Abstract
D-galacturonic acid is a potential platform chemical comprising the principal component of pectin in the citrus processing waste stream. Several enzyme activities are required for the enzymatic production of galacturonic acid from pectin, including exo- and endo-polygalacturonases. The gene TtGH28 encoding a putative GH28 polygalacturonase from Pseudothermotoga thermarum DSM 5069 (Theth_0397, NCBI# AEH50492.1) was synthesized, expressed in Escherichia coli, and characterized. Alignment of the amino acid sequence of gene product TtGH28 with other GH28 proteins whose structures and details of their catalytic mechanism have been elucidated shows that three catalytic Asp residues and several other key active site residues are strictly conserved. Purified TtGH28 was dimeric and hyperthermostable, with K 0.5t = 86.3 °C. Kinetic parameters for activity on digalacturonic acid, trigalacturonic acid, and polygalacturonic acid were obtained. No substrate inhibition was observed for polygalacturonate, while the K si values for the oligogalacturonides were in the low mM range, and K i for product galacturonic acid was in the low μM range. Kinetic modeling of the progress of reaction showed that the enzyme is both fully exo- and fully non-processional.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pectins comprise a group of heteropolysaccharides that are plant middle lamellae and primary cell wall constituents which, depending on their backbone structure, are classified as xylogalacturonan, rhamnogalacturonan I, rhamnogalacturonan II, modified hairy region, or homogalacturonan (HG) [1]. Pectin from citrus peel waste comprises mainly HG that is highly methyl esterified [2], and conversion of this food processing waste to biofuels and value-added products is the subject of much current interest [3–5]. In nature, pectin-degrading enzymes occur in plants, fungi, bacteria, and insects [6, 7], where they are involved in physiological processes such as plant cell wall modeling, fruit ripening [8], plant wound response [9], fungal and bacterial plant pathogenic processes [10–12], and insect herbivory [13–16]. Pectinolytic enzymes are important industrial enzymes used primarily in various food and fiber industries including fruit juice extraction and clarification, poultry feed additives [17–19], and in the production of pectic oligosaccharides with potential prebiotic health benefits [20, 21]. The predominant pectin saccharide D-galacturonic acid is a potentially valuable biomass-derived platform chemical, and as such Aspergillus niger has been metabolically engineered to convert D-galacturonic acid to L-ascorbic acid [22] and L-galactonic acid [23]. Additionally, D-galacturonic acid can be enzymatically converted to the di-acid galactaric acid [24], which may find application as a Ca-sequestering agent [25] and as a platform chemical for polymers [26, 27].
The enzymatic degradation of HG pectin proceeds by initial side chain removal by pectin methylesterases (EC 3.1.1.11) in carbohydrate esterase family 8 in the carbohydrate active enzyme (CAZY) database [28] since the HG backbone must be exposed before depolymerization by polygalacturonase [29]. Pectins can be depolymerized by two mechanistically distinct enzyme types: pectate lyases which proceed via β-elimination, cleaving the glycosidic bond at the C-4 position and eliminating H from C-5 to yield a ∆ 4:5 unsaturated product [30], and polygalacturonases, which hydrolyze the α-1,4 glycosidic bonds of polygalacturonic acid and HG regions of pectin in an exo- or endo-manner, that based on amino acid sequence homology are grouped in CAZY database glycosyl hydrolase family 28 (GH28). Glycosyl hydrolases catalyze the nucleophilic attack of glycosidic bonds, resulting in either retention of anomeric configuration from double-displacement reaction or inversion of configuration via a single displacement reaction [31, 32]. The GH28 enzymes comprise several different pectinolytic enzymes, which are divergent in substrate specificity but are expected to have the same catalytic mechanism. Both endo-(EC 3.2.1.15) and exo-(EC 3.2.1.67 and EC 3.2.1.82) GH28 polygalacturonases from Aspergillus have been shown to catalyze the hydrolysis with inversion of configuration [33, 34]. The endo-polygalacturonases (3.2.1.15) yield oligogalacturonides, while the exo-polygalacturonases hydrolyze polygalacturonic acid from the non-reducing end, releasing either digalacturonic acid (diGalUA) (EC 3.2.1.82; exo-poly-α-galacturonosidase) or galacturonic acid (GalUA) (EC 3.2.1.67; galacturan 1,4-α-galacturonosidase). Exo-polygalacturonases from both eukaryotic [15, 35–40] and prokaryotic [41–47] sources have been isolated and characterized, and we describe here the recombinant expression and characterization of a hyperthermostable GH28 exo-polygalacturonase (EC 3.2.1.67) from Pseudothermotoga thermarum DSM 5069 (Theth_0397, NCBI# AEH50492.1) termed TtGH28.
Materials and Methods
Materials
TtGH28 enzyme (calculated ε 280 nm 46.85 mM/cm [48]) was stored in 50 mM sodium phosphate, pH 7.0, with 150 mM NaCl and 10 % glycerol at −80 °C. Prior to initiating reactions, enzyme was diluted into 10 mM sodium phosphate, pH 7.0, and 0.1 mg/mL bovine serum albumin (BSA). Polygalacturonic acid (polyGalUA), MW 25–50 kDa, was from Sigma-Aldrich (St. Louis, MO), and an average molecular weight of 37.5 kDa was used in calculations of molarity. HEPES and Bis–Tris propane were obtained from Fisher Scientific (Pittsburgh, PA). 4-Nitrophenyl-α-d-xylopyranoside and 4-nitrophenyl-β-d-mannopyranoside were obtained from Toronto Research Chemicals (Toronto, ON, Canada). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO). Water was purified by a Milli-Q Academic A10 unit (EMD Millipore; Billerica, MA).
Gene Synthesis, Cloning, Expression, and Purification of Enzymes
The synthetic gene encoding TtGH28 was designed for expression in Escherichia coli based on the amino acid sequence (Supplementary Fig. 1; GenScript USA Inc., Piscataway, NJ), with the stipulation that NdeI and XhoI restriction sites be avoided to allow for subcloning into pET29b(+) (EMD Millipore). The expressed native peptide C-terminal residues were followed by Leu-Glu-(His)6 to allow for affinity purification, and the protein was expressed using E. coli BL21(DE3) CodonPlus cells (Agilent Technologies, Inc., Santa Clara, CA). A glycerol stock stab was used to inoculate 10 mL Terrific Broth containing 30 μg/mL kanamycin (TBkan) that was incubated overnight (37 °C, 250 rpm). 0.4 L TBkan was then seeded and grown to OD600nm ~2 before inducing protein expression with 1 mM IPTG, and protein expression occurred overnight at 25 °C. Cells were lysed using ~13 mL/g cell pellet lysis solution: CelLytic B reagent, 0.5 mg/mL hen egg-white lysozyme, 2 mM β-mercaptoethanol, 5 U/mL Benzonase (all from Sigma-Aldrich), and 1 μL/mL CalBiochem protease inhibitor cocktail III (EMD Millipore) for 2 h at room temp then centrifuged 10 min at 9000 × g. The supernatant was heated 10 min at 70 °C and centrifuged a second time. His-tagged TtGH28 was partially purified using Ni–NTA resin according to manufacturer’s instructions (Qiagen, Valencia, CA), followed by final purification using size exclusion chromatography on a Superdex 200 16/60 column (GE Healthcare Bio-Sciences, Pittsburg, PA, USA). Purity of the protein was assessed by PAGE using Novex Nupage 4–12 % Bis–Tris protein gels and Novex Sharp protein size standards according to manufacturer’s instructions (ThermoFisher Scientific, Waltham, MA), with absorbance at 280 nm being used to measure protein concentrations.
Oligomeric State Determination
Size Exclusion Chromatography with Multi-Angle Light Scattering detection (SEC-MALS) was used to determine the oligomeric state of 1.41 mg/mL TtGH28 in 100 mM succinate, pH 5.0. Size separation was performed on a Superdex 200 10/300 GL column (GE Healthcare Bio-Sciences). The column was mounted on an Agilent 1200 LC HPLC system equipped with the following on-line detectors: DAD UV-absorbance detector (Agilent Technologies), Wyatt Dawn® Heleos™II 18-angle MALS light scattering detector, Wyatt QELS™ quasi-elastic (dynamic) light scattering (QELS) detector, and Wyatt Optilab® T-rEX™ refractive index detector (Wyatt Technology Corp, Santa Barbara, CA) equipped with a 120-mW solid-state laser operating at 658 nm. The column was pre-equilibrated overnight in 100 mM succinate buffer, pH 5, at flow rate of 0.5 mL/min and 25 °C. The protein solution was equilibrated at 25 °C for 1 h and then filtered through Millipore MC-GV 0.22 μm centrifugal filters. Absorbance at 280 nm was measured from the protein solution before and after filtration using a Perkin Elmer Lambda 1050 spectrophotometer (Perkin Elmer, Waltham, MA). The UV–Vis absorbance spectrum of the filtered protein solution was recorded to verify no significant absorbance occurs at 658 nm. 150 μL was injected onto the column and elution simultaneously monitored with DAD UV (230, 260, 280, 320 nm), MALS, QELS, and RI detectors. Molecular weight was determined using the Debye fitting method (Fig. 1, Astra software package v. 5.3.4.20, Wyatt Technology Corp) as described [49]. The Wyatt Optilab® T-rEX™ refractive index detector was used as the concentration detector; the refractive index increment dn/dc value of 0.187 was determined using 1.5 mg/mL Bovine Serum Albumin (BSA) standard. Hydrodynamic radius was calculated from the diffusion coefficient measured by the QELS detector. The temperature-corrected solvent viscosity constant was set to 8.945 × 10−3 g/(cm-sec), based on the BSA standard run.
Debye plot for data collected at nine scattering angles (from 61.2° to 148.8°); a first order linear fit is shown; the Mw is calculated from the y–intercept [49]
K 0.5t Determination
A PCR machine (MJ Research thermocycler, Bio-Rad, Hercules, CA) was used to establish a temperature gradient from 79 °C to 93 °C, and aliquots of TtGH28 in 125 mM succinate, pH 5.0 containing 0.1 % BSA were incubated for 1 h, followed by 1 h at 30 °C for refolding. Assays for residual activity were performed by adding 5 μL aliquots of enzyme mixture to 75 μL succinate buffer with 20 μL 1 % polyGalUA substrate, and incubating 10 min at 80 °C, followed by cooling to 12 °C for 1 min before quenching by adding 100 μL dinitrosalicylic acid (DNSA) solution [50]. K 0.5t , the temperature at which ½ activity remains after incubation for 1 h, was calculated by fitting the fraction folded data to Boltzmann sigmoidal Eq. 6.
Separation and Quantification of Reaction Products
Products from TtGH28-catalyzed hydrolysis of diGalUA, triGalUA, or polyGalUA were separated and quantified using a DX500 HPLC system with an ED40 electrochemical detector (pulsed amperometry), AS3500 autosampler, PA-100 (4 × 250 mm) anion exchange column, and Chromeleon software (Dionex, Sunnyvale, CA) [51]. Samples (25 μL) were injected onto the column equilibrated with 0.1 M NaOH and developed with a 5-min linear gradient (0.05–0.45 M sodium acetate in 0.1 M NaOH) at ~25 °C and a flow rate of 1.0 mL/min. Several concentrations of the products of interest were used to establish standard curves on the same day experimental samples were run.
Steady-state Kinetics of TtGH28 Acting on diGalUA, triGalUA, or polyGalUA at pH 6.0 and 40 °C
Initial-rate reactions (0.5-mL) at 40 °C contained varied diGalUA concentrations (0.0341–8.52 mM), varied triGalUA concentrations (0.0317–9.51 mM), or varied polyGalUA concentrations (5.20–52.0 μM) in 100 mM MES pH 6.0, I (ionic strength) = 0.1 M adjusted using NaCl. Reactions were run in duplicate. Before (time = 0 min) and after (time = 2–4 min) initiating reactions with enzyme (7.56 nM), 100 μL aliquots of reaction mixtures were removed and quenched to pH 9.0 with NaOH, diluted into 10 mM Bis–Tris propane pH 9.0, and kept on wet ice or the HPLC autosampler at 5 °C until analysis. Products monoGalUA and diGalUA were separated and quantified.
MonoGalUA Inhibition of TtGH28 Acting on triGalUA at pH 5.0 and 40 °C
For product inhibition studies, initial-rate reactions (0.5 mL) at 40 °C were run as those of TtGH28 acting on triGalUA, described above, except that reactions contained varied triGalUA concentrations (0.037–0.22 mM) and varied monoGalUA concentrations (0–0.28 mM) in 100 mM sodium acetate pH 5.0, I = 0.1 M.
Progression of TtGH28 Acting on triGalUA at pH 6.0 and 40 °C
The reaction (3 mL) at 40 °C contained 99.1 μM triGalUA in 100 mM MES pH 6.0, I = 0.1 M. The reaction was run as previously described for steady-state kinetics of TtGH28 acting on triGalUA, with 100 μL aliquots removed and quenched before (time = 0 min) and after (time = 2–120 min) initiating the reaction with enzyme (7.56 nM). The substrate triGalUA and products diGalUA and monoGalUA were separated and quantified by HPLC. The data were fit to a model assuming a fully exo- and fully non-processive mechanism with KinTek Explorer [52] using the steady-state kinetic and equilibrium parameters of the enzyme acting on triGalUA and diGalUA (determined in this work).
Steady-state Kinetics of TtGH28 Acting on polyGalUA at Varied pH and 40 °C
Control experiments were performed to determine which buffers were non-inhibitory and appropriate for use in determining kinetic parameters: buffers used were 10 mM citrate (pH 3.0 and 4.0), 100 mM MES (pH 5.0, 5.5, and 6.0), 100 mM HEPES (pH 7.0 and 8.0), and 50 mM Bis–Tris propane (pH 9.0). All buffers were adjusted to 0.1 M ionic strength with NaCl. The following buffers were inappropriate for use in determining kinetic parameters: acetate, succinate, phosphate, and pyrophosphate (all of which were weakly inhibitory), and tricine (which interfered with HPLC quantification). We also ran a pH stability profile for the enzyme. TtGH28 (1.33 μM) was preincubated for 10 min at each pH in appropriate buffer equilibrated at 40 °C. Immediately following preincubation, enzyme was used to initiate reactions with 20.8 μM polyGalUA in 100 mM MES pH 6.0 and 40 °C, as above. From pH 3.0 to 9.0 (inclusive), in the buffers above that were found to be appropriate for use, the enzyme was fully stable for 10 min, so no corrections or omissions of pH conditions were necessary. Initial-rate reactions of TtGH28 acting on polyGalUA at varied pH were run as described above. Product monoGalUA was quantified by HPLC.
TtGH28 Acting on Nitrophenyl-substituted Substrates at 40 °C
TtGH28 activity was determined for potential nitrophenyl substrates. Reactions (0.8-mL) at 40 °C contained 1.0 mM substrate and either 20 mM sodium acetate, pH 5.0, or 20 mM sodium phosphate, pH 7.0. Reactions were initiated by addition of TtGH28 (0.464 nmol in 7 μL), and 0.25 mL aliquots of reaction mixture were quenched after 20 h by addition of 0.75 mL 100 mM sodium carbonate, pH 11.5. Control reactions with no enzyme were run under the same conditions to account for background absorbance of substrates. Product 4-nitrophenyl (or 2-nitrophenyl for 2-nitrophenyl-β-d-xylopyranoside substrate) was quantified by absorbance at 400 nm, ε = 18.3 mM/cm (or at 420 nm for 2-nitrophenyl, ε = 4.5 mM/cm). Data were collected in triplicate.
Equations
Data were fit to the following equations where ν is the observed initial (steady-state) rate of catalysis; E T is the total enzyme concentration; k cat is the turnover number of catalysis; S is the substrate concentration; K m is the Michaelis constant; K si is the substrate inhibition constant, the dissociation constant for S from ESS; I is the inhibitor concentration; K i is the inhibition constant; p is the determined parameter at a single pH; P is the pH-independent value of the parameter; P L is the lower limit of the parameter; H+ is the proton concentration; K a1 is the proton concentration where p is half of P for the first group(s) affecting P; K a2 is the proton concentration where p is half of P for the second group(s) affecting P; FF is enzyme fraction folded for a two-state thermal denaturation process: Folded = Unfolded; and K 0.5t is the temperature at which 50 % of the enzyme remains folded after incubation at temperature T for 1 h. Data were fit to non-linear equations using GraFit [53] or GraphPad Prism (GraphPad Software, Inc., LaJolla, CA):
Results and Discussion
Amino Acid Sequence Analysis
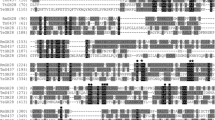
The amino acid sequence of TtGH28 upon which gene synthesis was based was identified in the CAZy database from functional annotation of a genomic sequence for Pseudothermotoga thermarum DSM 5069 (AEH50492.1) deposited in the NCBI database by the DOE Joint Genome Institute (Walnut Creek, CA USA). It should be pointed out that the predicted functional annotation as an endo-type GH28 enzyme has not been borne out by experiments in this report (vide infra). It is thought that members of a given GH family are evolutionarily related, with differences arising from tuning of enzyme substrate specificity and kinetic and biophysical parameters for a given physiological function, leaving the basic catalytic core and mechanism intact. A catalytic mechanism for an Aspergillus GH28 endopolygalacturonase consistent with anomeric inversion has been developed based on crystal structure and site-directed mutagenesis [54], and comparison to the suggested catalytic mechanism of tailspike rhamnosidase [55, 56]. The mechanism involves three GH28 conserved Asp residues (Asp271, Asp292, and Asp293, TtGH28 numbering, filled star in Fig. 2), where Asp292 acts as the catalytic acid and Asp271 and Asp293 activate the attacking water nucleophile. Crystal structures have been determined for exo-polygalacturonases from both Yersinia enterocolitica (EC 3.2.1.82; YeGH28) [57] (19 % identical, 29 % identical or similar with TtGH28) and Thermotoga maritima (EC 3.2.1.67; PelB, Tm0437) [58] (25 % identical, 37 % identical or similar with TtGH28). The core structure of GH28 exoPGases has been shown to consist of a right-handed parallel β-helix, with twelve turns in Tm0437 exoGH28 [58] and ten turns in YeGH28 [57], and this motif is also found in other enzymes involved in binding carbohydrate polymers and hydrolyzing glycosidic linkage enzymes such as pectate lyase C [59] and phage 22 tailspike protein [55]. There are three catalytic aspartates in YeGH28 (D381, D402, D403), where D402 is catalytic acid, and the other two aspartates interact with the water nucleophile, and these three residues align with TtGH28 D271, D292, and D293. Indeed, all eight residues conserved in YeGH28 (EC 3.2.1.82) [50] and endo-polygalacturonases (EC 3.2.1.15) [54, 57, 60, 61] determined to be involved in substrate binding and catalysis (filled square in Fig. 2) are also strictly conserved in TtGH28 (EC 3.2.1.67). In addition, amino acid sequence alignment (Vector NTI, ThermoFisher Scientific, Waltham, MA) of TtGH28 to the analogous enzyme Tm0437 (25 % identity overall) shows that cis peptide G332-S333 and residues Y391, K359, and E334 shown in the crystal structure to bind the -1 carboxylate group are strictly conserved. Moreover, 10 out of 14 additional residues shown to be in the Tm0437 active site (filled circle in Fig. 2) are either strictly conserved in TtGH28 or an Asp/Glu substitution in one case.
Oligomeric State
TtGH28 eluted in a single peak with polydispersity 1.001 and a fitted average Mw of 121.51 KDa (Fig. 3), within 5 % of the expected value (117.01 KDa) for dimeric TtGH28. The measured hydrodynamic radius was 4.1 nm. No protein peaks of higher or lower Mw forms were detected in the chromatogram by any of the three detectors (data for RI detector only, Fig. 3), and the protein was electrophoretically pure as determined by SDS-PAGE (Supplementary Fig. 2), and no large aggregates were present since absorbance readings before and after filtration were indistinguishable, together indicating that TtGH28 is a stable dimer under these solution conditions. Interestingly, the related exo-polygalacturonase Tm0437 from Thermotoga maritima [58] was shown to be a tetramer by dynamic light scattering, exhibiting decreased polydispersity with increasing temperature indicative of lessening non-specific aggregation at higher temperatures.
K 0.5t Determination
Thermal stability is a desirable property for industrially relevant enzymes due to increased turnover, lower viscosity, and diminished contamination issues. Both Tm0437 and TtGH28 were identified in the genomes of hyperthermophilic Thermotoga species, and as such can be expected to demonstrate useful thermostability profiles. Indeed, K 0.5t for TtGH28 of 86.3 °C was determined (Fig. 4), indicating hyperthermal stability. The thermal stability of Tm0437 has previously been determined using calorimetry, and its melting temperature was 105 °C [45]. We determined for Tm0437 a value of ~103 °C for K 0.5t from a partial denaturation curve obtained under the same conditions used to measure K 0.5t for TtGH28 (curve not shown), indicating marked thermostability differences between these closely related, functionally identical enzymes.
Steady-state Kinetics of TtGH28 Acting on Three Substrates
Steady-state kinetic parameters of TtGH28 acting on three substrates (diGalUA, triGalUA, and polyGalUA) were determined and compared to other GH28 EC 3.2.1.67 exo-polygalacturonases similarly characterized (Table 1; Fig. 5). The k cat/K m values of diGalUA and triGalUA are similar and the k cat and K m values for diGalUA are about 50 % higher than for triGalUA. Substrate inhibition is observed for diGalUA and triGalUA at high concentrations (K si values 3.63 mM and 17.2 mM, respectively). Substrate inhibition is not observed for the polymer. k cat and K m values reported for diGalUA and triGalUA for eukaryotic exo-polygalacturonases RPG15 and RPG16 from Rhizopus determined at 30 °C [36] were similar to the TtGH28 values (Table 1). Kinetic parameters have been determined at 80 °C for the closely related Tm0437 enzyme from Thermotoga maritima [45] (Fig. 2; Table 1), which in contrast to TtGH28 shows significantly higher k cat values when proceeding from di- to tri- to polyGalUA. The turnover rates of Tm0437, NfPG4, and A. giganteus PG for polyGalUA appear to be considerably higher than observed for TtGH28, above that expected considering the 40 °C temperature difference at which the reactions were performed, given a Q10 of 2. A Q10 of 2 is an increase of rate by a factor of 2 for a 10° increase in temperature (°C). Also, the Tm0437 km values reported are significantly higher for diGalUA and triGalUA. The K m for polyGalUA is lower than reported for the di- and tri-oligosaccharides for both TtGH28 and Tm0437. MonoGalUA competitively inhibits TtGH28 acting on triGalUA at pH 5.0 and 40 °C, with a K i value of 7.9 ± 1.6 μM (Fig. 6; Table 1). In contrast, the K i (GalUA) of the exo-polygalacturonases from Rhizopus was determined to be on the order of two magnitudes greater than observed for TtGH28 (Table 1).
MonoGalUA inhibition of TtGH28 acting on triGalUA at pH 5.0 and 40 °C. Initial-rate data are fit to Eq. 3: K m 11.7 ± 2.7 μM; k cat 9.62 ± 0.36 s−1, k cat/K m 819 ± 164 s−1mM−1, K monoGalUAi 7.9 ± 1.6 μM
Time Progression of TtGH28 Acting on TriGalUA at pH 6.0 and 40 °C
By following the progression of the enzyme acting on triGalUA, it is demonstrated that as the reaction proceeds, TtGH28 acts on the product diGalUA. The collected data for the triGalUA and the two products with time were fit to a model assuming a fully exo- and fully non-processive mechanism using the kinetic parameters presented in Table 1 and the K i (monoGalUA) of 7.9 μM (Fig. 7). The goodness of the fit supports the conclusion that TtGH28 is fully exo- in nature and fully non-processive.
pH Dependence of TtGH28 Acting on polyGalUA at 40 °C
Following ten minutes of preincubation between pH 3.0 and 9.0 (inclusive), TtGH28 is fully stable. From steady-state saturation curves determined at each pH, kinetic parameters of TtGH28 acting on polyGalUA at 40 °C and varied pH (3.0–9.0) were determined. It was hoped that in addition to determination of the pK a values of catalytic groups of the enzyme, that it could be determined whether the ionization state of the substrate influenced the reaction. From the literature, the pK a value of monoGalUA is 3.51 ± 0.01 at 20 °C [62]; more importantly, we determined that the pK a of the second carboxyl group to ionize of diGalUA is not much higher (if any) than that of the first, since we found that the pK a values of both carboxyl groups of the dimer are less than 4.0 (Stoller and Jordan, unreported results). It is likely the polymer behaves similarly with pK a values below 4.0.
The pH dependences of all three kinetic parameters (k cat, k cat/K m, and 1/K m) describe bell-shaped curves (Fig. 8). Relevantly, only one pK a is seen on the acidic side and the basic side of the bell curves, which are attributed to the catalytic acid and base, and no pK a is attributed to the substrate. In each case, the pK a values of the catalytic acid and catalytic base are closer together than 3 pH units, which precludes accurate determinations [63]. The approximate pK a values for the catalytic acid and catalytic base are, respectively, 7 and 5 for k cat (which represents the substrate-occupied enzyme), 6 and 4.5 for k cat/K m (which represents the substrate-unoccupied or free enzyme) and 6 and 4 for 1/K m. One detail is that k cat and 1/K m pH profiles do not have a zero endpoint on the acid side, and this may be attributable to participation of the low pH as a catalytic acid.
pH dependence of kinetic parameters of TtGH28 acting on polyGalUA at 40 °C. Initial-rate data were fit to Eq. 2 to obtain the kinetic parameters shown. a k cat. At pH 8.0 (hollow circle), k cat was not well determined; the error in k cat at pH 8.0 was large (16 ± 30 s−1). The filled data points were fit to Eq. 4: pK a1 5.46 ± 0.19, pK a2 7.77 ± 0.41, pH-independent parameter 14.5 ± 1.9 s−1, and lower limit 1.18 ± 0.67 s−1. b k cat/K m. Data were fitted to Eq. 5: pK a1 4.15 ± 0.69, pK a2 6.62 ± 0.69, and pH-independent parameter 91,000 ± 141,000 s−1mM−1. c 1/K m. Data were fitted to Eq. 4: pK a1 5.05 ± 0.11, pK a2 5.15 ± 0.11, pH-independent parameter 6230 ± 9370 mM−1, and lower limit 64.3 ± 23.4 mM−1
Survey of Potential Nitrophenyl Substrates
In search of a suitable synthetic substrate for use in a spectrophotometric assay of TtGH28, activity of TtGH28 was determined acting on sixteen nitrophenyl-substituted potential substrates: 4-nitrophenyl-α-l-arabinofuranoside, 4-nitrophenyl-α-l-arabinopyranoside, 4-nitrophenyl-β-l-arabinopyranoside, 2-nitrophenyl-β-d-xylopyranoside, 4-nitrophenyl-α-d-xylopyranoside, 4-nitrophenyl-β-d-xylopyranoside, 4-nitrophenyl-α-l-fucopyranoside, 4-nitrophenyl-β-d-fucopyranoside, 4-nitrophenyl-β-l-fucopyranoside, 4-nitrophenyl-α-d-galactopyranoside, 4-nitrophenyl-β-d-galactopyranoside, 4-nitrophenyl-β-d-glucopyranoside, 4-nitrophenyl-α-d-glucopyranoside, 4-nitrophenyl-α-d-mannopyranoside, 4-nitrophenyl-β-d-mannopyranoside, and 4-nitrophenyl-α-l-rhamnopyranoside. For each of the sixteen potential nitrophenyl substrates at pH 5.0 and 7.0 and 40 °C, the activity of TtGH28 was less than one-millionth that of TtGH28 acting on triGalUA. Thus, no suitable nitrophenyl substrate for use in a spectrophotometric assay of TtGH28 was found.
To conclude, TtGH28 from Pseudothermotoga thermarum DSM 5069 is potentially useful for conversion of food processing waste pectinic biomass to value-added products. The expressed enzyme was characterized and found to be a hyperthermostable dimer with K 0.5t 86.3 °C. Kinetic modeling of the reaction progress is consistent with the enzyme being fully exo and fully non-processive, releasing monoGalUA as sole product from polyGalUA (EC 3.2.1.67). We show here that the enzyme is subject to substrate inhibition by diGalUA and triGalUA, with K si in the low mM range. The enzyme has a low K m for polyGalUA of ~9 μM and appears to be quite sensitive to product inhibition by GalUA with a K i in the low μM range.
References
Wong, D. (2008). Enzymatic deconstruction of backbone structures of the ramnified regions in pectin. Protein Journal, 27, 30–42.
Yapo, B. M., Lerouge, P., Thibault, J.-F., & Ralet, M.-C. (2007). Pectins from citrus peel cell walls contain homogalacturonans homogenous with respect to molar mass, rhamnogalacturonan I and rhamnogalacturonan II. Carbohydrate Polymers, 69, 426–435.
Benz, J. P., Protzko, R. J., Andrich, J. M. S., Bauer, S., Dueber, J. E., & Somerville, C. R. (2014). Identification and characterization of a galacturonic acid transporter from Neurospora crassa and its application for Saccharomyces cerevisiae fermentation processes. Biotechnology for Biofuels, 7, 20.
Lin, C. S. K., Pfaltzgraff, L. A., Herrero-Davila, L., Mubofu, E. B., Abderrahim, S., Clark, J. H., et al. (2013). Food waste as a valuable resource for the production of chemicals, materials and fuels. Current situation and global perspective. Energy & Environmental Science, 6, 426–464.
Pourbafrani, M., Forgács, G., Horváth, I. S., Niklasson, C., & Taherzadeh, M. J. (2010). Production of biofuels, limonene and pectin from citrus wastes. Bioresource Technology, 101, 4246–4250.
Kirsch, R., Gramzow, L., Theißen, G., Siegfried, B. D., Ffrench-Constant, R. H., Heckel, D. G., & Pauchet, Y. (2014). Horizontal gene transfer and functional diversification of plant cell wall degrading polygalacturonases: Key events in the evolution of herbivory in beetles. Insect Biochemistry and Molecular Biology, 52, 33–50.
Strong, F. E., & Kruitwagen, E. C. (1968). Polygalacturonase in the salivary apparatus of Lygus hesperus (Hemiptera). Journal of Insect Physiology, 14, 1113–1119.
Hobson, G. E. (1964). Polygalacturonase in normal and abnormal tomato fruit. Biochemical Journal, 92, 324–332.
Bergey, D. R., Orozco-Cardenas, M., De Moura, D. S., & Ryan, C. A. (1999). A wound- and systemin-inducible polygalacturonase in tomato leaves. Proceedings of the National Academy of Sciences of the United States of America, 96, 1756–1760.
Albersheim, P., & Anderson, A. J. (1971). Proteins from plant cell walls inhibit polygalacturonases secreted by plant pathogens. Proceedings of the National Academy of Sciences of the United States of America, 68, 1815–1819.
Cook, B. J., Clay, R. P., Bergmann, C. W., Albersheim, P., & Darvill, A. G. (1999). Fungal polygalacturonases exhibit different substrate degradation patterns and differ in their susceptibilities to polygalacturonase-inhibiting proteins. Molecular Plant-Microbe Interactions, 12, 703–711.
D’Ovidio, R., Mattei, B., Roberti, S., & Bellincampi, D. (2004). Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochimica et Biophysica Acta-Proteins and Proteomics, 1696, 237–244.
Doostdar, H., McCollum, T. G., & Mayer, R. T. (1997). Purification and characterization of an endo-polygalacturonase from the gut of west indies sugarcane rootstalk borer weevil (Diaprepes abbreviatus L.) larvae. Comparative Biochemistry and Physiology B Biochemistry and Molecular Biology, 118, 861–867.
Kirsch, R., Heckel, D. G., & Pauchet, Y. (2016). How the rice weevil breaks down the pectin network: Enzymatic synergism and sub-functionalization. Insect Biochemistry and Molecular Biology, 71, 72–82.
Pauchet, Y., Kirsch, R., Giraud, S., Vogel, H., & Heckel, D. G. (2014). Identification and characterization of plant cell wall degrading enzymes from three glycoside hydrolase families in the cerambycid beetle Apriona japonica. Insect Biochemistry and Molecular Biology, 49, 1–13.
Paz Celorio-Mancera, M., Allen, M. L., Powell, A. L., Ahmadi, H., Salemi, M. R., Phinney, B. S., et al. (2008). Polygalacturonase causes lygus-like damage on plants: Cloning and identification of western tarnished plant bug (Lygus hesperus) polygalacturonases secreted during feeding. Arthropod-Plant Interactions, 2, 215–225.
Kashyap, D. R., Vohra, P. K., Chopra, S., & Tewari, R. (2001). Applications of pectinases in the commercial sector: A review. Bioresource Technology, 77, 215–227.
Jayani, R. S., Saxena, S., & Gupta, R. (2005). Microbial pectinolytic enzymes: A review. Process Biochemistry, 40, 2931–2944.
Pedrolli, D. B., Monteiro, A. C., Gomes, E., & Carmona, E. C. (2009). Pectin and pectinases: Production, characterization and industrial application of microbial pectinolytic enzymes. Open Biotechnology Journal, 3, 9–18.
Gullón, B., Gómez, B., Martínez-Sabajanes, M., Yáñez, R., Parajó, J. C., & Alonso, J. L. (2013). Pectic oligosaccharides: Manufacture and functional properties. Trends in Food Science & Technology, 30, 153–161.
Hotchkiss, A. T., Olano-Martin, E., Grace, W. E., Gibson, G. R., & Rastall, R. A. (2003). Pectic oligosaccharides as prebiotics. In G. Eggleston & G. L. Cote (Eds.), Oligosaccharides in food and agriculture (pp. 54–62). Washington, DC: ACS Publications.
Kuivanen, J., Penttilä, M., & Richard, P. (2015). Metabolic engineering of the fungal D-galacturonate pathway for l-ascorbic acid production. Microbial Cell Factories, 14, 2.
Kuivanen, J., Dantas, H., Mojzita, D., Mallmann, E., Biz, A., Krieger, N., et al. (2014). Conversion of orange peel to L-galactonic acid in a consolidated process using engineered strains of Aspergillus niger. AMB Express, 4(33), 1–8.
Wagschal, K., Jordan, D. B., Lee, C. C., Younger, A., Braker, J. D., & Chan, V. J. (2015). Biochemical characterization of uronate dehydrogenases from three Pseudomonads, Chromohalobacter salixigens, and Polaromonas naphthalenivorans. Enzyme and Microbial Technology, 69, 62–68.
Abbadi, A., Gotlieb, K. F., Meiberg, J. B. M., Peters, J. A., & Van Bekkum, H. (1999). New Ca-sequestering materials: Based on the oxidation of the hydrolysis products of lactose. Green Chemistry, 1, 231–235.
Kiely, D. E., Chen, L., & Lin, T. H. (2000). Synthetic polyhydroxypolyamides from galactaric, xylaric, D-glucaric, and D-mannaric acids and alkylenediamine monomers-some comparisons. Journal of Polymer Science Part A: Polymer Chemistry, 38, 594–603.
Lavilla, C., Alla, A., Martínez De Ilarduya, A., Benito, E., García-Martín, M. G., Galbis, J. A., & Muñoz-Guerra, S. (2011). Carbohydrate-based polyesters made from bicyclic acetalized galactaric acid. Biomacromolecules, 12, 2642–2652.
Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M., & Henrissat, B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Research, 42, D490–D495.
Jansen, E. F., & MacDonnell, L. R. (1945). Influence of methoxyl content of pectic substances on the action of polygalacturonase. Archives of Biochemistry, 8, 97–112.
Yadav, S., Yadav, P. K., Yadav, D., & Yadav, K. D. S. (2009). Pectin lyase: A review. Process Biochemistry, 44, 1–10.
Koshland, D. E. (1953). Stereochemistry and mechanism of enzyme reactions. Biological Review of the Cambridge Philosophical Society, 28, 416–436.
Sinnott, M. L. (1990). Catalytic mechanisms of enzymic glycosyl transfer. Chemical Reviews, 90, 1171–1202.
Biely, P., Benen, J., Heinrichová, K., Kester, H. C. M., & Visser, J. (1996). Inversion of configuration during hydrolysis of α-1,4-galacturonidic linkage by three Aspergillus polygalacturonases. FEBS Letters, 382, 249–255.
Pitson, S. M., Mutter, M., van den Broek, L. A. M., Voragen, A. G. J., & Beldman, G. (1998). Stereochemical course of hydrolysis catalysed by α-L-rhamnosyl and α-D-galacturonosyl hydrolases from Aspergillus aculeatus. Biochemical and Biophysical Research Communications, 242, 552–559.
Li, K., Meng, K., Pan, X., Ma, R., Yang, P., Huang, H., et al. (2015). Two thermophilic fungal pectinases from Neosartorya fischeri P1: Gene cloning, expression, and biochemical characterization. Journal of Molecular Catalysis. B: Enzymatic, 118, 70–78.
Mertens, J. A. (2013). Kinetic properties of two Rhizopus Exo-polygalacturonase enzymes hydrolyzing galacturonic acid oligomers using isothermal titration calorimetry. Applied Biochemistry and Biotechnology, 170, 2009–2020.
Mertens, J. A., & Bowman, M. J. (2011). Expression and characterization of fifteen Rhizopus oryzae 99-880 polygalacturonase enzymes in Pichia pastoris. Current Microbiology, 62, 1173–1178.
Maller, A., da Silva, T. M., Damásio, A. R. D. L., Hirata, I. Y., Jorge, J. A., Terenzi, H. F., & Polizeli, M. L. T. M. (2013). Functional properties of a manganese-activated exo-polygalacturonase produced by a thermotolerant fungus Aspergillus niveus. Folia Microbiologica, 58, 615–621.
Tari, C., Dogan, N., & Gogus, N. (2008). Biochemical and thermal characterization of crude exo-polygalacturonase produced by Aspergillus sojae. Food Chemistry, 111, 824–829.
Kester, H. C. M., Kusters-Van Someren, M. A., Müller, Y., & Visser, J. (1996). Primary structure and characterization of an exopolygalacturonase from Aspergillus tubingensis. European Journal of Biochemistry, 240, 738–746.
Kar, S., & Ray, R. C. (2011). Purification, characterization and application of thermostable exo-polygalacturonase from Streptomyces erumpens mtcc 7317. Journal of Food Biochemistry, 35, 133–147.
Sawada, K., Suzumatsu, A., Kobayashi, T., & Ito, S. (2001). Molecular cloning and sequencing of the gene encoding an exopolygalacturonase of a Bacillus isolate and properties of its recombinant enzyme. Biochimica et Biophysica Acta-General Subjects, 1568, 162–170.
Chen, Y., Sun, D., Zhou, Y., Liu, L., Han, W., Zheng, B., et al. (2014). Cloning, expression and characterization of a novel thermophilic polygalacturonase from Caldicellulosiruptor bescii DSM 6725. International Journal of Molecular Sciences, 15, 5717–5729.
Liao, C. H., Revear, L., Hotchkiss, A., & Savary, B. (1999). Genetic and biochemical characterization of an exopolygalacturonase and a pectate lyase from Yersinia enterocolitica. Canadian Journal of Microbiology, 45, 396–403.
Kluskens, L. D., Van Alebeek, G. J. W. M., Walther, J., Voragen, A. G. J., De Vos, W. M., & Van Der Oost, J. (2005). Characterization and mode of action of an exopolygalacturonase from the hyperthermophilic bacterium Thermotoga maritima. FEBS Journal, 272, 5464–5473.
Parisot, J., Langlois, V., Sakanyan, V., & Rabiller, C. (2003). Cloning expression and characterization of a thermostable exopolygalacturonase from Thermotoga maritima. Carbohydrate Research, 338, 1333–1337.
Pedrolli, D. B., & Carmona, E. C. (2010). Purification and characterization of the exopolygalacturonase produced by Aspergillus giganteus in submerged cultures. Journal of Industrial Microbiology and Biotechnology, 37, 567–573.
Gill, S. C., & von Hippel, P. H. (1989). Calculation of protein extinction coefficients from amino acid sequence data. Analytical Biochemistry, 182, 319–326.
Wyatt, P. J. (1993). Light scattering and the absolute characterization of macromolecules. Analytica Chimica Acta, 272, 1–40.
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Analytical Chemistry, 31, 426–428.
Jordan, D. B. (2008). B-D-Xylosidase from Selenomonas ruminantium: Catalyzed reactions with natural and artificial substrates. Applied Biochemistry and Biotechnology, 146, 137–149.
Johnson, K. A., Simpson, Z. B., & Blom, T. (2009). Global Kinetic explorer: A new computer program for dynamic simulation and fitting of kinetic data. Analytical Biochemistry, 387, 20–29.
Leatherbarrow, R. J. (2004). GraFit 5. Surrey: Erithacus Software Ltd.
Van Santen, Y., Benen, J. A. E., Schröter, K. H., Kalk, K. H., Armand, S., Visser, J., & Dijkstra, B. W. (1999). 1.68-Å crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis. Journal of Biological Chemistry, 274, 30474–30480.
Steinbacher, S., Baxa, U., Miller, S., Weintraub, A., Seckler, R., & Huber, R. (1996). Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proceedings of the National Academy of Sciences of the United States of America, 93, 10584–10588.
Steinbacher, S., Miller, S., Baxa, U., Budisa, N., Weintraub, A., Seckler, R., & Huber, R. (1997). Phage P22 tailspike protein: Crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. Journal of Molecular Biology, 267, 865–880.
Abbott, D. W., & Boraston, A. B. (2007). The structural basis for exopolygalacturonase activity in a family 28 glycoside hydrolase. Journal of Molecular Biology, 368, 1215–1222.
Pijning, T., van Pouderoyen, G., Kluskens, L., van der Oost, J., & Dijkstra, B. W. (2009). The crystal structure of a hyperthermoactive exopolygalacturonase from Thermotoga maritima reveals a unique tetramer. FEBS Letters, 583, 3665–3670.
Yoder, M. D., Keen, N. T., & Jurnak, F. (1993). New domain motif: The structure of pectate lyase C, a secreted plant virulence factor. Science, 260, 1503–1507.
Pickersgill, R., Smith, D., Worboys, K., & Jenkins, J. (1998). Crystal structure of polygalacturonase from Erwinia caratovora ssp. carotovora. Journal of Biological Chemistry, 273, 24660–24664.
Shimizu, T., Nakatsu, T., Miyairi, K., Okuno, T., & Kato, H. (2002). Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution. Biochemistry, 41, 6651–6659.
Kohn, R., & Kováč, P. (1978). Dissociation constants of D-galacturonic and D-glucuronic acid and their O-methyl derivatives. Chem Zvesti, 32, 478–485.
Cook, P. F., & Cleland, W. W. (2007). Enzyme kinetics and mechanism. New York: Garland Science.
Acknowledgments
This work was supported by the United States Department of Agriculture (USDA) CRIS 5325-41000-049-00 and the National Institute of Food and Agriculture grant 2012-03998 (V.C., C.C.L., K.W.), and USDA CRIS 3620-41000-118-00D (D.B.J. and J.R.S.). This research was also supported in part by an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the USDA (J.R.S.). ORISE is managed by Oak Ridge Associated Universities (ORAU) under DOE contract number DE-AC05-06OR23100. All opinions expressed in this paper are the authors’ and do not necessarily reflect the policies and views of USDA, ARS, DOE, or ORAU/ORISE. The Keck Biophysics Facility was supported in part by NCI grant CCSG P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center at Northwestern University (A.A.G.). The mention of firm names or trade products does not imply that they are endorsed or recommended by the USDA over other firms or similar products not mentioned. USDA is an equal opportunity provider and employer.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
12033_2016_9948_MOESM2_ESM.tif
PAGE of SEC purified TtGH28. Lanes 1,7 Novex Sharp protein standards, lane 2 0.625 μg TtGH28, lane 3 1.25 μg TtGH28, lane 4 2.5 μg TtGH28, lane 5 5 μg TtGH28, lane 6 10 μg TtGH28. Supplementary material 2 (TIFF 9701 kb)
Rights and permissions
About this article
Cite this article
Wagschal, K., Rose Stoller, J., Chan, V.J. et al. Expression and Characterization of Hyperthermostable Exo-polygalacturonase TtGH28 from Thermotoga thermophilus . Mol Biotechnol 58, 509–519 (2016). https://doi.org/10.1007/s12033-016-9948-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-016-9948-8