Abstract
SLFN11 is abnormally expressed and associated with survival outcomes in various human cancers. However, the role of SLFN11 in clear cell renal cell carcinoma (ccRCC) remains unclear. This study aimed to investigate the clinical value and potential functions of SLFN11 in ccRCC. Comprehensive bioinformatics analyses were performed using online databases. Quantitative real-time PCR (qPCR) and western blotting were used to validate the expression data. CCK8, flow cytometry analysis, and EdU staining were performed to determine the level of cell proliferation. Flow cytometry analysis was also used to detect cell apoptosis. Wound-healing assay and Transwell assays were performed to assess cell migration and invasion capability, respectively. SLFN11 was overexpressed and was an independent prognostic factor in ccRCC. SLFN11 knockdown inhibited cell proliferation, migration, and invasion and promoted apoptosis. Functional and pathway enrichment analyses suggested that SLFN11 may have an impact on tumorigenesis in ccRCC through regulation of the inflammatory response, the PI3K/AKT signaling pathway and other effectors. Furthermore, SLFN11 knockdown inhibited the phosphorylation of the PI3K/AKT signaling pathway and could be activated by 740 Y-P. Finally, we demonstrated that miR-183 may specifically target SLFN11, and miR-183 expression was correlated with predicted survival. SLFN11 may play a critical role in ccRCC progression and may serve as a novel prognostic biomarker in ccRCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Renal cell carcinoma (RCC) is the most lethal urological malignancy that arises from renal tubular epithelial cells, accounting for approximately 90% of renal malignancies and 3% of adult malignancies [1,2,3]. While RCC encompasses various histological subtypes, clear cell renal cell carcinoma (ccRCC) is the most frequent subtype, being responsible for approximately 75% of all diagnosed cases of RCC and accounting for approximately 90% of renal malignancies and up to 4% of all newly diagnosed cancer cases in 2018 worldwide [4, 5]. RCC may develop without typical symptoms, making clinical diagnosis and effective treatment exceptionally challenging, particularly in the early stages [6]. Currently, surgical resection is the mainstay of treatment for patients with RCC. RCC can be surgically removed in most patients. However, the high rate of local or distant metastasis can lead to a dismal prognosis with short survival times in RCC patients [7, 8]. Therefore, accurate diagnosis and prompt commencement of therapy are of significant value to improve outcomes for patients with renal clear cell carcinoma.
The pathogenesis of ccRCC is a complex process consisting of diverse molecular and cellular events and is controlled by complex signaling pathways. Currently, the development and progression of ccRCC are not fully understood and the underlying mechanism has not been clarified. To this end, high-throughput analyses have been performed under a variety of conditions, which may revolutionize our understanding of RCC [9,10,11,12]. By comparing the expression profiles in tumor tissues and adjacent nontumor tissues, distinct gene expression patterns, including up-or down-egulation of mRNAs and noncoding RNAs (e.g., miRNAs and lncRNAs) were found to be related to prognosis for patients with ccRCC. However, despite the unmet need to provide timely targeting therapy, no major improvements in the diagnosis of ccRCC have been implemented in the clinic. Thus, the identification of potential biomarkers for preliminary screening and prognosis is urgently needed.
Schlafen 11 (SLFN11) belongs to the group 3 Schlafen family and it halts the cell cycle progression when it expression results in the accumulation of DNA damage and replication stress [13]. SLFN11 is abnormally expressed in a variety of malignancies and is associated with prognosis in human cancers [14,15,16]. In recent years, SLFN11 was found to be associated with alterations in sensitivity to anticancer drugs. Junya et al. found that SLFN11 was a crucial factor in the anticancer activity of trabectedin in human sarcoma cells [17]. Moreover, SLFN11 was identified as a predictive biomarker of sensitivity to poly (ADP-ribose) polymerase (PARP) inhibitors in small-cell lung cancer, and to platinum-based chemotherapy in prostate cancer [18, 19]. SLFN11 is also significantly overexpressed in ccRCC tissues and cell lines [20]. However, the underlying role and mechanism of SLFN11 in the development and progression of ccRCC remain unclear. A previous study showed that changes in the DNA damage response (DDR) pathways were associated with the increased recurrence of locally advanced ccRCC [21]. Moreover, it has been shown that the sensitivity of cancer cells to DNA-damaging agents (DDAs) could be enhanced by SLFN11 in lung, colorectal, and ovarian cancers, and high SLFN11 expression was positively correlated with tumor-free survival in ovarian and colorectal cancer patients [14, 22, 23]. Therefore, we hypothesized that SLFN11 may be involved in ccRCC pathogenesis.
In this paper, we sought to investigate the role of SLFN11 and its potential as a diagnostic and prognostic biomarker in ccRCC. We first compared the expression of the SLFN11 gene in ccRCC and normal renal tissues using data retrieved from the Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) databases and analyzed the correlation between SLFN11 expression and overall survival (OS) rates. We also performed Cox regression analysis to define the prognostic role of SLFN11 in ccRCC. According to our findings, SLFN11 significantly inhibited ccRCC cell proliferation, triggered apoptosis, and repressed migration and invasion. Moreover, we found that SLFN11 can promote tumorigenesis through the PI3K/AKT pathway. Furthermore, we screened genes co-expressed with SLFN11 using LinkedOmics and cBioPortal and predicted the potential function of SLFN11 in ccRCC through Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. We then identified SLFN11-related signaling pathways in ccRCC using gene set enrichment analysis (GSEA). Finally, we assessed the internal interaction relationship between hsa-miR-183 and SLFN11 using computational prediction and expression relationship analyses. Taken together, these results together suggest that SLFN11 participates in ccRCC tumorigenesis and may represent a putative diagnostic and prognostic biomarker for ccRCC.
Materials and methods
Data collection
The data used in this paper are available from public datasets. Gene expression data and clinical information were downloaded from the Gene Expression Omnibus (GEO database, http://www.ncbi.nlm.nih.gov/geo) (accession nos. GSE68417, GSE66272, GSE53000, GSE46699, and GSE36895) and the UCSC Xena browser (http://xena.ucsc.edu/). The expression distribution of SLFN11 in a body map was explored using the GEPIA database (http://gepia.cancer-pku.cn/), a newly established platform and visualization site based on Genotype-Tissues Expression (GTEx) and the Cancer Genome Atlas (TCGA) data integration. A multitumor SLFN11 expression box plot was extracted from the Cancer Cell Line Encyclopedia project (CCLE, https://portals.broadinstitute.org/ccle), and the genetic alteration data (copy number alterations and mutations) of the SLFN11 gene were retrieved from cBioPortal (http://www.cbioportal.org).
Data processing and study design
We first identified genes that were significantly differentially expressed in each GEO dataset with the limma R package and then combined the results and eliminated the batch difference with the robustrankaggreg R package. After filtering out samples missing clinical information, subsequently, RNA-seq data of 523 ccRCC patients and matched clinical annotations from TCGA were acquired from the UCSC Xena browser for further analysis. Survival and Cox regression analyses, were conducted in R 3.4.3 with the R package survival, and forest plots were constructed. Kaplan–Meier plots were drawn using the R package survminer. All other plots were generated using the R package ggplot2 and the online tool LinkedOmics. The design and workflow of this study are illustrated as a flowchart in Fig. 1.
GO and KEGG analyses
The co-expression functions of the SLFN11 gene in ccRCC were further analyzed by LinkedOmics (http://www.linkedomics.org). Genes with a Pearson’s correlation values > 0.6 were applied to identify genes co-expressed with SLFN11. To provide a first overview of genes co-expressed with SLFN11, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations were performed using DAVID 6.8 (https://david.ncifcrf.gov/home.jsp). Biological process GO terms and KEGG pathways with enrichment values of p < 0.05 were considered significant.
Gene set enrichment analysis (GSEA). Gene set enrichment analysis (GSEA) was performed using GSEA 3.0 software from the Broad Institute (https://software.broadinstitute.org/gsea/). A total of 523 ccRCC specimens from TCGA were dichotomized into lower and higher SLFN11-expression groups according to the median expression levels. To obtain the functions and potential mechanism of SLFN11 in ccRCC, GSEA was then conducted to determine the enrichment significance of experimentally defined ccRCC-related gene sets or other tumor-associated gene sets. The expression of SLFN11 was used as the phenotypic label, and 1000 phenotype-based permutations were computed. The a priori-defined sets of genes we used were from KEGG pathways (c2.cp.Kegg,v7.2.symbols.gmt). All other parameters were set at their defaults. Gene sets with nominal values of p < 0.05 were considered significant.
Cell culture
Human renal clear cell carcinoma A-498, CAKI-1, and 786-O cells as well as the normal human kidney epithelial cell line HK-2 were purchased from the American Type Culture Collection (ATCC). OS-RC-2 cells were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). A-498, CAKI-1, OS-RC-2, and 786-O cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with 10% fetal bovine serum (FBS, Invitrogen), while HK-2 cells were cultured in DMEM/F12 medium containing 10% FBS. Cells were grown at 37 °C with 5% CO2.
RNA extraction and quantitative real-time polymerase chain reaction (qPCR)
Total RNA was extracted using an RNA-quick purification kit (ES Science) and then was subjected to synthesize cDNA synthesis using a reverse transcriptase kit (TaKaRa). Real-time qPCR was carried out with SYBR Premix Ex Taq (TaKaRa), and the mRNA and miRNA expression levels were normalized to β-actin and U6, respectively. All samples were run in triplicate. Relative expression was calculated by the comparative ∆∆Ct method. The primer sequences were as follows: SLFN11 forward: CAGCCTGACAACCGAGAAATG; SLFN11 reverse: GGCCCACTAGATAGACTCAGC; miR-183 forward: GCGGTGAATTACCGAAGGG; miR-183 reverse: AGTGCAGGGTCCGAGGTATT; RT primer: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTTATGG.
Western blotting
Analysis of protein expression was performed using western blotting. In brief, RIPA buffer (including PMSF) was added to cells after they were washed with PBS on ice. Cell scrapers were used to collect the cell lysates, which were vortexed for 30 s before being used. The lysates were then centrifuged at 4 °C at 12,000 rpm for 20 min, and the supernatants were collected. To detect the protein concentrations, a BCA protein assay was used (Beyotime). In this experiment, 50 ug of total protein sample was separated by 10% SDS‒PAGE and transferred to polyvinylidene fluoride membranes. After transfer, the PVDF membranes were blocked for 30 min at room temperature with the rapid blocking liquid. Then, the membranes were incubated overnight with a primary antibody at 4 °C. The following antibodies were used: SLFN11 (1:1000) (Abcam, Cat No: ab271354), PI3K (1:1000) (Cell Signaling Technology, Cat No: #4257), p-PI3K (1:1000) (Abcam, Cat No: ab271354), AKT (1:1000) (Proteintech, Cat No: 10176-2-AP ), p-AKT (1:2000) (Cell Signaling Technology, Cat No: #4060), and β-actin (1:20000) ( Proteintech, Cat No: 81115-RR).
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) (Boster, China) assay was used to determine cell proliferation, following the instructions provided by the manufacturer. In brief, cells were placed onto 96-well plates at a density of 1.5 × 103 cells per well and kept at 37 °C for incubation. Following the addition of 10 µL of CCK-8 reagents, the cells were incubated without interruption for 2 h. A microplate luminometer reader was utilized to measure the absorbance at 450 nm.
EdU staining was performed to assess cell proliferation using the EdU Assay/ BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 555. The detection procedure followed a standard protocol. An inverted fluorescence microscope (Nikon Corporation) was used to acquire the images of the plates.
Transwell migration/invasion assay
Cell migration and invasion abilities were determined using Biofil Transwell insert chambers (8 mm pore size; Biofil). Matrigel was purchased from Corning. We digested the cells with trypsin, added the suspension of cells (2 × 104) to the upper chamber of Transwell, and then added the medium to the lower chamber, and maintained the culture. The upper membrane was incubated for 12 h, after which the cells remaining on the membrane were removed, fixed with 4% paraformaldehyde, stained with crystal violet, and imaged, with the white cells counted.
Wound-healing assay
The cells were seeded in 6-well plates, and a wound-healing assay was performed. After cell confluence reached 80–90% the following day, pipette tips were used to make scratches perpendicular to the horizontal scratches. Then, the cells were rinsed with PBS and examined using a microscope. We observed and photographed the cells at 0, 12, and 24 h, and we calculated the scratch healing rate by using image analysis software.
Lentiviral infection
Stable cell lines were established with by transfection with lentiviral vectors. Lentiviral-based small hairpin RNA (shRNA) targeting SLFN11 was constructed by TSINGKE Biological Technology (Beijing, China). The target sequence of shSLFN11 was 5′-CAGAGGACTAGAGGAGTTAAT-3′.
Flow cytometry for cell cycle & apoptosis analyses
For cell cycle analysis, after the stable cells were digested, centrifuged, and purified, they were counted and seeded in a 6-well culture plate. After 48 h of incubation, the cells were then harvested, washed with ice-cold PBS, and fixed with 70% alcohol overnight at 4 °C. Then, cell cycle analysis was performed at the Laboratory of Flow Cytometry, School of Life Science, Chongqing Medical University.
For cell apoptosis, EDTA-free trypsin was used to digest the cells, which were washed twice with PBS after the digestion. Subsequently, the cells were resuspended in 500 µL of PBS and immediately sent to the Laboratory of Flow Cytometry, School of Life Science, Chongqing Medical University.
Statistical analysis
The statistical significance of SLFN11 expression between ccRCC specimens and adjacent normal tissues was calculated using a two-tailed Student’s t test. Pearson’s correlation coefficient was applied to assess correlations between gene expression. Associations between variables were evaluated by the chi-square test and the Kruskal-Wallis test for categorical and continuous variables. The log-rank test was used to compare the survival curves between groups of patients. Multivariate Cox regression analysis was adjusted according to age, sex, origin, tumor grade, tumor stage, T stage, M stage, and SLFN11 expression. All statistical analyses were two-sided and were performed using SPSS 26.0 and GraphPad Prism 8.0 software. A valued of p < 0.05 was considered statistically significant.
Results
SLFN11 was differentially expressed in diverse cancer types. To identify genes upregulated in ccRCC, a total of 5 GEO datasets (GSE68417, GSE66272, GSE53000, GSE46699, and GSE36895) were examined and analyzed for investigation. After quality assessment and data preprocessing, we found that SLFN11 was highly expressed in ccRCC based on data from GEO databases (Fig. 2A). The body map from GEPIA showed that SLFN11 was broadly expressed in multiple human tissues (Fig. 2B). In addition, we observed alterations in SLFN11 expression in different cancer cell lines based on analysis of the CCLE database (Fig. 2C). We next analyzed SLFN11 expression across multiple tumors and paired adjacent normal tissues with data from TCGA and found that SLFN11 was differentially expressed in diverse cancer types (Fig. 2D).
Expression profile of SLFN11 in multiple cancers. A The heatmap of expression differences (LogFC) of SLFN11 in 5 GEO ccRCC datasets. (p < 0.05). B SLFN11 gene expression map in human body. The median expression of SLFN11 was distributed in tumor tissue (red) and normal tissue (green). C Expression of SLFLN11 in human cancer cell lines. D Expression profile of SLFN11 in tumor samples and normal tissues in different cancers
Clinical and prognostic value of SLFN11 expression in ccRCC
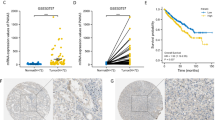
To further validate the high expression of SLFN11 in ccRCC, we analyzed the expression pattern of SLFN11 in the TCGA database. The results indicated that SLFN11 was significantly upregulated in ccRCC patients (p < 0.001, Fig. 3A). Similarly, SLFN11 expression was increased in cancer tissues compared to that in adjacent noncancerous tissues from the same patient (p < 0.001, Fig. 3B). Using median expression as the cutoff, 523 ccRCC specimens from TCGA were dichotomized into lower and higher SLFN11-expression groups. We observed that high SLFN11 expression was associated with poor overall survival (p < 0.001, Fig. 3C) in ccRCC patients. Consistent with the results in the TCGA cohort, the SLFN11 mRNA and protein expression levels were significantly higher in renal cancer cell lines (Fig. 3D and E). Moreover, SLFN11 expression was associated with age, T stage, M stage, and clinical tumor stage (p < 0.05) (Fig. 3G; Table 1). Cox regression analysis identified SLFN11 expression as an independent prognostic factor in ccRCC (HR (95% CI): 1.28 (1.03–1.60); p < 0.05) (Fig. 3H).
The clinical and prognostic value of SLFN11 in ccRCC. A SLFN11 expression in ccRCC tumor samples (red) and normal tissues (blue) from TCGA. B SLFN11 expression in ccRCC tumor samples (red) and adjacent paired normal tissues (blue) from TCGA. C High expression of SLFN11 predicted poor prognosis of ccRCC patients. (p < 0.001). D, E Validation of the SLFN11 expression in renal carcinoma cell lines (786-O, A-498, OS-RC-2, CAKI-1) and normal kidney epithelial cell (HK2) by qPCR and WB. F Relationships between expression levels of SLFN11 with age, tumor stage, T stage and M stage. G Univariate Cox and Multivariate Cox regression model for prediction of overall survival
SLFN11 facilitates cell proliferation, migration, and invasion, and inhibits apoptosis in vitro
According to the qPCR and WB analyses, SLFN11 expression levels were highest in the 786-O cell line. Therefore, using the lentivirus, we successfully created a stable ccRCC line in 786-O cells by knocking down SLFN11, and the knockdown efficiency was verified using qPCR and WB (Fig. 4A and B).
SLFN11 knockdown alters 786-O cell proliferation, invasion, migration and apoptosis. A, B Transfection efficiency of SLFN11 in 786-O. C CCK8 assay was used to evaluate cell proliferation. D Flow cytometry was performed to detect 786-O apoptosis. E Flow cytometry was performed to determine cell cycle. F EdU staining (red) was used to detect cell proliferation. G The scratch-wound assay revealed the effects of transfection on the migration of 786-O cells. H Transwell assay analysis of cell invasion and migration
To further examine the functional role of SLFN11 in ccRCC progression. The CCK8 assay showed that knockdown of SLFN11 inhibited the proliferation of ccRCC cells (Fig. 4C). Analysis by flow cytometry indicated that 786-O cells exhibited increased apoptosis ratios after SLFN11 knockdown (Fig. 4D). Additionally, SLFN11 knockdown interfered with the transition from the G0/G1 to the S phase during the cell cycle, thus inhibiting 786-O cell proliferation (Fig. 4E). Similarly, the EdU proliferation assay showed that the proliferation of 786-O cells was largely inhibited after SLFN11 knockdown (Fig. 4F). We also confirmed that the migration of 786-O cells was inhibited after SLFN11 knockdown by a cell wound-healing assay (Fig. 4G). Furthermore, the results of Transwell migration and invasion assays confirmed that SLFN11 knockdown also inhibited the migration and invasion of ccRCC cells (Fig. 4H).
Identification of genes co-expressed with SLFN11 and the potential function of SLFN11
The cBioPortal database was accessed to analyze the genetic alterations of SLFN11 in ccRCC (Kidney Cancer, TCGA, PanCancer Atlas). The results suggested that 6% (22/352) of ccRCC cases showed SLFN11 alterations, including 1 case of missense mutation, 1 case of amplification, 13 cases of mRNA upregulation, and 7 cases of mRNA downregulation (Fig. 5A). To gain further insight into the mutation profiles of SLFN11 in the 352 ccRCC samples, we next decided to focus on mutational hotspots of SLFN11 in the ccRCC. For our subsequent detailed mutational analysis, we found that SLFN11 had only one mutation type (K346T), which had a somatic mutation frequency of 0.3% (Fig. 5B). We next examined 20159 genes associated with SLFN11 expression and visualized the positive correlations (8715 genes) and negative correlations (4980 genes) in a gene heatmap (Pearson’s correlation analysis, p < 0.05) through an analysis of the TCGA database using the LinkedOmics (Fig. 5C–E). In total, 211 genes with Pearson’s correlation coefficient > 0.5 were selected as genes co-expressed with SLFN11, and top 5 co-expressed genes are C6orf150, FAM102B, CSGALNACT2, IFI16 and ZEB2 (Fig. 5F). Among the top 5 co-expressed genes of SLFN11, IFI16 was found to be significantly correlated with OS (p < 0.05) (Fig. 5G) in ccRCC patients. Under certain conditions, genes with similar expression patterns may be functionally related. To further explore the underlying mechanisms of SLFN11 in ccRCC, genes co-expressed with SLFN11 were submitted for GO analysis of gene function (Table 2) and KEGG pathway analysis in DAVID 6.8 (Table 3). According to the GO enrichment analysis, the genes co-expressed with SLFN11 were related to biological processes such as inflammatory response, signal transduction, adaptive immune response, innate immune response, defense response to virus, T-cell receptor signaling pathway, immune response, T-cell differentiation, hemopoiesis, and positive regulation of gene expression. Similarly, according to the KEGG analysis, the genes co-expressed with SLFN11 were mainly related to signaling pathways such as measles, the Jak-STAT signaling pathway, cytokine-cytokine receptor interaction, the PI3K-Akt signaling pathway, platelet activation, osteoclast differentiation, and the T-cell receptor signaling pathway, etc.
Identification of SLFN11 co-expressed genes and the potential function of SLFN11. A The alteration and expression heatmap of SLFN11 from cBioPortal database. B Mutational Hotspots of SLFN11 in the ccRCC from the cBioPortal database (TCGA, PanCancer Atlas). C–E Identification of SLFN11 co-expressed genes in ccRCC using Linkedomics database. F Scatter plot of correlation between expression levels of SLFN11 and top 5 co-expressed genes. G Kaplan–Meier analysis of IFI16 levels with overall survival in ccRCC. H Significantly enriched KEGG pathways by GSEA. I The levels of p-PI3K, PI3K, p-AKT, and AKT were determined by Western blot, and were analyzed after treatment with PI3K inhibitor (740 Y-P, 10ng/mL) in 786-O cells
To further delineate pathways that SLFN11 may regulate or influence, we utilized GSEA with curated gene sets. Specimens from TCGA were dichotomized into lower and higher SLFN11-expression groups according to the median of the expression level. Together, these data suggested that the pathways associated with tumorigenesis and progression were significantly altered with the abnormal expression of SLFN11, including “renal cell carcinoma”, “regulation of actin cytoskeleton”, “toll-like receptor signaling pathway”, “natural killer cell-mediated cytotoxicity”, and “nod like receptor signaling pathway”, and “Jak-STAT signaling pathway”, were significantly altered with abnormal SLFN11 expression (Fig. 5H). In conclusion, SLFN11 may be involved in the tumorigenesis of ccRCC through these signaling pathways.
To investigate whether SLFN11 inhibits 786-O cell proliferation and promotes apoptosis through the PI3K/Akt signaling pathway, we performed WB experiments to verify the effect of SLFN11 on the PI3K/Akt signaling pathway. We observed that the phosphorylation levels of PI3K and AKT were markedly suppressed in the SLFN11-knockdown cells, with an unaltered expression of PI3K and AKT between the groups (Fig. 5I). Furthermore, the PI3K agonist 740 Y-P was subsequently used to treat SLFN11-knockdown cells, in which SLFN11 had been silenced to further investigate the relationship between SLFN11 and the PI3K/AKT signaling pathway in ccRCC progression. Our data showed that 740 Y-P could restore the PI3K and AKT phosphorylation in SLFN11 knockdown 786-O cells (Fig. 5J).
miR-183 may directly target SLFN11
To identify miRNAs associated with SLFN11 expression, we explored the correlation between the expression of SLFN11 and miRNAs (Pearson’s correlation analysis, p < 0.05) (Fig. 6A–C). Then, we searched for miRNAs that may directly target SLFN11 via “TargetScan”, " miRBridge”, “RNAhybrid”, “miRanda” and “miRDB” databases, respectively. A Venn diagram was constructed to analyze the overlapping miRNAs in the five databases, and 53 overlapping miRNAs were identified (Fig. 6D). Then, the 53 overlapping miRNAs were crossed with the top 30 negatively correlated miRNAs in the Venn diagram, leaving only miR-183 (Fig. 6E). As with the previous prediction, low expression of miR-183 in human renal cancer cell lines was confirmed by qPCR using U6 as an internal control (Fig. 6F).
miR-183 may target SLFN11 expression. A Funnel plot demonstrating the distribution of positively and negatively correlated miRNAs with SLFN11. B, C Heatmap of expression of miRNAs positively (B) and negatively (C) correlated with SLFN11 expression (top 50). D Venn diagram showing the overlap of miRNAs that potentially interact with SLFN11. Different colors represented different databases; E Venn diagram showing miRNA overlapping in “53 Common miRNAs” and “SLFN11 negatively correlated miRNAs (top 30)”. F miR-183 expression was assessed by qRT-PCR. (p < 0.05). G Scatter plot of correlation between expression levels of SLFN11 and miR-183. H–J Correlation between miR-183 expression and clinical characteristics
We next used the LinkedOmics database to analyze the relationship between the expression of hsa-miR-183 and the prognosis of ccRCC patients. We found that miR-183 was negatively correlated with SLFN11 expression (R = − 0.4361, p < 0.001, Fig. 6G), and miR-183 expression was significantly correlated with overall survival (p = 0.046), pathological stage (p = 0.002), T stage (p = 0.002,) and N stage (p = 0.003) in ccRCC patients (Fig. 6H–K). Altogether, these results suggested that miR-183 may serve as a negative regulator of SLFN11 and could be used as a prognostic marker in ccRCC patients.
Discussion
SLFN11 is abnormally expressed in a variety of malignancies and is linked to prognosis in human cancers [14,15,16]. Using RNA-seq, Vincenza et al. found that SLFN11 was highly expressed in metastatic castration-resistant prostate cancer, and confirmed its overexpression in lung and ovarian cancers [18, 19, 23]. Moreover, high SLFN11 expression was associated with better prognosis in patients with ovarian cancer and esophageal squamous cell carcinoma [15, 23], and could predict better survival in colorectal cancer patients expressing wild-type KRAS exon 2 receiving oxaliplatin-based adjuvant chemotherapy [14]. However, the diagnostic and prognostic value of SLFN11 in ccRCC and the potential molecular mechanism of SLFN11 in the pathogenesis of ccRCC have not yet been reported.
In the present study, we have identified for the first time that SLFN11 may be a potential oncogene and a novel prognostic marker of poor prognosis in ccRCC in humans. To our knowledge, this is the first study to comprehensively investigate the expression and clinical value of SLFN11 in ccRCC. We found that SLFN11 was highly expressed in ccRCC, and its upregulation predicted a poor prognosis. These results were further confirmed by qPCR, indicating that SLFN11 may play a critical role in ccRCC.
To further explore the clinical significance of SLFN11 in ccRCC, we investigated the relationship between SLFN11 expression and clinicopathological variables of ccRCC with data from the TCGA database. We found that SLFN11 expression was related to malignant phenotypes of ccRCC, including tumor stage, T stage, and M stage (p < 0.05). Cox regression analysis also revealed an association of SLFN11 expression with poorer outcomes (HR (95% CI): 1.28 (1.03–1.60); p < 0.05). Additionally, we identified C6orf150, FAM102B, CSGALNACT2, IFI16, and ZEB2 as the top 5 genes co-expressed with SLFN11 in LinkedOmics analysis. Among them, IFI16 was found to be overexpressed in ccRCC and was associated with lower disease-free survival and overall survival [24]. ZEB2, an epithelial-to-mesenchymal transition (EMT) mediator, was also reported to be an independent biomarker for poor prognosis in patients with ccRCC [25]. These findings are consistent with our data, which indicated that SLFN11 expression was positively correlated with IFI16/ZEB2 expression and was related to poor prognosis in ccRCC. Furthermore, in vitro experiments, our findings showed that SLFN11 likely functions as a tumor promoter in ccRCC progression. Therefore, we suggest that SLFN11 may be a predictor of poor prognosis for ccRCC.
To facilitate the understanding of SLFN11 in ccRCC, GO and KEGG analyses of genes co-expressed with SLFN11 were performed. The results suggested that SLNF11 may participate in biological processes such as inflammatory response, signal transduction, adaptive immune response, innate immune response, defense response to virus, T-cell receptor signaling pathway, immune response, and T-cell differentiation. GSEA indicated that SLFN11 was involved in the regulation of various pathways, including natural killer cell-mediated cytotoxicity and the T-cell receptor signaling pathway. Interestingly, a recent study showed that SLFN11 mediates T-cell-mediated cytotoxicity in a tissue-dependent manner and could mediate the sensitivity of tumor cells to cytotoxic T-cell attack through the native immune response [26], suggesting that SLFN11 may be of value in T and NK cell-based based cancer immunotherapies. In addition, GSEA also found that SLFN11 expression was positively correlated with multiple ccRCC-related signaling pathways, including renal cell carcinoma, the Jak-STAT signaling pathway, the PI3K/AKT signaling pathway, and the Toll-like signaling pathway, etc. Thus, we found that SLFN11 inhibited the activation of PI3K/AKT signaling pathways by downregulating PI3K expression and AKT phosphorylation, and that it was activated using the PI3K agonist 740 Y-P.
Altogether, our results demonstrate that SLFN11 may act as an oncogene in ccRCC. MicroRNAs (miRNAs), which function as negative regulators of gene expression, have been reported to play an important physiological and pathological roles in ccRCC [27]. Evidence has suggested novel functional roles of miRNAs in ccRCC, including miR-124, miR-203 [28], and miR-122 [29], etc. In this study, we found that miR-183 was downregulated in ccRCC cell lines. Furthermore, we found that SLFN11 was a potential target of miR-183, while SLFN11 expression was negatively correlated with the level of miR-183 in ccRCC. Simultaneously, miR-183 expression was significantly associated with tumor stage, T stage, N stage, and overall survival in ccRCC patients. An increasing body of evidence has revealed the aberrant expression and essential roles of miR-183 in various types of human cancers [30, 31]. Yuan et al. found that miR-183 inhibits starvation-induced autophagy and apoptosis by targeting UVRAG in human gastric cancer cells [32]. However, the role of miR-183 has not been reported in ccRCC. Here, our study revealed that miR-183 may negatively target SLFN11, and play an anticancer role through SLFN11 in ccRCC.
We acknowledge that this study does have some limitations, including that the expression levels of SLFN11 and miR-183 were verified only in ccRCC cell lines. Further studies will be necessary to validate the results in a large number of clinical samples. In addition, biochemical and functional studies are needed to elucidate the exact function of SLFN11 and confirm this hypothesis.
In conclusion, our study revealed the abnormal expression and clinical value of SLFN11 in ccRCC through bioinformatics analysis. SLFN11 might have potential clinical value in the diagnosis and prognosis prediction for ccRCC as well as in targeted anticancer therapy for ccRCC patients.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Huang Q, Sun Y, Ma X, Gao Y, Li X, Niu Y, et al. Androgen receptor increases hematogenous metastasis yet decreases lymphatic metastasis of renal cell carcinoma. Nat Commun. 2017;8:918. https://doi.org/10.1038/s41467-017-00701-6.
Singh A, Singh I, Singh N, Puzanov I. Optimal management of first-line advanced renal cell carcinoma: focus on pembrolizumab. Onco Targets Ther. 2020;13:4021–34. https://doi.org/10.2147/OTT.S215173.
Bedke J, Gauler T, Grünwald V, Hegele A, Herrmann E, Hinz S, et al. Systemic therapy in metastatic renal cell carcinoma. World J Urol. 2017;35:179–88. https://doi.org/10.1007/s00345-016-1868-5.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. https://doi.org/10.3322/caac.21492.
Stitzlein L, Rao P, Dudley R. Emerging oral VEGF inhibitors for the treatment of renal cell carcinoma. Expert Opin Investig Drugs. 2019;28:121–30. https://doi.org/10.1080/13543784.2019.1559296.
Kang YT, Kim YJ, Lee TH, Cho YH, Chang HJ, Lee HM. Cytopathological study of the circulating tumor cells filtered from the cancer patients’ blood using hydrogel-based cell block formation. Sci Rep. 2018;8:15218. https://doi.org/10.1038/s41598-018-33464-1.
Dimitrieva S, Schlapbach R, Rehrauer H. Prognostic value of cross-omics screening for kidney clear cell renal cancer survival. Biol Direct. 2016;11:68. https://doi.org/10.1186/s13062-016-0170-1.
Grange C, Brossa A, Bussolati B. Extracellular vesicles and carried miRNAs in the progression of renal cell carcinoma. Int J Mol Sci. 2019;20:1832. https://doi.org/10.3390/ijms20081832.
Zhang H, Wei P, Lv W, Han X, Yang J, Qin S, et al. MELK is upregulated in advanced clear cell renal cell carcinoma and promotes disease progression by phosphorylating PRAS40. Cell Transplant. 2019;28:37S-50S. https://doi.org/10.1177/0963689719890860.
Morgan TM, Mehra R, Tiemeny P, Wolf JS, Wu S, Sangale Z, et al. A multigene signature based on cell cycle proliferation improves prediction of mortality within 5 Yr of radical nephrectomy for renal cell carcinoma. Eur Urol. 2018;73:763–9. https://doi.org/10.1016/j.eururo.2017.12.002.
Song W, Shao Y, He X, Gong P, Yang Y, Huang S, et al. IGFLR1 as a novel prognostic biomarker in clear cell renal cell cancer correlating with immune infiltrates. Front Mol Biosci. 2020. https://doi.org/10.3389/fmolb.2020.565173.
Lu J, Zhu L, Zheng LP, Cui Q, Zhu HH, Zhao H, et al. Overexpression of ULK1 represents a potential diagnostic marker for clear cell renal carcinoma and the antitumor effects of SBI-0206965. EBioMedicine. 2018;34:85–93. https://doi.org/10.1016/j.ebiom.2018.07.034.
Murai J, Tang SW, Leo E, Baechler SA, Redon CE, Zhang H, et al. SLFN11 blocks stressed replication forks independently of ATR. Mol Cell. 2018. https://doi.org/10.1016/j.molcel.2018.01.012.
Deng Y, Cai Y, Huang Y, Yang Z, Bai Y, Liu Y, et al. High SLFN11 expression predicts better survival for patients with KRAS exon 2 wild type colorectal cancer after treated with adjuvant oxaliplatin-based treatment. BMC Cancer. 2015;15:833. https://doi.org/10.1186/s12885-015-1840-6.
Kagami T, Yamade M, Suzuki T, Uotani T, Tani S, Hamaya Y, et al. The first evidence for SLFN11 expression as an independent prognostic factor for patients with esophageal cancer after chemoradiotherapy. BMC Cancer. 2020;20:1123. https://doi.org/10.1186/s12885-020-07574-x.
He T, Zhang M, Zheng R, Zheng S, Linghu E, Herman JG, et al. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics. 2017;9:849–62. https://doi.org/10.2217/epi-2017-0019.
Iwasaki J, Komori T, Nakagawa F, Nagase H, Uchida J, Matsuo K, et al. Schlafen11 expression is associated with the antitumor activity of trabectedin in human sarcoma cell lines. Anticancer Res. 2019;39:3553–63. https://doi.org/10.21873/anticanres.13501.
Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, et al. PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res. 2017;23:523–35. https://doi.org/10.1158/1078-0432.CCR-16-1040.
Conteduca V, Ku SY, Puca L, Slade M, Fernandez L, Hess J, et al. SLFN11 expression in advanced prostate cancer and response to platinum-based chemotherapy. Mol Cancer Ther. 2020;19:1157–64. https://doi.org/10.1158/1535-7163.MCT-19-0926.
Liu Y, Zhang Z, Fu S, Wang S, Cheng X, Lei K, et al. Study of clinical predictive value and immune characterization of SLFN11 in clear cell renal cell carcinoma. Int J Gen Med. 2021;14:6741–54. https://doi.org/10.2147/IJGM.S336823.
Na JC, Nagaya N, Rha KH, Han WK, Kim IY. DNA damage response pathway alteration in locally advanced clear-cell renal-cell carcinoma is associated with a poor outcome. Clin Genitourin Cancer. 2019;17:299–305. https://doi.org/10.1016/j.clgc.2019.05.004.
Tang SW, Thomas A, Murai J, Trepel JB, Bates SE, Rajapakse VN, et al. Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin Cancer Res. 2018;24:1944–53. https://doi.org/10.1158/1078-0432.CCR-17-0443.
Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, et al. Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A. 2012;109:15030–5. https://doi.org/10.1073/pnas.1205943109.
Yu B, Zhang J, Sun Z, Cao P, Zheng X, Gao Z, et al. Interferon-inducible protein 16 may be a biomarker and prognostic factor in renal cell carcinoma by bioinformatics analysis. Medicine (Baltimore). 2021;100: e24257. https://doi.org/10.1097/MD.0000000000024257.
Mezzadra R, de Bruijn M, Jae LT, Gomez-Eerland R, Duursma A, Scheeren FA, et al. SLFN11 can sensitize tumor cells towards IFN-γ-mediated T cell killing. PLoS ONE. 2019;14: e0212053. https://doi.org/10.1371/journal.pone.0212053.
Fang Y, Wei J, Cao J, Zhao H, Liao B, Qiu S, et al. Protein expression of ZEB2 in renal cell carcinoma and its prognostic significance in patient survival. PLoS ONE. 2013;8: e62558. https://doi.org/10.1371/journal.pone.0062558.
He YH, Chen C, Shi Z. The biological roles and clinical implications of microRNAs in clear cell renal cell carcinoma. J Cell Physiol. 2018;233:4458–65. https://doi.org/10.1002/jcp.26347.
Chen J, Zhong Y, Li L. miR-124 and miR-203 synergistically inactivate EMT pathway via coregulation of ZEB2 in clear cell renal cell carcinoma (ccRCC). J Transl Med. 2020;18:69. https://doi.org/10.1186/s12967-020-02242-x.
Jingushi K, Kashiwagi Y, Ueda Y, Kitae K, Hase H, Nakata W, et al. High miR-122 expression promotes malignant phenotypes in ccRCC by targeting occludin. Int J Oncol. 2017;51:289–97. https://doi.org/10.3892/ijo.2017.4016.
Zhou J, Zhang C, Zhou B, Jiang D. miR-183 modulated cell proliferation and apoptosis in ovarian cancer through the TGF-β/Smad4 signaling pathway. Int J Mol Med. 2019;43:1734–46. https://doi.org/10.3892/ijmm.2019.4082.
Bian W, Zhang H, Tang M, Zhang S, Wang L, Liu L, et al. Potential role of microRNA-183 as a tumor suppressor in hepatocellular carcinoma. Cell Physiol Biochem. 2018;51:2065–72. https://doi.org/10.1159/000495825.
Yuan Y, Zhang Y, Han L, Sun S, Shu Y. miR-183 inhibits autophagy and apoptosis in gastric cancer cells by targeting ultraviolet radiation resistance-associated gene. Int J Mol Med. 2018;42:3562–70. https://doi.org/10.3892/ijmm.2018.3871.
Acknowledgements
The authors thank Mr. Li Lei for their help with the bioinformatics analysis.
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
HW and ZZ contributed to the study concept and design. HW, XD, ZZ, and SP led the data collection and interpretation. HW, ZZ and HX performed laboratory and statistical analyses. HW and ZZ wrote the first draft of the manuscript. All authors revised the draft critically and gave final approval for the manuscript to be published.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest regarding this study.
Additional information
Publisher’s Note
Springer nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, HX., Zhao, ZP., Du, XY. et al. SLFN11 promotes clear cell renal cell carcinoma progression via the PI3K/AKT signaling pathway. Med Oncol 41, 54 (2024). https://doi.org/10.1007/s12032-023-02262-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-023-02262-9