Abstract
Background
K-RAS and recently N-RAS gene mutation testing are mandatory requirements prior to anti-epidermal growth factor receptor (EGFR) monoclonal antibody treatment of metastatic CRC. Mutation prevalence and distribution in Indonesian colorectal cancer (CRC) are not known.
Methods
Combined methods of PCR high-resolution melt (HRM), restriction fragment length polymorphism (RFLP), and direct DNA sequencing were used to genotype exons 2, 3, and 4 of both K-RAS and N-RAS genes for routine clinical testing of CRC patients. Descriptive analytical review of 595 consecutive CRC patients (years 2013 to 2016) was performed to find associations between gene mutations and clinicopathologic features.
Results
This retrospective study revealed overall K-RAS gene mutation in exon 2 (codon 12 and 13) rates being 34.9%. Women (42.5%), stages I and II (43.4%), and well and moderate differentiations (37.7%) had higher frequency of K-RAS exon 2 mutations than men (29%, p = 0.006), stages (III and IV 31.9%, p = 0.05), and poor differentiation (11.8%, p = 0.002), respectively. At later period (2015–2016), 121 of 595 patients were genotyped for the remaining exons 3 and 4 of K-RAS as well as exons 2, 3, and 4 of N-RAS mutations resulting in overall RAS mutation prevalence of 41%. Mucinous histology had highest frequency of N-RAS mutation.
Conclusions
Combination of PCR HRM with either RFLP or direct DNA sequencing was useful to detect K-RAS exon 2 and extended RAS mutations, respectively. Frequency of all RAS mutations in stage IV Indonesian (41%) was similar among Asians (41–49%), which tend to be lower than western (55%) CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is one of the most common and one of the leading causes of cancer-related death, especially in developing countries. Mean incidence rate in southeast Asian countries has been estimated to be 6.5 in 100,000 that is lower than developed nation such as UK [1]. The rising CRC incidence rates in Asia, especially among the young age group, have also raised concern [2].
CRC is among the most frequent malignancy in Indonesia with estimated age standardized rates per 100,000 of 19 in men and 15 in women [3]. Hospital-based data in Indonesia suggests that 35.2% of CRC patients were under 40 years old [4]. The underlying molecular mechanisms in these young patients are not entirely clear. Expression patterns of mismatch repair proteins (MLH1 and MSH2) were similar in native young and old Indonesian CRC patients [4].
Introduction of anti epidermal growth factor receptor (EGFR)-targeted therapy (e.g., cetuximab and panitumumab) has improved survival of metastatic CRC in patients with normal or wild-type alleles of both K-RAS and N-RAS genes [5]. K-RAS and N-RAS mutation testing has been endorsed by Asian clinical guidelines [6]. Beginning in 2011, Indonesian authority has mandated national health insurance agency to reimburse the cost of anti-EGFR therapy in metastatic CRC patients who have wild-type K-RAS gene.
Polymerase chain reaction-high resolution melt (PCR-HRM) method has been used to genotype K-RAS and N-RAS genes [7–9]. In this study, we used similar approaches and demonstrated utility HRM melt curve analyses in combination with restriction fragment length of polymorphism (RFLP) to genotype common mutations of K-RAS exon 2 (codons 12 and 13). In addition, PCR HRM melt curve and direct DNA sequencing had been used to genotype exons 3 and 4 of K-RAS and exons 2, 3, and 4 of N-RAS genes as routine methods of molecular testing serving patients living in major Indonesian cities. Our study also demonstrated association of K-RAS and N-RAS gene mutations with several clinicopathologic features of patients. These associations may fill in gaps of baseline information and understanding of underlying molecular pathways of Indonesian CRC.
Material and Methods
Study Population and Cell Lines
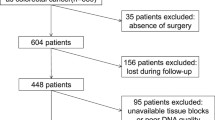
Records of test request forms and genotyping results were retrieved and reviewed of all colorectal cancer patients (January 2013 to March 2016) referred to Kalbe Genomics Laboratory for K-RAS mutations in codons 12 and 13. This central laboratory is accredited for ISO15189 and registered in the European Society of Pathology K-RAS external quality control scheme (http://K-RAS.eqascheme.org). Patients who had failed test results were excluded. Average failure rate was 10% mostly due to insufficient tumor contents and poor DNA quality causing PCR failure. Average turn around time was seven workdays.
Successful K-RAS mutations in codons 12 and 13 test results of 395 patients (during the years 2013–2016) were included in this study as summarized in Table 1. In the later period of March 2015 to 2016, there were 121 of 395 colorectal cancer patients who had been tested for extended RAS encompassing both K-RAS and N-RAS gene mutations in all exons 2, 3, and 4 paying particular attentions to codons 12, 13, 61, 117, and 146. This study was determined to be exempt by the Institutional Review Board and performed in accordance to 1964 Helsinki Declaration and its later amendments. This retrospective study was approved by Institutional Review Board (IRB) of Stem-cell and Cancer Institute (SCI). Because of the retrospective nature of this study, informed consent was waived. Patient identities had been anonymized. Cell lines were obtained as gifts from Genomic Institute of Singapore (GIS) BT549 (K-RAS gene wild type), A549 (K-RAS gene G12S homozygous), and HCT116 (K-RAS gene G13D heterozygous).
Molecular Profiling
Tumor section from FFPE tissue samples were stained with hematoxylin-eosin (HE) and reviewed by two experienced pathologists. Clinicopathological information was obtained by reviewing the request forms to extract the following clinicopathology information such as age, gender, tumor sites, histological types, differentiation grades, and AJCC TNM stages. Right-sided tumors were defined as specimens originated in cecum, ascending colon to transverse colon, and left-sided tumors in splenic flexure, descending colon to rectosigmoid junction.
DNA Extraction
To obtain maximal tumor DNA, senior pathologists chose and marked areas of tumor components that were greater than 30%. DNA was then extracted from these tumor-enriched areas using the Qiagen QIAamp DNA Micro Kit (cat no. 56304, Qiagen, Germany) following the manufacturer’s protocol.
High-Resolution Melting-Polymerase Chain Reaction
Polymerase chain reaction-high resolution melting (HRM) protocol was adopted from previous publication [10] with some modifications to screen for mutations in exon 2 (codons 12 and 13) of K-RAS gene. Commercial RAS SplitSCAN (KalgenDNA, Jakarta) HRM primers were used to screen hotspot mutation of exons 3 and 4 of K-RAS and exons 2, 3, and 4 of N-RAS genes. Samples were tested in duplicate using Rotor-Gene 6000™ (Corbett Life Science, Australia) or Rotor-gene Q (Qiagen, Germany). Melting curve was generated and scan for the presence of “split peaks” indicating mutations (Fig. 1). Controls containing DNA of K-RAS wild type (BT549) and K-RAS mutant (HCT116) were always included for melt curve comparisons. Samples showing split peak patterns similar to HCT116 were then genotyped using PCR-RFLP and/or direct DNA sequencing. Gel electrophoresis was performed on 2.5% (w/v) agarose to confirm the presence of single band of PCR amplicon.
Analytical sensitivity of high-resolution melt, direct DNA sequencing, and restriction fragment length polymorphism (RFLP) to detect mutations in KRAS exon 2 codon 12 and 13. HCT116 cell line harboring heterozygous KRAS mutant in codon 13 and BT549 cell line carrying normal KRAS gene was used as wild-type control. Serial dilutions were performed in order to obtain a mutant to wild-type allele ratio of 100, 50, 25, 12.5, 6.25, and 0%. a Melt curve graph. Single peak representing KRAS wild-type allele was shown in black line. Split peaks were shown in different colors representing different ratios of KRAS mutant to wild-type alleles. b Direct sequencing. Red arrows pointed to position of mutation KRAS allele due to single nucleotide substitution. c RFLP analyses. Digestion of PCR products by restriction enzyme led to either presence or absence of 156 and 128 base pairs (bp) representing mutated and wild-type KRAS alleles
PCR-RFLP Analysis
Polymerase chain reaction-restriction fragment length polymorphism (RFLP) to detect mutations in codons 12 and 13 of K-RAS gene was conducted as described [11].
Sanger DNA Sequencing
Sequencing primers were adopted from previous publication [12]. PC amplification products were purified using the HighPure PCR Product Purification Kit (Roche), according to the manufacturer’s protocol. Sequencing analysis was performed on an Applied Biosystem 3500 Genetic Analyzer.
Analytical Sensitivity Test
K-RAS gene mutant control DNA (extracted from both A549 and HCT116 cell lines) were mixed with wild-type DNA (extracted from BT549 cell line) and serially diluted as 50, 25, 12.5, and 6.25% mutant to wild-type DNA ratios. These mixtures were used for sensitivity detection of K-RAS gene mutant by PCR-HRM split peak, PCR-RFLP analysis, and Sanger sequencing.
Statistical Analysis
The data were processed using www.graphpad.com (GraphPad software, Inc., La Jolla, CA, USA). Individual information and baseline characteristics were summarized using descriptive statistics. The chi-square (χ2) test or, where appropriate, the Fisher’s exact test was used to compare the proportion of gene mutations among group with different clinicopathological parameters. Online calculator (http://vassarstats.net/kappa.html) was used to get kappa value and 95% CI as measure of diagnostic concordance between PCR HRM and RFLP and direct DNA sequencing methods.
Results
Analytical Sensitivity of PCR HRM
A simple melt curve of PCR amplicons of K-RAS gene showed different patterns for wild-type (single peak) and mutated allele (separated or split peak). As shown in Fig. 1a, black tracer line representing wild-type K-RAS from BT549 cell line was noticeably different from orange line (mutant codon 13 of K-RAS gene HCT116 cell line). Titration experiment (Fig. 1a) showed that split patterns of mutant alleles were detected as low as 6.25% HCT116. As expected, DNA sequencing ability to detect mutated K-RAS dropped off after 25% HCT116 (Fig. 1b) and completely failed to detect mutations at 6.25% mutant allele. Both PCR HRM and PCR RFLP (Fig. 1c) had shown better limit of detection than direct DNA sequencing.
To determine diagnostic sensitivity and specificity PCR HRM/RFLP, we used 147 archived specimens comprising of 92 K-RAS wild-type and 55 mutant cases that had been genotyped using direct DNA sequencing. PCR HRM/RFLP detected K-RAS mutations in 6 of 92 wild-type specimens and misgenotyped 1 of 55 mutated cases. Therefore, diagnostic sensitivity and specificity was 98.9% (95%CI 93.8–99.8) and 90% (95% CI 79.9–95.3) and concordance rate estimated by Kappa value being 0.9 (95% CI = 0.8281 to 0.9721).
K-RAS Exon 2 Mutation Association with Clinicopathology Parameters
Combined PCR HRM and RFLP were used during routine genotyping of consecutive 395 colorectal cancer patients (year 2013–2015) for presence of mutations in K-RAS codons 12 and 13. As shown in Table 2, overall mutation rate of K-RAS mutations was 34.9%. High rate of K-RAS mutation in codons 12 and 13 was found in females (42.5% vs males 29%, p = 0.006), well and moderate differentiation (37.7% vs poor 11.8%, p = 0.002), and early stages I and II (43.4% vs stage III 31.9% vs stage IV 30%, p = 0.05). Frequency of K-RAS mutation was also higher in right colon (39.6%) than left colorectal (32.6%) although the difference rate did not reach statistical significance.
N-RAS Mutation Association with Clinicopathology Parameters
Beginning 2015, extended RAS genotyping comprised of K-RAS exons 3 and 4 and N-RAS exons 2, 3, and 4 were performed on 121 out of 395 consecutive colorectal cancer patients using PCR HRM and direct DNA sequencing. Examples of PCR HRM split peak tracers were represented in Fig. 2. As shown in Table 3, extended RAS mutations (in exons 3 and 4 of K-RAS mutations and exons 2, 3, and 4 of N-RAS mutations) contributed additional 10.7 to 30.6% of K-RAS mutation in exon 2 (codons 12 and 13), yielding an overall mutation rate of both K-RAS and N-RAS being 41.3% in 121 colorectal cancer patients. N-RAS mutation may be more frequent in patients with mucinous component (21.4%) than non-mucinous patients (3%, p = 0.026). There were no other clinicopathology parameters that held significant associations with N-RAS mutations.
Presence of split peaks in melt curve graphs to detect mutations in exons 2, 3, and 4 of KRAS and NRAS genes. Representative melt curve graphs of individual exons of RAS gene were shown together with sequencing traces (upper left corner) and gel electrophoresis of PCR products (lower right corner). Arrows pointed to positions of mutated nucleotides in sequencing traces. In the melt curve graphs, split peaks (red lines) and single peaks (green lines) indicated mutant alleles and normal/wild-type alleles, respectively. Gel electrophoresis images of PCR amplicons showed specific single band of PCR products
Types of Mutations
One hundred twenty-one patients were genotyped using direct DNA sequencing for both K-RAS and N-RAS genes. Mutant-type K-RAS gene G12D (12.3%) was detected more frequently than G13D (4.9%) of K-RAS gene in exon 2. Mutation frequencies of K-RAS exons 3 (Q61H) and 4 (K117 N, A146T) were 0.8 and 5%, respectively. NRAS mutations in exons 2 (G12C, G12S), 3 (Q61R, Q61K), and 4 (A146T) each contributed 1.7%.
Discussions
We had validated PCR HRM coupled with RFLP and/or direct DNA sequencing to genotype K-RAS and N-RAS genes in consecutive Indonesian CRC patients. As previously observed, derivative melt graph of PCR HRM was the simpler method to scan and score mutation probability than were difference or normalized graphs [9]. We had observed and confirmed that specimens having mutant alleles showed split peaks when high-resolution melt data were analyzed using derivative melt graph. Split peaks were due to mixture of two populations (wild-type and mutant alleles) of PCR products having identical lengths (shown as single band in gel electrophoresis) but harboring single nucleotide variant due to substitutions or point mutations [9]. We also showed that PCR HRM analytical sensitivity to detect low-abundance mutant alleles (<10%) was better than direct DNA sequencing requiring 10–30% mutant alleles as summarized in recent meta analyses [13]. Moreover, HRM is also an attractive method for routine clinical testing because of high sensitivity and specificity, as well as rapid turn-around time by reducing burden of direct DNA sequencing [13]. In our experience, we saved up to 60% of incoming routine samples from genotyping K-RAS or N-RAS. Using these combination approaches (HRM, RFLP, and direct DNA sequencing), we were also able to gain several insights about associations of K-RAS and N-RAS with clinicopathology features of Indonesian colorectal cancer patients.
High prevalence of K-RAS mutation in women CRC patients of our population was also seen in Brazil in certain age group (40–60 years old) [14] and in Japan [15]. Tumors with well and moderate differentiation also harbored more frequent K-RAS mutations than tumors with poor differentiation as observed in past studies [8, 16], but not others [17, 18].
Using the HRM method, several laboratories in Europe and Japan report overall K-RAS mutation (mostly exon 2 and codons 12 and 13) between 41 and 45% [7–9, 19]. However, an earlier Japanese study using direct DNA sequencing involving more than 5000 patients shows overall prevalence K-RAS exon 2 mutations being 37% [15]. While genotyping analytical sensitivities do contribute to variation in KRAS prevalence, recent study using mass spectrometry-based genotyping method demonstrates consistent trend of low frequency of KRAS mutation (Asian, 39%; Black, 53%; White, 43%) [20]. K-RAS mutation frequency (36%) in our cohort seemed to be similar within Asian populations in spite of using techniques (HRM and RFLP) that were more sensitive than DNA sequencing.
Alternatively, variation K-RAS mutation rate in Asian vs western cohort may be explained partly by incidence of proximal and distal tumors. K-RAS mutations tend to occur in the right or proximal colon instead of the left or distal colon [21]. As observed previously [22], the left or distal colorectal is more common in Asian than in western population.
The low prevalence of K-RAS mutation in stage IV disease (30–33%) was even more apparent in this study and other Southeast Asian patients [18, 23] than in western patients (40–44%) [24]. It is not clear whether “westernized” dietary habits, environmental exposures, or inherent genetic susceptibility influence spectrum of gene mutation. A study in Japan and Singapore comparing K-RAS mutations among patients diagnosed in 1960s (0–25%) vs 1990s (28–36%) also revealed increasing trend of K-RAS mutations [25, 26] in Asia which mirrors rising incidence of CRC itself. Inherent difference of colorectal cancer genetic profile may also be reflected by relatively low frequency of BRAF mutation, a downstream effector of RAS signaling pathways in different ethnic groups [20].
In our cohort, stage III CRC had higher frequency of K-RAS mutations than advance stage IV. In our routine testing populations, stage III patients made up to 34%. Although K-RAS and N-RAS testing are primarily mandated for stage IV patients, there is consideration to test at earlier stage during routine clinical practice to anticipate progress to distant disease [27].
Five percent frequency of N-RAS mutation in our CRC cohort is also observed by many Asian studies with 2–5% range [8, 16, 28]. Combining K-RAS and N-RAS (extended or All RAS) mutation frequency in this study (41%) has similar rates with other Asian sites such as Korea (41%) [28], Japan (45%) [8], and China (49%) [16].
Sites of primary tumor have been shown as important prognostic factor of colorectal cancer [29]. Moreover, maximum benefits of targeted therapies (anti-EGFR or anti-angiogenesis) may also depend on primary tumor sites. K-RAS wild-type patients whose primary tumor sites were in the left sides had better survival than the ones in the right sides as shown in the Asian population [30]. While the frequency of K-RAS in our cohort seemed to be higher in the right side of the colon than in the left side of colorectal in our study, a large study in Japan suggests frequent K-RAS mutation in primary tumors located in the right side vs left side [15].
Conclusion
Melting graph analysis is a simple, rapid, cost-effective, and highly sensitive method for K-RAS and N-RAS mutation screening. To accompany HRM mutation screening, the RFLP method was useful to confirm mutation, but direct sequencing was still required for deeper and specific analysis. Overall rates of K-RAS and N-RAS mutations in Indonesian colorectal cancer patients were similar to other Asian populations, which tend to be lower than those in patients of European or western descents.
References
Kokki I, Papana A, Campbell H, Theodoratou E. Estimating the incidence of colorectal cancer in South East Asia. Croat Med J. 2013;54:532–40.
Pourhoseingholi MA. Increased burden of colorectal cancer in Asia. World J Gastrointest Oncol. 2012;4(4):68–70.
Presario. Molecular profile of colorectal cancer in Indonesia: is there another pathway? Gastroenterol Hepatol Bed Bench. 2012;5:71–8.
Sudoyo AW, Hernowo B, Krisnuhoni E, Reksodiputro AH, Hardjodisastro D. Colorectal cancer among young native Indonesians: a clinicopathological and molecular assessment on microsatellite instability. Medical Journal of Indonesia. 2010;19:245–51.
Atreya CE, Corcoran RB, Kopetz S. Expanded RAS: refining the patient population. J Clin Oncol. 2015;33:682–5.
Taniguchi H, Yamazaki K, Yoshino T, Muro K, Yatabe Y, Watanabe T, et al. Japanese Society of Medical Oncology clinical guidelines: RAS (KRAS/NRAS) mutation testing in colorectal cancer patients. Cancer Sci. 2015;106:324–7.
Negru S, Papadopoulou E, Apessos A, Stanculeanu DL, Ciuleanu E, Volovat C, et al. KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: a cohort study. BMJ Open British Medical Journal Publishing Group. 2013;4 e004652–2
Ogura T, Kakuta M, Yatsuoka T, Nishimura Y, Sakamoto H, Yamaguchi K, et al. Clinicopathological characteristics and prognostic impact of colorectal cancers with NRAS mutations. Oncol Rep. 2014;32(1):50–6.
Borràs E, Jurado I, Hernan I, Gamundi MJ, Dias M, Martí I, et al. Clinical pharmacogenomic testing of KRAS, BRAF and EGFR mutations by high resolution melting analysis and ultra-deep pyrosequencing. BMC Cancer BioMed Central Ltd. 2011;11:406.
Krypuy M, Newnham GM, Thomas DM, Conron M, Dobrovic A. High resolution melting analysis for the rapid and sensitive detection of mutations in clinical samples: KRAS codon 12 and 13 mutations in non-small cell lung cancer. BMC Cancer. BioMed Central. 2006;6:295.
Nollau P, Moser C, Weinland G, Wagener C. Detection of K-ras mutations in stools of patients with colorectal cancer by mutant-enriched PCR. Int J Cancer. Wiley Subscription Services, Inc., A Wiley Company1996;66:332–6.
Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, et al. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19:157–63.
Liu Y-P, Wu H-Y, Yang X, Xu H-Q, Chen D, Huang Q, et al. Diagnostic accuracy of high resolution melting analysis for detection of KRAS mutations: a systematic review and meta-analysis. Sci Rep. Nature Publishing Group2014;4:7521.
Ferreira CG, Aran V, Zalcberg-Renault I, Victorino AP, Salem JH, Bonamino MH, et al. KRAS mutations: variable incidences in a Brazilian cohort of 8,234 metastatic colorectal cancer patients. BMC Gastroenterol. 2014;14:1–8.
Watanabe T, Yoshino T, Uetake H, Yamazaki K, Ishiguro M, Kurokawa T, et al. KRAS mutational status in Japanese patients with colorectal cancer: results from a Nationwide, multicenter, cross-sectional study. Oxford University Press. Jpn J Clin Oncol. 2013;43(7):706–12.
Zhang J, Zheng J, Yang Y, Lu J, Gao J, Lu T, et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Sci Rep Nature Publishing Group. 2015;5:1–8.
Liu X, Jakubowski M, Hunt JL. KRAS gene mutation in colorectal cancer is correlated with increased proliferation and spontaneous apoptosis. Am J Clin Pathol. 2011;135:245–52.
Phua LC, Ng HW, Yeo AHL, Chen E, Lo MSM, Cheah PY, et al. Prevalence of KRAS, BRAF, PI3K and EGFR mutations among Asian patients with metastatic colorectal cancer. Oncol Lett. 2015;10:2519–26.
Akiyoshi K, Yamada Y, Honma Y, Iwasa S, Kato K, Hamaguchi T, et al. KRAS mutations in patients with colorectal cancer as detected by high-resolution melting analysis and direct sequencing. Anticancer Res. 2013;33:2129–34.
Hanna MC, Go C, Roden C, Jones RT, Pochanard P, Javed AY, et al. Colorectal cancers from distinct ancestral populations show variations in BRAF mutation frequency. PLoS One. 2012;8:e74950–0.
Missiaglia E, Jacobs B, D’Ario G, Di Narzo AF, Soneson C, Budinska E, et al. Distal and proximal colon cancers differ in terms of molecular, pathological, and clinical features. Ann Oncol. 2014;25:1995–2001.
Cheung DY, Kim TH, Kim CW, Kim JI, Cho SH, Park S-H, et al. The anatomical distribution of colorectal cancer in Korea: evaluation of the incidence of proximal and distal lesions and synchronous adenomas. Intern Med. 2008;47:1649–54.
Chaiyapan W, Duangpakdee P, Boonpipattanapong T, Kanngern S, Sangkhathat S. Somatic mutations of K-ras and BRAF in Thai colorectal cancer and their prognostic value. Asian Pac J Cancer Prev. 2013;14:329–32.
Peeters M, Kafatos G, Taylor A, Gastanaga VM, Oliner KS, Hechmati G, et al. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: a pooled analysis of randomised controlled trials. Eur J Cancer Elsevier. 2015;51:1704–13.
Tang WY, Elnatan J, Lee YS, Goh HS, Smith DR. c-Ki-ras mutations in colorectal adenocarcinomas from a country with a rapidly changing colorectal cancer incidence. Br J Cancer. Nature Publishing Group1999;81:237–41.
Kinoshita H, Yanagisawa A, Watanabe T, Nagawa H, Oya M, Kato Y, et al. Increase in the frequency of K-ras codon 12 point mutation in colorectal carcinoma in elderly males in Japan: the 1990s compared with the 1960s. Cancer Sci. 2005;96:218–20.
Webster J, Kauffman TL, Feigelson HS, Pawloski PA, Onitilo AA, Potosky AL, et al. KRAS testing and epidermal growth factor receptor inhibitor treatment for colorectal cancer in community settings. Cancer Epidemiol Biomarkers Prev American Association for Cancer Research. 2013;22:91–101.
Lee W-S, Lee JN, Baek J-H, Park YH. RAS status in Korean patients with stage III and IV colorectal cancer. Clin Transl Oncol Springer Milan. 2015;17:751–6.
Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. Oxford University Press. J Natl Cancer Inst. 2015;107(3):dju427.
Chen K-H, Shao Y-Y, Chen H-M, Lin Y-L, Lin Z-Z, Lai M-S, et al. Primary tumor site is a useful predictor of cetuximab efficacy in the third-line or salvage treatment of KRAS wild-type (exon 2 non-mutant) metastatic colorectal cancer: a nationwide cohort study. BMC Cancer BioMed Central. 2016;16:327.
Acknowledgements
This research effort was funded by an internal grant for PT Kalbe Farma Tbk. We thanked Fitria Yunida, Dewi Nawangwulan, and Siska Yogiwanti for excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study was determined to be exempt by the Institutional Review Board and performed in accordance to 1964 Helsinki Declaration and its later amendments. This retrospective study was approved by Institutional Review Board (IRB) of Stem-cell and Cancer Institute (SCI).
Conflict of interest
ML, GP, FS, and AU are employees of PT Kalbe Farma; GW and WG are senior pathologists receiving honoraria from PT Bifarma Adiluhung, Kalbe Farma; and all other authors have declared no conflicts of interests.
Rights and permissions
About this article
Cite this article
Levi, M., Prayogi, G., Sastranagara, F. et al. Clinicopathological Associations of K-RAS and N-RAS Mutations in Indonesian Colorectal Cancer Cohort. J Gastrointest Canc 49, 124–131 (2018). https://doi.org/10.1007/s12029-016-9901-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-016-9901-x