of their abundance, functionality and role in genome evolution. Do TEs embrace the concept of useless and nonfunctional ‘junk’ DNA? Definitely not for the whole genome fraction of TEs. First of all, many TEs are functional. Their DNA is biochemically active, encode proteins, bind proteins, synthesize regulatory RNA, thus meets the ENCODE criterion of functional elements. Second, many TEs are not ‘junk’ as there is a plenty of ways (see below) through which they provide a benefit to their host genomes. There is, however, a substantial fraction of decaying dead TEs generated from active TEs in the evolutionary past. It is suggested that different loads of such TE relics mostly explains the C-value paradox with larger genomes having a larger fraction of them. One should accept that there is a portion of DNA that seems to serve little useful purpose for the organism. However, I would like to point out on the inappropriateness of referring to DNA sequence as ‘junk’ unless we do not completely understand or have characterized it. DNA that appears useless today may provide a reservoir of sequences from which potentially advantageous new genes can emerge in future (Burt and Trivers 2006). In this way, it may be an important genetic basis for evolutionary innovation. After all, there might be a small amount of DNA that is a true junk. This DNA may just act as a protective buffer against the accumulation of harmful mutations.
In parallel to the interplay between genome size variation and TE accumulation, a recent study explored the link between TE diversity (types and number of TEs) and genome expansion (Elliot and Gregory 2015). The authors showed that this correlation is straightforward only to a certain point of genome size (specifically, around 500 Mbp), and then manifested by either a lack of relationship in animals or a negative correlation in plants. The likely common scenario may be that TE diversity and abundance increase as genomes expand up to a moderate size, whereas further genomic growth beyond this point is driven by a proliferation and divergence of a small subset of TE superfamilies. For example, 50% of the barley genome is made up of just 14 families, 12 of which long-terminal repeats (LTR) retrotransposons (Wicker et al. 2009). Also, retrotransposons BARE-1, Wis and Angela account for more than 10% of the Triticeae genomes. Consistently, differences in BARE-1 abundance primarily explain genome size variation among Hordeum species (Vicient et al. 1999). One point that should be emphasized, however, is that the establishment of a more or less stable equilibrium between genome size and TE proliferation within and among taxa is influenced by several other selection factors like population size, mating system, polyploidization events and ecogeographical distribution. The complexity of the interacting factors means that many comparative studies need to be done before patterns of TE abundance and diversity versus taxon-specificity and genome size can be described and understood.
Epigenetic silencing drives the ups and downs of transposable elements
Why TEs, apparently useless and potentially damaging, are widely spread in higher organisms? Generally, the abundance and accumulation rates of TEs result from a balance between two main forces: TE transposition leading to an increase in copy number and, from the other side, the elimination and inactivation of TEs mediated by mutations in their sequences or through the process of ectopic recombination between TE copies at nonhomologous loci (Charlesworth and Charlesworth 1983). Recombination events between TEs at nonhomologous chromosome locations lead to the generation of truncated inactive elements thus reducing their functional activity and accumulation rate. This balance is widely achieved by epigenetic silencing of TEs by siRNA-directed DNA methylation (Ito 2012). The mechanism consists of the synthesis of small TE-specific noncoding RNAs that guide DNA methylation and silencing of homologous DNA sequences at posttranscriptional level (Lister et al. 2008). Differential expression of TEs influenced by siRNA-directed DNA methylation was observed between different plant genomes (Alzohairy et al. 2014). For instance, differences in TE accumulation and genome size are largely influenced by the extent of TE-silencing as shown in a comparison study between Arabidopsis lyrata and A. thaliana (Hollister et al. 2011). Fedoroff (2012) argued that actually the evolution of epigenetic mechanisms, that control homology-dependent recombination, is driving the accumulation rate of TEs in a long-term aspect. The methylation in prokaryotes represents a nuclear defense system to limit the destructive potential of ‘parasitic sequences’ including TEs (Yoder et al. 1997). Consistently, the epigenetic control is not so efficient in prokaryotes and lower eukaryotes, and ectopic recombination among dispersed TEs would rapidly eliminate them either directly by deletions or indirectly by creating nonviable chromosomes. As a result, these processes keep the genomes of prokaryotes and many lower eukaryotes small. In contrast, higher eukaryotes have larger genomes due to more stringent epigenetic control and lower recombination rates that allow the accumulation of TEs.
Transposable elements are capacitors of genome dynamics
It is obvious that the high frequency of TEs, their capacity to change the location within the host genome and to induce chromosome aberrations (e.g. deletions, duplications and insertions) would confer a potentially negative impact of TEs on genome integrity. Transposable elements can affect genome dynamics with possible effects on phenotypes through multiple mechanisms, depending on the TE itself and its insertion site. Some examples of the effects of TE insertions on gene structure and function are shown in figure 1.
Effects of TE insertions on gene structure and function. (a) Insertion of LTR retrotransposon upstream of the Ruby gene in the blood orange provides a new promoter controlling the expression of a gene for flesh colouration of the fruit (Butelli et al. 2012). (b) Insertion of TE into the rugosa locus encoding a starch-branching enzyme in pea results in a wrinkled seed (Bhattacharyya et al. 1990). Insertion of TE into a gene that produces anthocyanin pigments leads to its inactivation and changes in the colour: yellow seeds in maize. (c) White sectors in Petunia hybrida (d) Kroon et al. (1994). Revertant sectors (dark-spotted seeds or blue stripes) appear when the transposon spontaneously excises from that gene in certain cells and the pigment production is restored. (e) siRNA-controlled methylation of a TE insertion influence the expression of the FLC locus (a gene delaying flowering) and leads to earlier flowering in A. thaliana (Liu et al. 2004). (f) Alterations in the transcription levels of genes by antisense transcription from adjacent TE insertions as observed for the agouti colour gene in mice (Morgan et al. 1999).
Transposable elements-mediated alterations in the structure and function of genes
The most obvious TE-induced change is gene disruption leading to observable loss of function. A classical example are mutations in the gene coding for the colour of the maize kernels provoked by the Ac element detected by Barbara McClintock (figure 1c). Similarly, the wrinkled seed phenotype studied by Mendel in pea was found to result from an insertion of a TE into a locus encoding a starch-branching enzyme (Bhattacharyya et al. 1990). Beside plant genomes, ∼0.3% of all human mutations are caused by TE insertions or rearrangements (Cordaux and Batzer 2009). The currently active nonLTR retrotransposons, L1 (LINE 1), SVA and Alu (SINE), are reported to be the causative factors of many genetic disorders, such as haemophilia A, Apert syndrome, familial hypercholesterolaemia, colon and breast cancer, muscular dystrophy etc. (for review see O’Donnell and Burns 2010; Ayarpadikannan and Kim 2014). Alu elements (named for the enzyme used to identify it) are short sequences (∼300 bp) that occur almost a million times in the human genome and comprise up to 3.5% of the total DNA. Transposition of Alu elements to sites in and near genes, or Alu-mediated ectopic recombination events can have occasional deleterious effects on genes. Indeed, many cloned genes were shown to harbour Alu elements in their sequences. On the other hand, Alu sequences likely have positive regulatory functions as mutations within them have been associated with cancer. Transposable elements can also disrupt existing regulatory motifs (repressors or enhancers) or to provide new regulatory information thereby influencing the gene expression and causing mutant phenotypes. Several examples about such effects of TEs are reported in Drosophila (Lerman et al. 2003; Lerman and Feder 2005; McCart and Ffrench-Constant 2008) and plants (Salvi et al. 2007; Studer et al. 2011; Butelli et al. 2012).
Transposable elements affect the epigenetic regulation of genes
A substantial part of TEs are targeted by siRNA-directed DNA methylation that is repressing their activity. This epigenetic silencing can spread to genes located in the vicinity of TE insertions and may generate stable epialleles of potential evolutionary relevance (Slotkin and Martienssen 2007; Wang et al. 2013). For instance, an insertion of a LTR retrotransposon near the agouti gene in mice alters the chromatin state and DNA methylation at this locus (Morgan et al. 1999). Variations in the epigenetic status of the retrotransposon ultimately influence the gene transcript level and the colour of the mice coat (figure 1f). Similarly, early flowering of the Ler ecotype of A. thaliana is controlled by a DNA transposon insertion in the FLC gene responsible for a delayed flowering. This insertion is targeted by TE-derived siRNAs which results in the epigenetic silencing of the gene (Liu et al. 2004).
Transposable elements can mediate genome restructuring
Recombination events between TE copies at nonhomologous sites in the genome (ectopic recombination) either on one or different chromosomes is important mechanism by which TE mediate genome restructuring (deletions, insertions, inversions, translocations and duplications), thus promoting chromosomal instability (Gray 2000). In addition, TEs can enhance genome reshuffling by capturing and transferring genes within genomes (Jiang et al. 2004; Morgante et al. 2005; Schaack et al. 2010).
The number and the variety of mutations induced by TEs is extraordinary and can be hardly embraced. One can still argue that TEs have predominantly negative impact and increase the genetic load and scientists ask the question: Do TEs, through the induction of mutations or regulatory functions, provide some benefit to the organism itself or just stand as pure ‘selfish’ and detrimental DNA?
TEs have important biological functions
During the last two decades, a major focus has been on the positive contribution of TEs to the evolution of gene regulation and diversification of host genes. The major break-through in this area was achieved with the advent of high-throughput sequence technologies and software platforms for the annotation of TEs. We have tremendously enriched our knowledge about the biology of TEs and their interaction with other components of the host genome. One way in which TEs contribute to evolution is that their sequences (e.g. genes, binding sites, and terminal repeats) can be coopted to perform functions beneficial for the proper functioning of the host genome (Sinzelle et al. 2009). This genomic coopting of a molecular parasite is often referred to as molecular domestication or exaptation (Gould and Vrba 1982; Miller et al. 1999). Here I present examples of the different ways that TEs have evolved from strictly parasitic elements to mutualistic sequences that benefit their host genomes (see also table 2).
Transposable elements as a source of novel regulatory networks
The human genome provides the majority of examples about TEs involved in domestication which has helped to spur remarkable evolutionary innovations. The initial analysis of the human genome has revealed that ∼25% of human promoter regions and ∼4% of human exons contain sequences derived from TEs (Lander et al. 2001; Nekrutenko and Li 2001; Jordan et al. 2003; Kapitonov and Jurka 2005; Jurka et al. 2007).
V(D)J recombination is a unique mechanism of genetic recombination that occurs only in developing lymphocytes during the early stages of T and B cell maturation. The process results in a highly diverse repertoire of antigen receptors in these cells and is a distinguished feature of the adaptive immune system in vertebrates. The V(D)J recombination is mediated by genes RAG1 and RAG2 that are evolutionarily derived from ancient insertions of Transib DNA transposons (Zhou et al. 2004; Ramsden et al. 2010). As expected, based on our knowledge about class II TEs transposition, this process involves the generation of double-strand DNA breaks in a way that is mechanistically similar to the ‘cut’ component of ‘cut and paste’ transposition.
Repeat-induced gene silencing involving L1 retroelements has been hypothesized for X-chromosome inactivation, which is necessary to maintain the proper gene dosage in females. Inactivation is initiated at the X-chromosome inactivation centre (XIC) from which the silencing signal spreads along the chromosome. According to one hypothesis, LINE retrotransposons might trigger the heterochromatization in XIC centre and boost the efficient spread of the silencing away this centre (reviewed in Lyon 2006). In support to this idea, the X chromosomes of many mammals, including humans, are rich in LINE elements, except in regions that are prone to escaping X inactivation (Ross et al. 2005). It is still unknown whether LINEs function in the spread of heterochromatin on the X chromosome or their enrichment may simply be a consequence of the heterochromatic nature of the inactive X. However, Cohen et al. (2007) have shown that short tandem repeats homologous to retrotransposons regulate X-chromosome inactivation by producing bidirectional transcripts in differentiating mouse cells, thus providing indirect evidence that TEs function in both the initiation and spread of X inactivation.
Transposable elements are also functionally implicated in the proper functioning of the mammalian embryo at earliest stages of its development (Macfarlan et al. 2012; Tomkins 2013). In this line, it was recently reported that a family of TEs in mammals provide enhancer sequences that modulate the gene expression in placental cells thus regulating the interaction between the mother and the offspring (Chuong et al. 2013). In humans, the highly conserved centromere-binding protein CENBP facilitates centromere formation and is derived from transposases of the pogo DNA transposon family (Casola et al. 2008).
Several examples about TE exaptation are available in the plant kingdom as well. In Arabidopsis, FHY3 and FAR3 are transcription factors, related to the MuDR family of transposases, that bind to promoter regions and activate several genes involved in far-red light and circadian clock signalling (Hudson et al. 2003; Lin et al. 2007).
A recent study in Arabidopsis has shown that the COPIA-R7 transposon, inserted into the plant disease resistance gene RPP7, enhances its host’s immunity to a pathogenic microorganism from a large group of fungus-like parasites that cause a number of plant diseases (Tsuchiya and Eulgem 2013). The Rim2 gene implicated in defence against fungal infection appears to have been directly exapted from part of a CACTA DNA TE element (He et al. 2000). The rice blast disease resistance gene Pit was refunctionalized by the recruitment of a copia-like LTR element as a promoter (Hayashi and Yoshida 2009).
Relatively few data are available about the direct role of TEs in the processes of species domestication. An insertion of the TE Hopscotch in the regulatory region of the maize domestication gene, teosinte branched1 (tb1), acts as an enhancer of its expression and confers the increased apical dominance in maize compared to its progenitor, teosinte (Studer et al. 2011). Insertion of a CACTA-like transposon into the promoter of the gene ZmCCT, which modulate photoperiod sensitivity, can suppress its expression thus enhancing the spread of maize to long-day temperate regions (Yang et al. 2013). Mustang and Sleeper gene families present in flowering plants have sequences derived from exapted transposases from Mutator-like and hAT DNA elements, respectively (Joly-Lopez et al. 2012; Knip et al. 2012). Mustang genes are present only in the angiosperm lineage and encode putative transcriptional regulators that play important roles in growth, flower development and reproduction.
Transposable elements maintain chromosome stability and functioning
The coexistence of TEs and the host genome has resulted in several regulatory pathways, including a combination of various epigenetic mechanisms, i.e. DNA methylation, small RNAs and histone posttranslational modifications. In A. thaliana, methylated TEs promote the epigenetic silencing and the formation of heterochromatin (Fagegaltier et al. 2009). TEs are abundant in heterochromatin-rich regions, centromeres (Casola et al. 2008; Mateo and González 2014) and telomeres (Levin and Moran 2011; Pardue and DeBaryshe 2011). Such phenomena might be the consequence of: (i) insertional preferences of TEs into heterochromatin; (ii) positive selection of TE maintenance into heterochromatin for genomic stability; or (iii) an induction of heterochromatin by TE sequences. An example of a host using the movement of a retrotransposon to its advantage was found in the telomere maintenance of Drosophila. Two retrotransposons, HeTA and TART, are present in multiple copies on the telomeres and maintain their length in replicating cells (Abad et al. 2004; Shpiz et al. 2007). A correlation between TEs activity and the maintenance of heterochromatin integrity was also demonstrated by the observation that the increase in TE expression and transposition leads to an age-related breakdown in heterochromatin structure and subsequent cellular dysfunction (Wood and Helfand 2013). St Laurent et al. (2010) argued that the stress-induced activation of LINE/L1 elements, particularly prevalent in mammals, leads to mutagenic insertions and DNA damage that accumulate with age and can cause genomic instability even in the absence of a successful transposition event. This fact shows that LINE elements may play an important role in mammalian ageing and evolution. In support to these data a study in budding yeast, using a chronological ageing model, observed that the yeast retrotransposon Ty1 showed increased mobility with age, and this was correlated with chromosome rearrangements and other hallmarks of genomic instability (Maxwell et al. 2011).
Matrix attachment regions (MARs) are DNA sequences that bind to the nuclear matrix forming functionally independent chromatin domains. Colocalization of TEs with MARs located in introns and 5 ′flanking regions of genes was observed which suggests the putative role of mobile elements and/or their products (transposases) in the formation of these structures (Avramova et al. 1998; Tikhonov et al. 2000; Pathak et al. 2014). For instance, the insulator protein BEAF-32 in Drosophila is entirely derived from a hAT transposase and is involved in chromatin functioning through interactions with the nuclear matrix (Pathak et al. 2007). A cross talk between the distribution and genome organization of BEAF-32 contributes to new gene-expression profiles and distinct phenotypes with a putative role in the evolution. Nevertheless, the functional link between MAR and retrotransposons remains to be comprehensively investigated.
Although, several domesticated TEs have been reported so far, their total number may be much higher than currently reported and many more domesticated TEs may await discovery in the near future. Traditional genetic methods may be insufficient to find such TEs and to assess their evolutionary and functional impact, e.g. due to functional redundancy. Systematic searches that exploit genomic signatures of natural selection have been employed to identify potential domesticated genes, but their predictions have yet to be experimentally verified.
TEs as capacitors of species adaptation to changing environments
Most TEs are usually in a transpositionally silent state but can be occasionally activated in response to different environmental stimuli (Grandbastien 1998), a phenomenon what Barbara McClintock called ‘genome shock’ (McClintock 1984). The major question is whether TE activity is just an undesired side effect of stress exposure or TE-induced genetic diversity accounts for microevolutionary processes such as rapid adaptive evolution and speciation in natural populations. A prevalent view among evolutionary biologists is that the vast majority of TE insertions are nearly neutral and unlikely to have a strong evolutionary impact. Some host forces do indeed select against TE insertions (due to the deleterious impact of insertions or of their effects through ectopic recombination) and to efficiently purge deleterious insertions. However, the selection on TEs accumulation at population level is affected by a complex of factors and the outcome is often difficult to be predicted. Nevertheless, several reports provide convincing evidence about a turnover of TEs closely matching ecogeographical distribution of gene pools. For instance, increase of full length copies of the retrotransposon BARE-1 was observed in barley populations in dry environments compared to those grown a few dozen metre apart in less stressful habitats (Kalendar et al. 2000). A good example of the capacity of TEs to elicit mutational consequences potentially helpful in adapting to new environments is provided by three diploid sunflower species, Helianthus anomalus, H. Deserticola and H. paradoxus. The three hybrid taxa, independently derived through hybridization events between the two parental taxa, H. annuus and H. petiolaris, encountered a rapid, retrotransposon-mediated genome expansion and all of them occupy habitats considered abiotically extreme relative to either parental species (Ungerer et al. 2006; Kawakami et al. 2010). Another example of a TEs involved in the adaptation is the early flowering of the Ler ecotype of A. thaliana controlled by a DNA transposon insertion (Liu et al. 2004). In addition, Gonzalez et al. (2010) have shown that at least 32% of the putatively adaptive insertions screened in natural population of D. melanogaster have a distribution consistent with selection by contrasted ecogeographical conditions. Most of these TE insertions, belonging to multiple TE families, were linked to functional genes with distinct phenotypic changes.
Insights offered by such reverse population genomic approaches pinpoint the importance of TEs as a source of adaptive variation. However, such surveys should be undertaken in a wider range of species to reliably estimate the impact of TEs on their evolutionary ecology.
Transposable elements are reliable DNA molecular markers
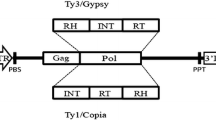
The high copy number, chromosome coverage and variable arrangement pattern even among closely related species give TEs advantage as informative markers to assess natural and stress-induced genetic diversity, and to enhance marker assisted selection (MAS) in plant breeding programmes. Both DNA transposons and retrotransposons can be utilized for the generation of markers, the latter ones being much more efficient. Retrotransposons have been found to comprise the most common class of TEs in eukaryotes, and represent up to 90% in plant genomes (Feschotte et al. 2002). They constitute for >50% of the maize and cereal genomes (Meyers et al. 2001; Brenchley et al. 2012) and 14% of the Arabidopsis genome (The Arabidopsis Genome Initiative 2000). Moreover, the presence of conserved domains at both ends (LTR) can be easily exploited for the design of PCR primers (Kalendar et al. 2011).
Major retrotransposon-based markers and type of inheritance
Retrotransposon-based molecular analysis relies on PCR amplification of DNA sequences (markers) using a primer corresponding to a retrotransposon and a primer matching a section of the neighbouring genome (e.g. microsatellite, restriction site or another TE copy). Among the wide diversity of retransposon-based techniques, the three most frequently used, as tools for diversity studies are sequence-specific amplified polymorphism (SSAP) (Waugh et al. 1997), interretrotransposon amplified polymorphism (IRAP) (Kalendar et al. 1999), retrotransposon microsatellite amplified polymorphism (REMAP) (Kalendar et al. 1999) and retrotransposon-based amplified polymorphism (RBIP) (Flavell et al. 1998). A schematic representation of the main marker methods is presented in figure 2. Several molecular marker systems based on the information available for the transposable elements sequences were developed to measure diversity, similarity and cladistic relationships in plants: barley (Kalendar et al. 2000; Vicient et al. 2001), citrus (Breto et al. 2001), genus Malus (Antonius-Klemola et al. 2006), rice (Branco et al. 2007), flax (Melnikova et al. 2014,), pea (Ellis et al. 1998; Pearce et al. 2000; Smýkal 2006), wheat (Queen et al. 2004; Melnikova et al. 2011), pear (Kim et al. 2012) and others. Transposable elements find substantial application as genetic markers in animal kingdom as well. For example, the method transposon display has been recently used to study the effect of interspecies hybridization on TEs dynamics in Drosophila (Vela et al. 2014). In addition, mobile elements are found to be active and to provide polymorphism in human populations (Mills et al. 2007). Consistently, SINE (Alu) and LINE (L1) elements have been used to trace human roots to Africa (Witherspoon et al. 2006). Also, L1 insertion polymorphisms have potential use in forensic analyses (Ray et al. 2007).
Main transposon-based marker methods tracking polymorphism in the insertion pattern of TEs. The techniques rely on simultaneous PCR amplification of sequences between copies of a candidate TE and adjacent genomic regions which can be other TE copies interretrotransposon amplified polymorphism (IRAP); microsatellite loci, retrotransposon-microsatellite amplified polymorphism (REMAP) or restriction sites, sequence-specific amplified polymorphism (SSAP). Red arrowheads indicate the primers for PCR amplification.
RBIP is the only retrotransposon-based method directing at polymorphism in the integration of an element at a single-copy locus and can distinguish between its heterozygous or homozygous state, thus having a codominant inheritance. Other marker techniques generally behave as dominant (i.e. presence/absence of a TE insertion) and do not allow one to discriminate the allelic state of the locus. Even though, it may be possible to map two polymorphisms to the same TE integration site but this is very tedious in practice and can not be determined directly. The difficlulty comes from the complexity of band profiles in multitarget PCR reactions and the less sufficient resolution provided by the commonly used standard agarose gel and polyacrylamid gel electrophoresis (PAGE) methods of amplicon separation. The development of high-throughput marker method for genetic studies, the fluorescent SSAP system, improved the available amplicon number and the accuracy in their scoring which allow to dicriminate alleles and to identify heterozygous loci at a resolution of a single nucleotide (Knox et al. 2009). Another way to overcome the dominant nature of the other marker systems is to use genetically homozygous material. For mapping populations, this can be achieved using double-haploid, recombinant inbred lines or haploid tissues like the endosperm of gymnosperms. The efficacy of double-haploid populations for the mapping of retrotransposon markers and genes has been well established (Waugh et al. 1997; Manninen et al. 2000).Advantages of retrotransposon-based markersThere are few properties of TEs that give them an advantage over other DNA molecular markers.
Transposable elements are prone to activation by stress
As mentioned earlier in this review, TEs are prone to activation by different stress factors. Beside the few examples of TEs dynamics at population level, a plethora of studies have also demonstrated the stress-induced escape from the silent state for many TE families under control experimental conditions. In plants, these activating events include artificial interspecific hybridization and allopolyploidization (for review see Parisod et al. 2009; Yaakov and Kashkush 2011), infection (Melayah et al. 2001; Grandbastien et al. 2005), abiotic stresses like drought (Aprile et al. 2009), high and low temperatures (Ivashuta et al. 2002; Young et al. 2005), protoplast isolation, cell culture, wounding, methyl jasmonate, CuCl 2 and salicylic acid (Hirochika 1993; Moreau-Mhiri et al. 1996; Mhiri et al. 1997; Takeda et al. 1998). The activation of wheat retrotransposons under light and salinity stresses has been also reported (Woodrow et al. 2010). Stress response may differ between host genotypes possibly reflecting an adaptive response of ancestral populations to different stimuli. Therefore, these and more studies show without doubt that TEs can be used as reliable DNA markers to assess genome response to stress factors both at individual and population levels.
Although, the exact process of transcription induction remains not completely elucidated, it has been shown that TEs are likely to become activated by mechanisms similar to those employed by host defense genes. Indeed, promoter sequences of both TEs and host defense genes share nucleotide similarities and are likely to bind to similar transcription factors (Casacuberta and Santiago 2003). For instance, Tnt1 retrotransposons from tobacco present regulatory regions with specific motifs that are commonly observed in genes induced by drought, anaerobic conditions or oxidative stresses (Grandbastien et al. 2005). Transposable elements could have captured promoters from normal stress-inducible genes or inversely, that they provided their own stress-inducible promoters to some plant defense genes (Grandbastien et al. 1997; Takeda et al. 1999).
Transposable elements display high level of insertional polymorphism
DNA marker techniques based on TEs are anonymous, producing fingerprints from multiple sites of retrotransposon insertions in the genome (Schulman et al. 2004). The outcome is a high degree of heterogeneity and insertional polymorphism observed both within and between species. One can still narrow the regions upon which one is looking for TE polymorphism. To achive this goal, one should rely on the biology and insertional pattern of TEs. For instance, high copynumber families like the active LTR retrotransposon BARE-1 tend to form clustered (i.e. adjacent insertions) and nested (i.e. insertions within one another) arrangements in intergenic regions of large genomes. In contrast, insertions of short TEs such as Ac/Ds, MITEs, LINEs, SINEs, but also low copy number LTR retrotransposons, seem overrepresented near or within genes in plants and in humans (i.e. mostly in introns, 5 ′ or 3 ′ UTR as well as in flanking regions; Wright et al. 2003; Majewski and Ott 2002). In addition, retrotransposons allows to detect large genome changes and seem to be more informative as a complement to convenient DNA markers like amplified fragment length polymorphism (AFLP), microsatellites (SSR) and single-nucleotide polymorphism (SNP) (Waugh et al. 1997; Ellis et al. 1998; Yu and Wise 2000; Porceddu et al. 2002; Tam et al. 2005) which mainly detect single nucleotide changes.
Retrotransposons are homoplasy-free phylogenetic markers
The regions of transposon insertions are thought to be more or less random. Thus, TE copies at exactly the same location in homeologous chromosomes within and among individuals appear unlikely. Retrotransposon integrations are also assumed to be irreversible events unless a chromosomal segment containing the repeat becomes deleted. An insertion at a specific genomic location suggests a derived character state, and species which share an insertion at a particular locus are grouped together on the tree. Lack of an insertion at an orthologous locus is regarded as an ancestral state and the corresponding individuals are considered basal in the phylogenetic tree. Thus, as opposed to reversible changes in DNA sequence composition, retrotransposon insertions have been claimed to generate homoplasy-free phylogenetic markers that provide an extremely accurate picture of evolutionary relationships and have been proven very successful in elucidating problematic phylogenies. For example, SINE elements were evaluated as reliable cladistic markers to resolve phylogenetic relationships among human and nonhuman primates, clinical diagnostics of diseases and forensic identification (for review see Schmitz et al. 2005; Ray 2007; Konkel et al. 2010; Ray and Batzer 2011).
Transposable elements as a tool for gene manipulation
Insertional mutagenesis
As discussed earlier, TEs can change their genomic location upon activation. If inserted inside the coding or regulatory sequence of a gene, disruption of the reading frame can lead to a loss of gene function. This phenomenon provides the platform for the development of the technique called transposon mutagenesis which has been used to induce mutations, identify and study the function of the responsive gene. The standard mutagenesis platform consists of crossing genetic lines with inactive or nonautonomous TEs with lines containing an active (autonomous) element. The offspring carrying an autonomous transposon, through its transcribing transposase, can mobilize the nonautonomous transposon. The reactivated transposon can further insert randomly into new genomic sites thus causing mutations in the offspring. Hundreds and thousands of individuals can then be screened for a new mutation of interest. If such a phenotype is found, then it can be assumed that TE insertion has inactivated the gene responsible for this phenotype. Because, the sequence of TE is known, the gene can be easily identified either by sequencing the whole genome and searching for the sequence or using PCR to amplify specifically that gene. Several examples are available on genes succesfully tagged using TEs in species like tobacco (Fitzmaurice et al. 1999), maize (Howard et al. 2014) and tomato (van der Biezen et al. 1996).
Transposable elements as gene delivery vehicles
Virtually any DNA sequence of interest can be placed between the terminal inverted repeats (TIRs) of a TE and mobilized in trans by supplementing the transposase gene in the form of an expression plasmid or through mRNA synthesized in vitro. This feature makes TEs natural and easily controllable DNA delivery vehicles to transfer genes to a host organisms’ chromosome for the purposes of introducing new traits and to discover new genes. The use of TEs as nonviral vehicles for persistent gene delivery has emerged with the discovery of DNA transposon ‘sleeping beauty’ as a tool for genetic modifications and persistent gene expression was demonstrated in a wide variety of vertebrate cell lines and species (Ammar et al. 2012). Several other synthetic DNA transposon system vehicles like the PiggyBac (PB) transposon with activity in mammalian cells have been studied and tested (for review see Skipper et al. (2013)). Transposon-based gene transfer is more efficient for stable expression of foreign genes in vertebrates compared to classic approaches as the latter ones rely on physical methods of gene constructs delivery: transfection, electroporation, sonoporation, or microinjection. The main drawbacks of these approaches are the low rates of genomic integration and unstable expression of the chromosomally integrated gene construct. This is believed to be associated with the phenomenon of concatemerization of the injected DNA before genomic integration and repeat-induced gene silencing (Henikoff 1998).
The ongoing investigations will certainly prompt new ideas and new designs to be developed in the expanding universe of TE-based technologies for genetic and cell engineering. Innovative aspect of these studies represents the evaluation of TE-based delivery systems as a potential approach for the therapy of human deseases. Despite the various examples of preclinical efficacy for in vivo gene therapy, the road to the clinic will wind through additional experimentation and evidence of therapeutic effects in large animal models.
Conclusions
Transposable elements occupy a significant portion of eukaryotic genomes and have been long time considered as noncoding ‘junk DNA’ with no beneficial functions for the host genome. With the advent of high-throughput sequence technologies, however, scientists are beginning to find that intrinsic relationships exist between TEs and the other part of the genome. Perhaps only a small proportion of these relationships evolve to become mutually beneficial over the long term. Transposable DNAs have been expertly integrated into the incredibly complex function of the genome as important coordinators in many biological processes such as maintenance of chromosome integrity and creation of novel regulatory networks. The number of evidences for the benefits of TEs is constantly increasing, however, this is perhaps the tip of the iceberg. At the end, TEs are the main provider of genetic diversity which is the raw material to promote eucaryotic genome flexibility and evolution. Transposable elements can therefore be viewed as important genomic symbionts and their study should go ‘hand-in-hand’ with the other part of the genome. The fraction of TEs is often neglected in genomewide studies. For instance, the common practice of scientists is to mask repeat sequences in next generation data focussing on the ‘good’ stuff: coding regions. One should appreciate, however, that by throwing out all data on TEs, we are turning our back on important findings on genome functioning and evolution. In short, the selfish, junk and beneficial DNA hypotheses are all relevant but by no means mutually exclusive and single label for these relationships is inappropriate and potentially misleading. One of the key take homes from the numerous sequenced plant and animal genomes is that we still have a lot to learn about the organization of genomes, function of genes to comprehensively recognize the diverse functional importance of the ‘junk’ DNA.
References
Abad J. P., De Pablos B., Osoegawa K., De Jong P. J., Martin-Gallardo A., Villasante A. et al. 2004 Genomic analysis of Drosophila melanogaster telomeres: Full-length copies of HeT-A and TART elements at telomeres. Mol. Biol. Evol. 21, 1613–1619.
Alzohairy A. M., Sabir J. S. M., Gyulai G., Younis R. A. A., Jansen R. K. and Bahieldin A. 2014 Environmental stress activation of plant LTR-retrotransposons. Funct. Plant Biol. 41, 557–567.
Aminetzach Y. T., Macpherson J. M. and Petrov D. A. 2005 Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309, 764–767.
Ammar I., Izsvák Z. and Ivics Z. 2012 The sleeping beauty transposon toolbox. Methods Mol. Bio. 859, 229–240.
Antonius-Klemola K., Kalendar R. and Schulman A. H. 2006 TRIM retrotransposons occur in apple and are polymorphic between varieties but not sports. Theor. Appl. Genet. 112, 999–1008.
Aprile A., Mastrangelo A. M., De Leonardis A. M., Galiba G., Roncaglia E., Ferrari F. et al. 2009 Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics 10, 279.
Avramova Z., Tikhonov A., Chen M. and Bennetzen L. 1998 Matrix attachment regions and structural colinearity in the genomes of two grass species. Nucleic Acids Res. 26, 761–767.
Ayarpadikannan S. and Kim H. S. 2014 The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 12, 98–104.
Bailey J. A., Carrel L., Chakravarti A. and Eichler E. 2000 Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc. Natl. Acad. Sci. USA 97, 6634–6639.
Baucom R., Estill J., Chaparro C., Upshaw N., Jogi A., Deragon J. et al. 2009 Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5, e1000732.
Bennett M. D. and Leitch I. J. 2012 Plant DNA C-values Database. (release 6.0, Dec. 2012), http://data.kew.org/cvalues/.
Bhattacharyya M., Smith A. M., Ellis T. H. N., Hedley C. and Martin C. 1990 The wrinkled-seed character of pea described by Mendel is caused by a transposon-like insertion in a gene encoding starch-branching enzyme. Cell 60, 115–122.
Branco C. J. S., Vieira E. A., Malone G., Kopp M. M., Malone E., Bernardes A. et al. 2007 IRAP and REMAP assessments of genetic similarity in rice. J. Appl. Genet. 48, 107–113.
Brenchley R., Spannagl M., Pfeifer M., Barker G. L. A., D’Amore R., Allen A. M. et al. 2012 Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710.
Breto M. P., Ruiz C., Pina J. A. and Asıns M. J. 2001 The diversification of Citrus clementina Hort. ex Tan., a vegetatively propagated crop species. Mol. Phylogenet. Evol. 21, 285–293.
Burt A. and Trivers R. 2006 Genes in conflict: the biology of selfish genetic elements. The Belknap Press of Harvard University Press, Cambridge, USA.
Butelli E., Licciardello C., Zhang Y., Liu J., Mackay S., Bailey P. et al. 2012 Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255.
Casacuberta J. M. and Santiago N. 2003 Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene 311, 1–11.
Casola C., Hucks D. and Feschotte C. 2008 Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 25, 29–41.
Charlesworth B. and Charlesworth D. 1983 The population dynamics of transposable elements. Genet. Res. 42, 1–27.
Chuong E. B., Rumi M. A., Soares M. J. and Baker J. C. 2013 Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 45, 325–329.
Cohen D. E., Davidow L. S., Erwin J. A., Xu N., Warshawsky D. and Lee J. T. 2007 The DXPas34 repeat regulates random and imprinted X inactivation. Dev. Cell 12, 57–71.
Cordaux R. and Batzer M. A. 2009 The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10, 691–703.
Darboux I., Charles J. F., Pauchet Y., Warot S. and Pauron D. 2007 Transposon-mediated resistance to Bacillus sphaericus in a field-evolved population of Culex pipiens (Diptera: Culicidae). Cell. Microbiol. 9, 2022–2029.
de Koning A. P., Gu W., Castoe T. A., Batzer M. A. and Pollock D. D. 2011 Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 7, e1002384.
Doolittle W. F. 2013 Is junk DNA bunk? A critique of ENCODE. Proc. Natl. Acad. Sci. USA 110, 5294–5300.
Doolittle W. F. and Sapienza C. 1980 Selfish genes, the phenotype paradigm and genome evolution. Nature 284, 601–603.
Eddy S. R. 2012 The C-value paradox, junk DNA and ENCODE. Curr. Biol. 22, 898–899.
Elliot T. A. and Gregory T. R. 2015 Do larger genomes contain more diverse transposable elements? BMC Evol. Biol. 15, 69.
Ellis T. H., Poyser S. J., Knox M. R., Vershinin A. V. and Ambrose M. J. 1998 Polymorphism of insertion sites of Ty1-copia class retrotransposons and its use for linkage and diversity analysis in pea. Mol. Gen. Genet. 260, 9–19.
ENCODE Project Consortium 2012 An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74.
Fagegaltier D., Bouge A. L., Berry B., Poisot E., Sismeiro O., Coppée J. -Y. et al. 2009 The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc. Natl. Acad. Sci. USA 106, 21258–21263.
Fedoroff N. V. 2012 Transposable elements, epigenetics, and genome evolution. Science 388, 758–767.
Feschotte C., Jiang N. and Wessler S. R. 2002 Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3, 329–341.
Fitzmaurice W. P., Nguyen L. V., Wernsman E. A., Thompson W. F. and Conkling M. A. 1999 Transposon tagging of the sulfur gene of tobacco using engineered maize Ac/Ds elements. Genetics 153, 1919–1928.
Flavell A. J., Knox M. R., Pearce S. R. and Ellis T. H. N. 1998 Retrotransposon-based insertion polymorphisms (RBIP) for high throughput marker analysis. Plant J. 16, 643–650.
Gall J. G. 1981 Chromosome structure and the C-value paradox. J. Cell Biol. 91, 3–14.
Garcia S., Leitch I. J., Anadon-Rosell A., Canela M. A., Galvez F., Garnatje T. et al. 2014 Recent updates and developments to plant genome size databases. Nucleic Acids Res. 42, 1159–1166.
Gonzalez J., Macpherson J. M. and Petrov D. A. 2009 A recent adaptive transposable element insertion near highly conserved developmental loci in Drosophila melanogaster. Mol. Biol. Evol. 26, 1949–1961.
Gonzalez J., Karasov T. L., Messer P. W. and Petrov D. A. 2010 Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 4, e1000905.
Gould S. J. and Vrba E. 1982 Exaptation – a missing term in the science of form. Paleobiology 8, 4–14.
Grandbastien M. A. 1998 Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 3, 181–187.
Grandbastien M. A., Lucas H., Morel J. -B., Mhiri C., Vernhettes S. and Casacuberta J. M. 1997 The expression of the tobacco Tnt1 retrotransposon is linked to the plant defense responses. Genetica 100, 241–252.
Grandbastien M. A., Audeon C., Bonnivard E., Casacuberta J. M., Chalhoub B., Costa A. -P. P et al. 2005 Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet. Genome Res. 110, 229–241.
Graur D., Zheng Y., Price N., Azevedo R. B. R., Zufall R. A. and Elhaik E. 2013 On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol. Evol. 5, 578–590.
Gray Y. H. 2000 It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 16, 461–468.
Gregory T. R. and DeSalle R. 2005 Comparative genomics in eukaryotes. In The evolution of the genome (T. R. Gregory), pp. 585–675. Elsevier, San Diego, USA.
Hayashi K. and Yoshida H. 2009 Refunctionalization of the ancient rice blast disease resistance gene Pit by the recruitment of a retrotransposon as a promoter. Plant J. 57, 413–425.
He Z. H., Dong H. T., Dong J. X., Li D. B. and Ronald P. C. 2000 The rice Rim2 transcript accumulates in response to Magnaporthe grisea and its predicted protein product shares similarity with TNP2-like proteins encoded by CACTA transposons. Mol. Gen. Genet. 264, 2–10.
Henikoff S. 1998 Conspiracy of silence among repeated transgenes. BioEssays 20, 532–535.
Hirochika H. 1993 Activation of tobacco retrotransposons during tissue culture. EMBO J. 12, 2521–2528.
Hollister J. D., Smith L. M., Guo Y. L., Ott F., Weigel D. and Gaut B. S. 2011 Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc. Natl. Acad. Sci. USA 108, 2322–2327.
Howard T. P., Hayward A. P., Tordillos A., Fragoso C., Moreno M. A., Tohme J. et al. 2014 Identification of the maize gravitropism gene lazy plant1 by a transposon-tagging genome resequencing strategy. PLoS One 9, e87053.
Hudson M. E., Lisch D. R. and Quail P. H. 2003 The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 34, 453–471.
Ibarra-Laclette E., Lyons E., Hernández-Guzmán G., Pérez-Torres C. A., Carretero-Paulet L., Chang T. H. et al. 2013 Architecture and evolution of a minute plant genome. Nature 498, 94–98.
Ito H. 2012 Small RNAs and transposon silencing in plants. Dev. Growth Differ. 54, 100–107.
Ivashuta S., Naumkina M., Gau M., Uchiyama K., Isobe S., Mizukami Y. et al. 2002 Genotype-dependent transcriptional activation of novel repetitive elements during cold acclimation of alfalfa (Medicago sativa). Plant J. 31, 615–627.
Jiang N., Bao Z., Zhang X., Eddy S. R. and Wessler S. R. 2004 Pack-MULE transposable elements mediate gene evolution in plants. Nature 431, 569–573.
Joly-Lopez Z., Forczek E., Hoen D. R., Juretic N. and Bureau T. E. 2012 A gene family derived from transposable elements during early angiosperm evolution has reproductive fitness benefits in Arabidopsis thaliana. PLoS Genet. 8, e100293.
Jordan I. K., Rogozin I. B., Glazko G. V. and Koonin E. V. 2003 Origin of a substantial fraction of human regulatory sequences from transposable elements. Trends Genet. 19, 68–72.
Jurka J., Kapitonov V. V., Kohany O. and Jurka M. V. 2007 Repetitive sequences in complex genomes: structure and evolution. Annu. Rev. Genomics Hum. Genet. 8, 241–259.
Kalendar R., Grob T., Regina M., Suoniemi A. and Shulman A. H. 1999 IRAP and REMAP: two new retrotransposon-based DNA fingerprinting techniques. Theor. Appl. Genet. 98, 704–711.
Kalendar R., Tanskanen J., Immonen S., Nevo E. and Schulman A. H. 2000 Genome evolution of wild barley (Hordeum spontaneum) by BARE-1 retrotransposon dynamics in response to sharp microclimatic divergence. Proc. Natl. Acad. Sci. USA 97, 6603–6607.
Kalendar R., Flavell A. J., Ellis T. H. N., Sjakste T., Moisy C. and Schulman A. H. 2011 Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 106, 520–530.
Kanazawa A., Liu B., Kong F., Arase S. and Abe J. 2009 Adaptive evolution involving gene duplication and insertion of a novel Ty1/copia-like retrotransposon in soybean. J. Mol. Evol. 69, 164–175.
Kapitonov V. V. and Jurka J. 2005 RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 3, e181.
Kawakami T., Strakosh S. C., Zhen Y. and Ungerer M. C. 2010 Different scales of Ty1/copia-like retrotransposon proliferation in the genomes of three diploid hybrid sunflower species. Heredity 104, 341–350.
Kim H., Terakami S., Nishitani C., Kurita K., Kanamori H., Katayose Y. et al. 2012 Development of cultivar-specific DNA markers based on retrotransposon-based insertional polymorphism in Japanese pear. Breed. Sci. 62, 53–62.
Knip M., de Pater S. and Hooykaas P. J. 2012 The SLEEPER genes: a transposase-derived angiosperm-specific gene family. BMC Plant Biol. 12, 192.
Knox M., Moreau C., Lipscombe J., Baker D. and Ellis N. 2009 High-throughput retrotransposon-based fluorescent markers: improved information content and allele discrimination. Plant Methods 5, 10.
Konkel M. K., Walker J. A. and Batzer M. A. 2010 LINEs and SINEs of primate evolution. Evol. Anthropol. 19, 236–249.
Kroon J., Souer E., de Graaff A., Xue U. Y., Mol J. and Koes R. 1994 Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J. 5, 69–80.
Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J. et al. 2001 Initial sequencing and analysis of the human genome. Nature 409, 860–921.
Lerman D. N. and Feder M. E. 2005 Naturally occurring transposable elements disrupt Hsp70 promoter function in Drosophila melanogaster. Mol. Biol. Evol. 22, 776–783.
Lerman D. N., Michalak P., Helin A. B., Bettencourt B. R. and Feder M. E. 2003 Modification of heat-shock gene expression in Drosophila melanogaster populations via transposable elements. Mol. Biol. Evol. 20, 135–144.
Levin H. L. and Moran J. V. 2011 Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627.
Li W., Zhang P., Fellers J., Friebe B. and Gill B. 2004 Sequence composition, organization and evolution of the core Triticeae genome. Plant J. 40, 500–511.
Lin R., Ding L., Casola C., Ripoll D. R., Feschotte C. and Wang H. 2007 Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318, 1302–1305.
Lister R., O’Malley R. C., Tonti-Filippini J., Gregory B. D., Berry C. C., Millar A. H. et al. 2008 Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536.
Liu R. and Bennetzen J. L. 2008 Enchilada redux: how complete is your genome sequence? New Phytol. 179, 249–250.
Liu J., He Y., Amasino R. and Chen X. 2004 siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes Dev. 18, 2873–2878.
Lyon M. F. 2006 Do LINEs have a role in X-chromosome inactivation? J. Biomed. Biotechnol. 2006, 59746.
Maxwell P. H., Burhans W. C. and Curcio M. J. 2011 Retrotransposition is associated with genome instability during chronological aging. Proc. Natl. Acad. Sci. USA 108, 20376–20381.
Macfarlan T. S., Gifford W. D., Driscoll S., Lettieri K., Rowe H. M., Bonanomi D. et al. 2012 Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487, 57–63.
Magwire M. M., Bayer F., Webster C. L., Cao C. and Jiggins F. M. 2011 Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet. 7, e1002337.
Majewski J. and Ott J. 2002 Distribution and characterization of regulatory elements in the human genome. Genome Res. 12, 1827–1836.
Manninen O., Kalendar R., Robinson J. and Schulman A. H. 2000 Application of BARE-1 retrotransposon markers to map a major resistance gene for net blotch in barley. Mol. Gen. Genet. 264, 325–334.
Mateo L. and González J. 2014 Pogo-like Transposases have been repeatedly domesticated into CENP-B-related proteins. Genome Biol. Evol. 6, 2008–2016.
McCart C. and Ffrench-Constant R. H. 2008 Dissecting the insecticide-resistance-associated cytochrome P450 gene Cyp6g1. Pest Manag. Sci. 64, 639–645.
McClintock B. 1950 The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 36, 344–355.
McClintock B. 1984 The significance of responses of the genome to challenge. Science 226, 792–801.
Melayah D., Bonnivard E., Chalhoub B., Audeon C. and Grandbastien M. A. 2001 The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J. 28, 159–168.
Melnikova N. V., Konovalov F. A. and Kudryavtsev A. M. 2011 Long terminal repeat retrotransposon Jeli provides multiple genetic markers for common wheat (Triticum aestivum). Plant Genet. Resour. 9, 163–165.
Melnikova N. V., Kudryavtseva A. V., Zelenin A. V., Lakunina V. A., Yurkevich O. Y., Speranskaya A. S. et al. 2014, Retrotransposon-based molecular markers for xnalysis of genetic diversity within the genus Linum. Bio. Med. Res. Int. 2014, 231589.
Meyers B. C., Tingey S. V. and Morgante M. 2001 Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676.
Mhiri C., Morel J. B., Vernhettes S., Casacuberta J. M., Lucas H. and Grandbastien M. A. 1997 The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol. Biol. 33, 257–266.
Michael T. P. and Jackson S. 2013 The first 50 plant genomes. The Plant Genome 6, 1–7.
Miller W. J., McDonald J. F., Nouaud D. and Anxolabéhère D. 1999 Molecular domestication – more than a sporadic episode in evolution. Genetica 107, 197–207.
Mills R. E., Bennett E. A., Iskow R. C. and Devine S. E. 2007 Which transposable elements are active in the human genome? Trends Genet. 23, 183–191.
Moreau-Mhiri C., Morel J. B., Audeon C., Ferault M., Grandbastien M. A. and Lucas H. 1996 Regulation of expression of the tobacco Tnt1 retrotransposon in heterologous species following pathogen-related stresses. Plant J. 9, 409–419.
Morgan H. D., Sutherland H. G., Martin D. I. and Whitelaw E. 1999 Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23, 314–318.
Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A. and Rafalski A. 2005 Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37, 997–1002.
Nekrutenko A. and Li W. H. 2001 Transposable elements are found in a large number of human protein-coding genes. Trends Genet. 17, 619–621.
Niu D. -K. and Jiang L. 2013 Can ENCODE tell us how much junk DNA we carry in our genome? Biochem. Biophys. Res. Commun. 430, 1340–1343.
O’Donnell K. A. and Burns K. H. 2010 Mobilizing diversity: transposable element insertions in genetic variation and disease. Mobile DNA 1, 21.
Orgel L. E. and Crick F. H. 1980 Selfish DNA: the ultimate parasite. Nature 284, 604–607.
Pardue M. -L. and DeBaryshe P. G. 2011 Retrotransposons that maintain chromosome ends. Proc. Natl. Acad. Sci. USA 108, 20317–20324.
Parisod C., Salmon A., Zerjal T., Tenaillon M., Grandbastien M. A. and Ainouche M. 2009 Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 184, 1003–1015.
Pathak R. U., Rangaraj N., Kallappagoudar S., Mishra K. and Mishra R. K. 2007 Boundary element-associated factor 32B connects chromatin domains to the nuclear matrix. Mol. Cell. Biol. 27, 4796–4806.
Pathak R. U., Srinivasan A. and Mishra R. K. 2014 Genome-wide mapping of matrix attachment regions in Drosophila melanogaster. BMC Genomics 15, 1022.
Pearce S. R., Knox M., Ellis T. H., Flavell A. J. and Kumar A. 2000 Pea Ty1-copia group retrotransposons: transpositional activity and use as markers to study genetic diversity in Pisum. Mol. Gen. Genet. 263, 898–907.
Porceddu A., Albertini E., Baracaccia G., Marconi G., Bertoli F. B. and Veronesi F. 2002 Development of S-SAP markers based on an LTR-like sequence from Medicago sativa L. Mol. Genet. Genomics 267, 107–114.
Queen R. A., Gribbon B. M., James C., Jack P. and Flavell A. J. 2004 Retrotransposon-based molecular markers for linkage and genetic diversity analysis in wheat. Mol. Genet. Genomics 271, 91–97.
Ramsden D. A., Weed B. D. and Reddy Y. V. 2010 V(D)J recombination: born to be wild. Semin. Cancer Biol. 20, 254–260.
Ray D. A. 2007 SINEs of progress: Mobile element applications to molecular ecology. Mol. Ecol. 16, 19–33.
Ray D. A. and Batzer M. A. 2011 Reading TE leaves: New approaches to the identification of transposable element insertions. Genome Res. 21, 813–820.
Ray D. A., Walker J. A. and Batzer M. A. 2007 Mobile element-based forensic genomics. Mutat. Res. 616, 24–33.
Ross M. T., Grafham D. V., Coffey A. J., Scherer S., McLay K., Muzny D. et al. 2005 The DNA sequence of the human X chromosome. Nature 434, 325–337.
Salvi S., Sponza G., Morgante M., Tomes D., Niu X., Fengler K. A. et al. 2007 Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc. Natl. Acad. Sci. USA 104, 11376–11381.
Schaack S., Gilbert C. and Feschotte C. 2010 Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 25, 537–546.
Schmidt J., Good R., Appleton B., Sherrard J., Raymant G. C., Bogwitz M. R. et al. 2010 Copy Number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 6, e1000998.
Schmitz J., Roos C. and Zischler H. 2005 Primate phylogeny: molecular evidence from retroposons. Cytogenet. Genome Res. 108, 26–37.
Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., Pasternak S. et al. 2009 The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115.
Schulman A. H., Flavell A. J. and Ellis T. H. N. 2004 The application of LTR retrotransposons as molecular markers in plants. Methods Mol. Biol. 260, 145–173.
Shpiz S., Kwon D., Uneva A., Kim M., Klenov M., Rozovsky Y. P. et al. 2007 Characterization of Drosophila telomeric retroelement TAHRE: transcription, transpositions, and RNAi-based regulation of expression. Mol. Biol. Evol. 24, 2535–2545.
Sinzelle L., Izsvák Z. and Ivics Z. 2009 Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol. Life Sci. 66, 1073–1093.
Skipper K. A., Andersen P. R., Sharma N. and Mikkelsen J. G. 2013 DNA transposon-based gene vehicles - scenes from an evolutionary drive. J. Biomed. Sci. 20, 92.
Slotkin R. K. and Martienssen R. 2007 Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285.
Smýkal P. 2006 Development of an efficient retrotransposon-based fingerprinting method for rapid pea variety identification. J. Appl. Genet. 47, 221–230.
St Laurent G., Hammell N. and McCaffrey T. A. 2010 A LINE-1 component to human aging: do LINE elements exact a longevity cost for evolutionary advantage? Mech. Ageing Dev. 131, 299–305.
Studer A., Zhao Q., Ross-Ibarra J. and Doebley J. 2011 Identification of a functional transposon insertion in the maize domestication gene tb1. Nat. Genet. 43, 1160–1163.
Takeda S., Sugimoto K., Otsuki H. and Hirochika H. 1998 Transcriptional activation of the tobacco retrotransposon Tto1 by wounding and methyl jasmonate. Plant Mol. Biol. 36, 365–376.
Takeda S., Sugimoto K., Otsuki H. and Hirochika H. 1999 A 13-bp cisregulatory element in the LTR promoter of the tobacco retrotransposon Tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant J. 18, 383–393.
Tam S. M., Mhiri C., Vogelaar A., Kerkveld M., Pearce S. R. and Grandbastien M. A. 2005 Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor. Appl. Genet. 110, 819–831.
The Arabidopsis Genome Initiative 2000 Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815.
Tikhonov A. P., Bennetzen J. L. and Avramova Z. V. 2000 Structural domains and matrix attachment regions along colinear chromosomal segments of maize and Sorghum. Plant Cell 12, 249–264.
Tomkins J. 2013 Transposable Elements Are Key to Genome Regulation. www.icr.org/article/7388/.
Tsuchiya T. and Eulgem T. 2013 An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA 110, E3535–E3543.
Ungerer M. C., Strakosh S. C. and Zhen Y. 2006 Genome expansion in three hybrid sunflower species is associated with retrotransposon proliferation. Curr. Biol. 16, R872–R873.
van der Biezen E. A., Brandwagt B. F., van Leeuwen W., Nijkamp H. J. and Hille J. 1996 Identification and isolation of the FEEBLY gene from tomato by transposon tagging. Mol. Gen. Genet. 251, 267–280.
Vela D., Fontdevila A., Vieira C. and García Guerreiro M. P. 2014 A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLoS One 9, e88992.
Vicient C. M., Suoniemi A., Anamthawat-Jónsson K., Tanskanen J., Beharav A., Nevo E. et al. 1999 Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11, 1769–1784.
Vicient C. M., Jaaskelainen M. J., Kalendar R. and Schulman A. H. 2001 Active retrotransposons are a common feature of grass genomes. Plant Physiol. 125, 1283–1292.
Wang Z., Pang X. M., Lv Y. F., Xu F., Zhou T. and Li X. 2013 A dynamic framework for quantifying the genetic architecture of phenotypic plasticity. Brief. Bioinform. 14, 82–95.
Waugh R., McLean K., Flavell A. J., Pearce S. R., Kumar A. and Thomas B. B. T. 1997 Genetic distribution of BARE-1-like retrotransposable elements in the barley genome revealed by sequence-specific amplifcation polymorphisms (S-SAP). Mol. Gen. Genet. 253, 687–694.
Werren J. H. 2011 Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 108, 10863–10870.
Wicker T., Sabot F., HuaVan A., Bennetzen J. L., Capy P., Chalhoub B. et al. 2007 A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982.
Wicker T., Krattinger S. G., Lagudah E. S., Komatsuda T., Pourkheirandish M., Matsumoto T. et al. 2009 Analysis of intraspecies diversity in wheat and barley genomes identifies breakpoints of ancient haplotypes and provides insight into the structure of diploid and hexaploid Triticeae gene pools. Plant Physiol. 149, 258–270.
Witherspoon D. J., Marchani E. E., Watkins W. S., Ostler C. T., Wooding S. P., Anders B. A. et al. 2006 Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum. Hered. 62, 30–46.
Wood J. G. and Helfand S. L. 2013 Chromatin structure and transposable elements in organismal aging. Front. Genet. 4, 274.
Woodrow P., Pontecorvo G., Fantaccione S., Fuggi A., Kafantaris I., Parasi D. et al. 2010 Polymorphism of a new Ty-1-copia retrotransposon in durum wheat under salt and light stresses. Theor. Appl. Genet. 121, 311–322.
Wright S. I., Agrawal N. and Bureau T. E. 2003 Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13, 1897– 1903.
Yaakov B. and Kashkush K. 2011 Methylation, transcription, and rearrangements of transposable elements in synthetic allopolyploids. Int. J. Plant Genomics 2011, 569826.
Yang Q, Li Z., Li W., Ku L., Wang C., Ye J. et al. 2013 CACTA-like transposable element in ZmCCT attenuated photoperiod sensitivity and accelerated the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 110, 16969–16974.
Yoder J. A., Walsh C. P. and Bestor T. H. 1997 Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13, 335–340.
Young L. W., Cross R. H., Byun-McKay S. A., Wilen R. W. and Bonham-Smith P. C. 2005 A high-and low-temperature inducible Arabidopsis thaliana HSP101 promoter located in a nonautonomous mutator-like elements. Genome 48, 547–555.
Yu G. X. and Wise R. P. 2000 An anchored AFLP- and retrotransposon-based map of diploid Avena. Genome 43, 736–749.
Zedek F., Smerda J., Smarda P. and Bureš P. 2010 Correlated evolution of LTR retrotransposons and genome size in the genus Eleocharis. BMC Plant Biol. 10, 265.
Zhou L., Mitra R., Atkinson P. W., Hickman A. B., Dyda F. and Craig N. L. 2004 Transposition of the hAT element links transposable elements and V(D)J recombination. Nature 432, 995–100.
Acknowledgements
I would like to thank the anonymous reviewers for the helpful suggestions and constructive criticism.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: N. G. Prasad
[Bonchev G. N. 2016 Useful parasites: the evolutionary biology and biotechnology applications of transposable elements. J. Genet. 95, xx–xx]
Rights and permissions
About this article
Cite this article
BONCHEV, G.N. Useful parasites: the evolutionary biology and biotechnology applications of transposable elements. J Genet 95, 1039–1052 (2016). https://doi.org/10.1007/s12041-016-0702-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-016-0702-6