Abstract
Multiple sclerosis (MS) as a chronic autoimmune disease of the central nervous system (CNS) has been associated with dysregulation of several genes including miRNAs. In the present study, we assessed transcript levels of seven miRNAs (miR-96-5p, miR-211-5p, miR-15a, miR-34a-5p, miR-204-5p, miR-501-5p, and miR-524-5p) in the peripheral blood of MS patients compared with healthy subjects in association with response to fingolimod treatment. Expression levels of miR-211-5p and miR-34a-5p were significantly decreased in MS patients compared with healthy subjects (P values of 0.002 and 0.47). While subgroup analysis showed downregulation of miR-211-5p in both fingolimod responders and non-responders, miR-34a-5p expression was only decreased in responders. Moreover, miR-204-5p was downregulated in non-responder male patients compared with male controls. The current study underscores the role of miRNAs in determination of response to fingolimod in MS patients.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Multiple sclerosis (MS) is a chronic autoimmune disease of the central nervous system (CNS) (Dastmalchi et al. 2018). Several studies have assessed genomic variants and transcript levels of multiple genes to explore their significance in conferring risk of this disorder or their participation in the response of patients to disease modifying therapies (DMTs) (Sayad et al. 2017; Rahimi et al. 2018). MicroRNAs (miRNAs) as principle regulators of immune response have been regarded as blood-based biomarkers for MS disease diagnosis and monitoring (Ebrahimkhani et al. 2017). A previous study assessed miRNA expression in the peripheral blood of patients with relapsing-remitting MS (RRMS), and healthy individuals has detected 165 differentially expressed miRNAs between patients and healthy subjects. An individual miRNA marker (hsa-miR-145) could differentiate disease status with a specificity of 89.5%, a sensitivity of 90.0%, and an accuracy of 89.7%. Assessment of transcript levels of a panel of 48 miRNAs significantly increased the diagnostic power to 96.3% (Keller et al. 2009). Other microarray studies have demonstrated downregulation of T cell suppressor miR-17 and miR-20a in whole blood of MS patients compared with healthy controls (Cox et al. 2010). Notably, assessment of expression profile of 364 miRNAs in the peripheral blood mononuclear cells (PBMC) of MS patients and healthy controls has shown the role of hsa-miR-18b and hsa-miR-599 in relapse phase and participation of hsa-miR-96 in remission phase (Otaegui et al. 2009). Moreover, a bioinformatics approach has identified miR-199a and miR-142-3p as critical genes in the pathogenesis of MS which modulate fundamental susceptibility genes, especially KRAS and IL7R (Luo and Fu 2018). Another study has demonstrated an association between upregulation of IFN-β-responsive genes and downregulation of numerous miRNAs (Hecker et al. 2013).
Expression of miR-15b, miR-23a, and miR-223 has been shown to be decreased in MS patients compared with healthy subjects. Notably, expression of these miRNAs significantly increased after 6 months of treatment with fingolimod (Fenoglio et al. 2016). Taken together, miRNAs have fundamental roles in conferring MS risk, determination of disease course, and patients’ response to DMTs. In the present study, we assessed transcript levels of seven miRNAs (miR-96-5p, miR-211-5p, miR-15a, miR-34a-5p, miR-204-5p, miR-501-5p, and miR-524-5p) in the peripheral blood of MS patients compared with healthy subjects in association with response to fingolimod treatment. The aim of this study was to find a miRNA signature which could predict presence of MS or response of patients to fingolimod.
Material and Methods
Study Participants
The current case–control study was performed on blood samples obtained from 78 RRMS patients from Farshchian Hospital, Hamadan, Iran. All patients have received fingolimod 0.5 mg orally once-daily. Diagnosis of patients was implemented according to Poser’s criteria (Fangerau et al. 2004). Following a 6-month treatment period, patients were classified to fingolimod responders (n = 49) and non-responders (n = 29). Responders had no increase in the Expanded Disability Status Scale (EDSS) score and no relapse during 2 years of follow-up. Patients who had at least one relapse during the follow-up period and one point increase in the EDSS score (that lasted for 6 months) were regarded as non-responders (Rio et al. 2006). A group of age-matched healthy subjects with no history of autoimmune disorder, chronic disease, or malignancy was recruited as control group (n = 79). The study protocol was approved by the local ethics committee of Hamadan University of Medical Sciences (IR.UMSHA.REC.1395.506). Written informed consent forms were obtained from all study participants. We confirmed that all methods were performed in accordance with the relevant guidelines and regulations.
Expression Analysis
RNA was extracted from blood samples using Hybrid-RTM Blood RNA Extraction Kit (Geneall Biotechnology Co Ltd., South Korea). TaqMan® MicroRNA Reverse Transcription Kit (Invitrogen) and TaqMan® MicroRNA Assays Kit (Applied Biosystems) were used for cDNA synthesis and calculation of transcript levels of miRNAs respectively. RNU6B gene was used as normalizer. All experiments were performed in rotor gene 6000 Corbett Real-Time PCR System in duplicate.
Statistical Analysis
Statistical analyses were performed in R3.4.2 software using “Quantreg” and “Brms” packages. Relative expression of miRNAs in MS patients were compared with controls using Ln (Efficiency−∆CT) values. The significance of difference in mean values of miRNA expressions between cases and controls was evaluated using Bayesian multilevel model using probabilistic programming language Stan. Two-sided 95% credible intervals (95% CrI) were used to make decisions about results. Spearman rank correlation coefficient was calculated to appraise correlations between relative expressions of genes. The receiver operating characteristic (ROC) curve was schemed to measure the diagnostic power of miRNA transcript levels for categorizing disease status or patients (responders vs. non-responders).
In Silico Identification of miRNA Targets
Target genes of each considered miRNA were predicted using mirDIP v4.1 target gene prediction tool. Genes with integrated scores upper than 0.7 were selected for protein-protein interaction (PPI) network analysis. By the use of the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/cgi/), interactions among the Target genes were identified. Interactions with confidence scores greater than 0.7 were retrieved, and the resulting PPI network for modules with Cytoscape-MCODE was screened. The following parameters were applied: degree cutoff = 2, node score cutoff = 0.2, k-score = 0.2, and maximum depth = 100. The DAVID online tool was applied to analyze the functional characteristics of target genes of each captured module through Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. The cutoff criterion P < 0.05 of the false discovery rate was used.
Results
General Demographic Data of Study Participants
Table 1 shows the summary of demographic and clinical data of study participants.
Relative Expression of miRNAs in MS Patients Compared with Healthy Subjects
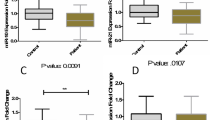
Bayesian multilevel model has shown significant downregulation of miR-211-5p and miR-34a-5p in MS patients compared with healthy subjects after adjusting the effects of age and sex (Table 2). However, expressions of other miRNAs were not significantly different between MS patients and healthy controls. For miR-204-5p, although Bayesian multilevel model showed no significant difference in gene expression after controlling the effects of age and sex, quantile regression analysis demonstrated significant difference in gene expression between male patients and male controls (P = 0.008) which means that the interaction between disease and sex is significant (P = 0.034).
Subsequently, we compared expression of miRNAs between healthy subjects and subgroups of MS patients (responders and non-responders) (Tables 3, 4, and 5). Subgroup analysis showed downregulation of miR-211-5p in both fingolimod responders and non-responders. However, miR-34a-5p expression was only decreased in responders. Moreover, miR-204-5p was downregulated in non-responder male patients compared with male controls. Finally, miR-211-5p was downregulated in responder female patients compared with female healthy subjects.
Expressions of miR-211-5p and miR-501-5p were inversely correlated with age in female participants. Expressions of other miRNAs were not correlated with age in any subgroup (Table 6).
Significant pairwise correlations were found between expressions of miRNAs in certain subgroups of patients (Table 7).
ROC Curves
We plotted the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings. The diagnostic power values of miR-211-5p and miR-34-5p in differentiation of MS patients from controls as measured by AUC were 0.68 and 0.62 respectively (Table 8). However, none of miRNAs could be regarded as appropriate biomarker for differentiation of responders from non-responders.
miRNA Targets as Identified by In Silico Methods
The list of miRNA targets are shown in Table 9. Figure 1 shows the PPI depicted for miR-211-5p and miR-34a-5p as the two miRNAs with differential expressions between study groups.
Discussion
In the current study, we assessed expression of seven miRNAs in peripheral the blood of MS patients and found differential expression of certain miRNAs in association with response to fingolimod. We demonstrated significant downregulation of miR-211-5p and miR-34a-5p in MS patients compared with healthy subjects.
Recently, investigations have revealed a novel layer of regulation in miRNA–mRNA interactions namely “target RNA-directed MicroRNA degradation” which results in miRNA 3′-end tailing by the adding of A/U nucleotides and very specific microRNA degradation (Fuchs Wightman et al. 2018). Consequently, downregulation of expression of mentioned miRNAs in MS patients might be due to upregulation of their targets. Future studies are needed to elaborate the mechanism.
Based on our in silico analysis, miR-211-5p in involved in neurotrophin signaling pathway. Such role might contribute in the pathogenesis of MS. miR-211 has been also among the previously reported downregulated miRNAs in the peripheral blood of patients with all MS subtypes including primary progressive, secondary progressive, and relapsing-remitting (RR) MS (Cox et al. 2010). However, miR-211 has been shown to target the nicotinic acetylcholine receptor the nicotinic acetylcholine receptor α7 (nAChRa7). This receptor has an anti-inflammatory effect (Bekenstein et al. 2017). Cholinergic system has an appreciated role in modulation of immune response through nAChRa7. Moreover, the vagus nerve delivers signals to the brain concerning inflammatory responses happening in the peripheral tissues (Rosas-Ballina and Tracey 2009). While miR-34a expression has been decreased in CD4+ T cells from PBMCs of RRMS patients (Lindberg et al. 2010), its expression has been increased in active brain lesions of MS patients (Junker et al. 2009). Its elevated expression in the brain tissue has been associated with downregulation of CD47 in local cells, liberating macrophages from inhibitory control leading to phagocytosis of myelin (Junker et al. 2009). We also detected significant difference in miR-204-5p expression between male patients and male controls. This miRNA has been previously shown to be downregulated in B lymphocytes from the peripheral blood samples of RRMS patients (Sievers et al. 2012). Consequently, the results of the current study regarding dysregulation of three miRNAs in MS patients are consistent with the previous studies.
We also detected associations between response to fingolimod and expression of miR-34a-5p and miR-204-5p in a way that responders had lower levels of miR-34a-5p and higher levels of miR-204-5p compared with non-responders. However, the latter reached significance level only in male subjects. The differential expression pattern of miR-34a-5p in responders vs. non-responders is consistent with the recently reported higher levels of this miRNA in relapsing phase of MS compared with controls and patients in remission phase (Ghadiri et al. 2018). In line with our data, higher miR-34a levels have been associated with lower function of regulatory T cells (Ghadiri et al. 2018). The role of miR-34 has been previously acknowledged in the process of aging where mir-34 deficiency induced expression of genes which were associated with enhanced brain aging, neuron degeneration, and a remarkable decrease in survival (Fuchs Wightman et al. 2018). Future studies are needed to explore whether this miRNA expression pattern is a result of fingolimod administration in responsive patients or itself determines the response of patients to this drug. Our study is among the first studies evaluating expression of mature miRNAs in fingolimod-treated patients. Fingolimod has been shown to change expression of several genes including miRNA precursors in peripheral CD4+ cells (Friess et al. 2017). Moreover, administration of first dose of fingolimod has resulted in elevation of extracellular vesicle concentration and a considerable alteration in their miRNA load (Saenz-Cuesta et al. 2018). Besides, others have demonstrated amelioration of dysregulated miRNA profile of MS patients following treatment with fingolimod (Fenoglio et al. 2016).
Assessment of correlation between miRNA levels and age revealed inverse correlation between expressions of two miRNAs (miR-211-5p and miR-501-5p) and age in female participants. Several age-dependent alterations have been demonstrated in MS disease course including decrease in regenerative potential of the brain and evolution of a progressive phase (Cerqueira et al. 2018). Future studies are needed to elaborate the associations between these miRNAs and age-related phenomena in MS. Notably, miRNAs that affect immune function have been among those with age-dependent expression profile (Huan et al. 2018).
We also demonstrated significant correlations between expressions of miRNAs in most subgroup analyses. Totally, except for some cases, correlations tend to be stronger in controls compared with MS subjects which might imply distortion of miRNA regulation in MS patients. The most prominent correlation was found between miR-34a-5p and miR-524-5p in healthy subjects. Both miRNAs affect expression of the Notch ligand Jagged-1 (Pang et al. 2010; Chen et al. 2012). Jagged-1 signaling has an established role in the expansion of regulatory T cells and mesenchymal stromal cells defense against inflammation (Cahill et al. 2015). Future studies are needed to clarify if weakening of correlation between expressions of these miRNAs in MS patients compared with healthy subjects is involved in the pathogenesis of MS.
Finally, we assessed the diagnostic power of miRNAs in differentiation of MS patients from controls or responders from non-responders. The diagnostic power values of miR-211-5p and miR-34-5p in differentiation of MS patients from controls as measured by AUC were 0.68 and 0.62 respectively. However, in spite of different expression patterns of certain miRNAs between responders and non-responders, none of miRNAs could be regarded as appropriate biomarker for differentiation of responders from non-responders which can be attributed to unequal trends in their sensitivity and specificity values, i.e., miR-204-5p was highly sensitive but not specific while miR-211-5p and miR-34-5p had the opposite situation.
In conclusion, we demonstrated dysregulation of certain miRNAs in MS patients compared with healthy subjects and different patterns of miRNA expression between responder and non-responders to fingolimod. Our study has a limitation regarding lack of assessment of expression patterns of miRNA targets in the peripheral blood of study participants.
References
Bekenstein U, Mishra N, Milikovsky DZ, Hanin G, Zelig D, Sheintuch L, Berson A, Greenberg DS, Friedman A, Soreq H (2017) Dynamic changes in murine forebrain miR-211 expression associate with cholinergic imbalances and epileptiform activity. Proc Natl Acad Sci U S A 114:E4996–E5005
Cahill EF, Tobin LM, Carty F, Mahon BP, English K (2015) Jagged-1 is required for the expansion of CD4+ CD25+ FoxP3+ regulatory T cells and tolerogenic dendritic cells by murine mesenchymal stromal cells. Stem Cell Res Ther 6:19
Cerqueira JJ, Compston DAS, Geraldes R, Rosa MM, Schmierer K, Thompson A, Tinelli M, Palace J (2018) Time matters in multiple sclerosis: can early treatment and long-term follow-up ensure everyone benefits from the latest advances in multiple sclerosis? J Neurol Neurosurg Psychiatr 89(8):844–850
Chen L, Zhang W, Yan W, Han L, Zhang K, Shi Z, Zhang J, Wang Y, Li Y, Yu S (2012) The putative tumor suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in glioma. Carcinogenesis 33:2276–2282
Cox MB, Cairns MJ, Gandhi KS, Carroll AP, Moscovis S, Stewart GJ, Broadley S, Scott RJ, Booth DR, Lechner-Scott J (2010) MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One 5:e12132
Dastmalchi R, Ghafouri-Fard S, Omrani MD, Mazdeh M, Sayad A, Taheri M (2018) Dysregulation of long non-coding RNA profile in peripheral blood of multiple sclerosis patients. Mult Scler Relat Disord 25:219–226
Ebrahimkhani S, Vafaee F, Young PE, Hur SSJ, Hawke S, Devenney E, Beadnall H, Barnett MH, Suter CM, Buckland ME (2017) Exosomal microRNA signatures in multiple sclerosis reflect disease status. Sci Rep 7:14293
Fangerau T, Schimrigk S, Haupts M, Kaeder M, Ahle G, Brune N, Klinkenberg K, Kotterba S, Mohring M, Sindern E (2004) Diagnosis of multiple sclerosis: comparison of the Poser criteria and the new McDonald criteria. Acta Neurol Scand 109:385–389
Fenoglio C, de Riz M, Pietroboni AM, Calvi A, Serpente M, Cioffi SM, Arcaro M, Oldoni E, Scarpini E, Galimberti D (2016) Effect of fingolimod treatment on circulating miR-15b, miR23a and miR-223 levels in patients with multiple sclerosis. J Neuroimmunol 299:81–83
Friess J, Hecker M, Roch L, Koczan D, Fitzner B, Angerer IC, Schroder I, Flechtner K, Thiesen HJ, Winkelmann A, Zettl UK (2017) Fingolimod alters the transcriptome profile of circulating CD4+ cells in multiple sclerosis. Sci Rep 7
Fuchs Wightman F, Giono LE, Fededa JP, De La Mata M (2018) Target RNAs strike back on microRNAs. Front Genet 9:435
Ghadiri N, Emamnia N, Ganjalikhani-Hakemi M, Ghaedi K, Etemadifar M, Salehi M, Shirzad H, Nasr-Esfahani MH (2018) Analysis of the expression of mir-34a, mir-199a, mir-30c and mir-19a in peripheral blood CD4+ T lymphocytes of relapsing-remitting multiple sclerosis patients. Gene 659:109–117
Hecker M, Thamilarasan M, Koczan D, Schroder I, Flechtner K, Freiesleben S, Fullen G, Thiesen HJ, Zettl UK (2013) MicroRNA expression changes during interferon-beta treatment in the peripheral blood of multiple sclerosis patients. Int J Mol Sci 14:16087–16110
Huan T, Chen G, Liu C, Bhattacharya A, Rong J, Chen BH, Seshadri S, Tanriverdi K, Freedman JE, Larson MG (2018) Age-associated micro RNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell 17:e12687
Junker A, Krumbholz M, Eisele S, Mohan H, Augstein F, Bittner R, Lassmann H, Wekerle H, Hohlfeld R, Meinl E (2009) MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain 132:3342–3352
Keller A, Leidinger P, Lange J, Borries A, Schroers H, Scheffler M, Lenhof HP, Ruprecht K, Meese E (2009) Multiple sclerosis: microRNA expression profiles accurately differentiate patients with relapsing-remitting disease from healthy controls. PLoS One 4:e7440
Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L (2010) Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol 40:888–898
Luo DL, Fu J (2018) Identifying characteristic miRNAs—genes and risk pathways of multiple sclerosis based on bioinformatics analysis. Oncotarget 9:5287–5300
Otaegui D, Baranzini SE, Armananzas R, Calvo B, Munoz-Culla M, Khankhanian P, Inza I, Lozano JA, Castillo-Trivino T, Asensio A, Olaskoaga J, Lopez De Munain A (2009) Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS One 4:e6309
Pang RT, Leung CO, Ye T-M, Liu W, Chiu PC, Lam KK, Lee K-F, Yeung WS (2010) MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis 31:1037–1044
Rahimi M, Aghabozorg Afjeh SS, Omrani MD, Arsang-Jang S, Ganji M, Noroozi R, Taheri M, Ghafouri-Fard S (2018) Soluble receptor for advanced glycation end products (sRAGE) is up-regulated in multiple sclerosis patients treated with interferon beta-1a. Cell Physiol Biochem 46:561–567
Rio J, Nos C, Tintore M, Tellez N, Galan I, Pelayo R, Comabella M, Montalban X (2006) Defining the response to interferon-beta in relapsing-remitting multiple sclerosis patients. Ann Neurol 59:344–352
Rosas-Ballina M, Tracey KJ (2009) Cholinergic control of inflammation. J Intern Med 265:663–679
Saenz-Cuesta M, Alberro A, Munoz-Culla M, Osorio-Querejeta I, Fernandez-Mercado M, Lopetegui I, Tainta M, Prada A, Castillo-Trivino T, Falcon-Perez JM, Olascoaga J, Otaegui D (2018) The first dose of fingolimod affects circulating extracellular vesicles in multiple sclerosis patients. Int J Mol Sci 19
Sayad A, Ghafouri-Fard S, Omrani MD, Noroozi R, Taheri M (2017) Myxovirus resistance protein A (MxA) polymorphism is associated with IFNbeta response in Iranian multiple sclerosis patients. Neurol Sci 38:1093–1099
Sievers C, Meira M, Hoffmann F, Fontoura P, Kappos L, Lindberg RL (2012) Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin Immunol 144:70–79
Funding
The current study was supported by grant number 9512177928 from Hamadan University of Medical Sciences.
Author information
Authors and Affiliations
Contributions
SGF wrote the manuscript. MT supervised the study. SAJ analyzed the data. MM was the clinical consultant. AK and RS conducted the bioinformatics analyses. MME performed the laboratory assessment.
Corresponding authors
Ethics declarations
The study protocol was approved by the local ethics committee of Hamadan University of Medical Sciences (IR.UMSHA.REC.1395.506). Written informed consent forms were obtained from all study participants. We confirmed that all methods were performed in accordance with the relevant guidelines and regulations.
Competing Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Eftekharian, M.M., Komaki, A., Mazdeh, M. et al. Expression Profile of Selected MicroRNAs in the Peripheral Blood of Multiple Sclerosis Patients: a Multivariate Statistical Analysis with ROC Curve to Find New Biomarkers for Fingolimod. J Mol Neurosci 68, 153–161 (2019). https://doi.org/10.1007/s12031-019-01294-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12031-019-01294-z