Abstract
Colorectal cancer (CRC) ranks among the most prevalent cancer types in both men and women. Screening of RAS (Kirsten rat sarcoma viral oncogene homolog (KRAS), neuro-blastoma RAS viral oncogene homolog (NRAS), and v-raf murine sarcoma viral oncogene homolog B1 (BRAF)) somatic mutations is necessary prior to considering anti-epidermal growth factor receptor (EGFR) therapies in CRC patients. Next-generation sequencing studies have confirmed that RAS gene panels could be used while developing treatment strategies for patients with CRC. The present study explored genetic mutations in KRAS, NRAS, and BRAF in CRC patients in the Telangana state of India. Patients with confirmed CRC (n = 100) who visited the Apollo hospitals were evaluated. Genomic DNA was extracted from formalin-fixed, paraffin-embedded tissues, and pyrosequencing analysis was performed. Patient DNA samples were screened for 54 different KRAS, NRAS, and BRAF mutations, which revealed 34 somatic mutations. Exon 11 of BRAF possessed 4 mutations with highest individuals documented with G469A mutation. Pyrosequencing, a reliable method for analyzing somatic mutations present in RAS, could aid in taking treatment decisions for patients with CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is the second most common multifactorial disease that leads to death worldwide [2], with colorectal cancer (CRC, OMIM #114500) being the fourth leading cause of cancer-related deaths globally [17, 19]. Based on epidemiological investigations, most CRC cases could be attributed to environmental factors [18, 28]. Genetic evidences have revealed that individuals with specific single nucleotide polymorphisms (SNPs) might be more cancer susceptible [11]. Somatic mutations in Kirsten rat sarcoma viral oncogene homolog (KRAS), neuro-blastoma RAS viral oncogene homolog (NRAS), and v-raf murine sarcoma viral oncogene homolog B1 (BRAF) genes have been confirmed in routine screening of patients with advanced-stage CRC by next-generation sequencing (NGS) [6]. In humans, 3 RAS gene mutation occurs in 90% of the pancreatic cancers, followed by colon (45%) and lung cancers (35%), respectively. RAS is a common target in oncology-related drug development [26], and its mutations act as important biomarkers for predicting the response to anti-epidermal growth factor receptor (EGFR) antibody-based therapy in CRC [3]. For CRC development and progression, EGFR signals play a major role [10]. Three RAS genes (KRAS, NRAS, and BRAF) code for kinases involved in the RAS-RAF-mitogen-activated protein kinase signaling pathway and their mutations lead to the resistance against anti-EGFR monoclonal antibody therapy. Most of the KRAS mutations (98.5%) occur at codons 12 and 13 of exon 2. NRAS mutations typically occur in exon 2-4. In BRAF, nearly all (81.9%) mutations occur in codon 600 (V600E) [32]. Only a few studies have included Indian populations for analyzing all the RAS (KRAS-NRAS-BRAF) mutations in CRC cases [1, 12]. Therefore, this study was conducted to assess the association of KRAS, NRAS, and BRAF somatic mutations with CRC risk in the population of Telangana, India.
Subjects and Methods
Patient Selection
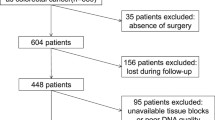
In this study, patients who had been diagnosed with CRC (n = 100) between January, 2014, and December, 2018, at the Department of Oncology, Apollo Hospitals, Jubilee Hills, Hyderabad, Telangana, India, were enrolled. The inclusion criteria were a confirmed diagnosis, based on histopathological examinations and primary CRC. Exclusion criteria included the presence of any other cancers, prior anti-cancer treatment, and non-adenocarcinomas of the colon. This study was conducted at the Department of Molecular Biology and Cytogenetics, Apollo Hospitals, Hyderabad, India.
DNA Isolation
CRC biopsy samples (100), placed in 10% buffer for formalin-fixed paraffin-embedded (FFPE) tissue(s) preparation were enrolled. DNA from FFPE tissues was extracted using Qiagen kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocol. DNA quality based on concentration and purity was assessed with a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Purified DNA was used for the pyrosequencing analysis.
Pyrosequencing Analysis
In this study, polymerase chain reaction (PCR) was performed with a pyrosequencing method using specific primers designed to detect KRAS, NRAS, and BRAF mutations. The target sequence covering the polymorphic site was amplified with one of the specific biotinylated primers. Pyrosequencing analysis was carried out as described previously [4, 24]. The total PCR volume and thermal cycler conditions were set in accordance with those previously described by Khan et al. [14], except for the annealing temperatures, which were modified. The primers, master-mix, and protocol were supplied by Qiagen. Table 1 provides a list of the tested mutations during screening.
Results
In this study, 100 CRC cases were screened and the clinicopathological characteristics of tumors or FFPE tissues (obtained from the patients) were evaluated. Among the CRC cases studies, 63% were men, whereas, 37% were women, with a mean patient age of 59.1 ± 11.92 years. It was noted that 33% of patients were smokers, while 28% consumed alcohol. All colon tissue samples were > 5 cm in size. Family histories were collected for only 22% of the CRC cases. Specific clinical pathological stages were not documented for all the patients, and 67% of cases were stage II (67/100). The baseline characteristics are given in Table 2. Pyrosequencing analysis was carried out to detect KRAS, BRAF, and NRAS mutations in patients with CRC. Overall, 13% of the patients possessed mutations in KRAS. Exon 2 of KRAS exhibited G12C (3%), G13D (2%), G12A (3%), G12D (2%), and G12S (1%) mutations, whereas, Q16L (1%) and Q16E (1%) mutations were detected in exon 3 of KRAS (Table 3). Fifteen (15%) patients exhibited mutations in exon 11 of BRAF, with G1397T (2%), G1406C (1%), G1391A (3%), G1405T (1%), G1406C (4%), G1406C (4%) (maximum), and G1391T (2%) mutations. In exon 15 of BRAF, G1798A (1%) and T1799A (1%) mutations were detected. In addition to this, 3 patients exhibited mutations in exon 3 of NRAS (G175A (2%) and A182T (1%) mutations) (Table 3; Fig. 1).
Discussion
Molecular testing has become more important in cancer-effected patients, due to the improvements in targeted therapy [16]. Well-documented genetic mutations in RAS (KRAS, NRAS, and BRAF) genes aid in evaluating patients with CRC and have a high rate of incidence and mortality, which is lowering because of the recent advancements in disease identification through genetic screening, and is useful in making treatment decisions. Biologically and epidemiologically, CRC is a heterogenous disease with multiple etiologies. This heterogeneity impacts its prognosis and clinical management, as the tumor tissue’s location does not accurately reflect KRAS mutation’s status in the entire tumor [20]. In 100 patients with CRC, 54 mutations were screened and 31 were detected in KRAS, NRAS, and BRAF. In KRAS, NRAS, and BRAF; 13, 38, and 3% of the mutations were detected, respectively.
The RAS gene family is well documented to be cancer associated with mutations in codons 12 and 13 of KRAS, observed in 35–45% of all CRC cases. Detecting KRAS mutational status has become a crucial tool for managing the patients with CRC [5]. These mutations are recognized as biomarkers for predicting stage IV CRC and indicate poor efficiency of anti-EGFR therapy. However, KRAS mutation’s prognostic value remains controversial, with some studies on patients with stage II and III CRC, revealing a positive association [15]. The subjects evaluated in the current study were mainly in stage II CRC. In mammalian cells, RAS gene encodes monomeric GTPases that are involved in signal transduction pathways and regulate cell proliferation and differentiation [31]. RAS gene has primarily been documented as a viral gene, and its mutations are detected in 30% of the human cancer cells. KRAS protein (also known as p21), a member of the RAS superfamily of proteins, is a membrane-anchored GTP/GDP-binding protein that is broadly expressed in human cells [29]. The use of monoclonal antibodies targeting EGFR (cetuximab and panitumumab) as combination chemotherapy has exhibited clinical benefits in patients, with a response rate of ~ 20% for metastatic CRC [27]. KRAS is a proto-oncogene, mainly involved in the RAS-MAPK signaling pathway to regulate cell growth, proliferation, and differentiation. Mutations frequently occur around KRAS gene codons, converting normal cells into cancerous. Whole genome sequencing studies confirmed that KRAS mutations are triggered in limited number of carcinomas, with CRC being one of them and KRAS mutations indicate a residual tumor [7]. KRAS was initially identified as a p21GTPase, and a high rate of somatic mutations in KRAS was observed in carcinomas such as leukemia, non-small cell lung cancer, pancreatic cancer, and CRC. Most of these mutations were present in the exons 2 and 3, with the common mutations including G12C, G12D, G12R, G12S, G12V, G13D, and Q16H, whereas, exon 4 displayed K117 and A146G mutations [31]. In our study, G12C, G13D, G12A, G12D, G12S, Q61L, and Q61E mutations in KRAS were detected. In general, KRAS mutational analysis is performed by Sanger sequencing analysis or pyrosequencing. Pyrosequencing technique is considered as a homogenous genotype that concerns KRAS mutation status in tumor cells. When KRAS mutations are left undetected by DNA sequencing, anti-EGFR therapy may fail to have antiproliferation effects; therefore, performing pyrosequencing analysis with KRAS mutation kit (Qiagen) is recommended. Codons 12 and 13 in exon 2 of KRAS alter the protein sequence. Changes in the first 2 base pairs result in amino acid substitution in KRAS protein, resulting in the treatment [34].
The frequency of other KRAS mutations at codons 16, 146, and 154 were found to be approximately 5%. However, among the somatic mutations in cancer database, 500 mutations (approximately) have been detected in KRAS gene in CRC samples [29]. Mutations in RAS genes such as KRAS and NRAS (in exons 2-4) indicate that these metastatic CRC patients might be resistant to anti-EGFR targeted therapy [13]. BRAF mutations are potential prognostic markers of CRC therapy. At codon 600, the conversion of amino acid valine to glutamic acid (V600E) is responsible for CRC development in 10% of the cases. A correlation between the combination of KRAS and BRAF mutations and the prognosis has been demonstrated and might be useful for developing cancer treatments [30].
The prevalence of KRAS, NRAS, and BRAF mutations have been evaluated in global studies; however, limited data is available regarding their respective frequencies in CRC [8,9,10, 21,22,23, 25, 33]. Results of the present study were not in agreement with that of other studies [8,9,10, 22, 25, 33], which might be due to ethnicity, modified lifestyle, and/or genetics (Table 4).
Pyrosequencing analysis was performed to detect the mutations in RAS genes in patients with CRC. We have predicted that CRC patients with KRAS mutations might not benefit from anti-EGFR therapy, which however, might treat metastatic CRC and the patients should be screened for KRAS mutations before therapy administration. Furthermore, the study also lacked control tissues and subjects, thereby, making the information of the clinical pathological stages incomplete. Additionally, studies examining large number of patients are required. The presence of somatic mutations in KRAS, NRAS, and BRAF genes has been confirmed in the routine screening of patients with advanced-stage CRC, with NGS [6].
Limited studies including RT-PCR, NGS, and Sanger sequencing, to rule out the disease, have been previously performed; however, in the present study, pyrosequencing was performed to detect CRC-associated RAS mutations. In this study, including 100 samples is an advantage which constitutes a high sample number when comparing with other ethnic population(s). The similarity of the current study with other global studies is the screening of similar mutations in RAS genes.
In conclusion, we confirmed and suggested RAS genes screening by pyrosequencing prior to anti-EGFR therapy administration in patients with CRC. Pyrosequencing analysis of the RAS genes revealed high clinical sensitivity, which could be used for determining the effective treatments for patients with CRC. Pyrosequencing could be performed by the pathologists/molecular biologists in order to obtain clinical confirmation that further aid in developing strategies for treating cancer.
References
Bagadi, S. B., Sanghvi, M., Nair, S. B., & Das, B. R. (2012). Combined mutational analysis of KRAS, NRAS and BRAF genes in Indian patients with colorectal carcinoma. The International Journal of Biological Markers, 27, 27–33.
Biswas, S., Reddy, N. D., Jayashree, B., & Rao, C. M. (2018). Evaluation of novel 3-hydroxyflavone analogues as HDAC inhibitors against colorectal cancer. Advances in Pharmacological Sciences, 2018.
Boussios, S., Ozturk, M. A., Moschetta, M., Karathanasi, A., Zakynthinakis-Kyriakou, N., Katsanos, K. H., Christodoulou, D. K., & Pavlidis, N. (2019). The developing story of predictive biomarkers in colorectal cancer. Journal of Perinatal Medicine, 9, 12.
Córdoba, E. E., Lacunza, E., Abba, M. C., Fernández, E., & Güerci, A. M. (2018). Single nucleotide polymorphisms in ATM, TNF-α and IL6 genes and risk of radiotoxicity in breast cancer patients. Mutation Research/Genetic Toxicology and Environmental Mutagenesis, 836, 84–89.
Dinu, D., Dobre, M., Panaitescu, E., Bîrlă, R., Iosif, C., Hoara, P., Caragui, A., Boeriu, M., Constantinoiu, S., & Ardeleanu, C. J. (2014). Prognostic significance of KRAS gene mutations in colorectal cancer-preliminary study. Journal of Medicine and Life, 7, 581.
Franczak, C., Dubouis, L., Gilson, P., Husson, M., Rouyer, M., Demange, J., Leroux, A., Merlin, J.-L., & Harlé, A. (2019). Integrated routine workflow using next-generation sequencing and a fully-automated platform for the detection of KRAS, NRAS and BRAF mutations in formalin-fixed paraffin embedded samples with poor DNA quality in patients with colorectal carcinoma. PLoS One, 14, e0212801.
Fu, Y., Duan, X., Huang, J., Huang, L., Zhang, L., Cheng, W., Ding, S., & Min, X. (2019). Detection of KRAS mutation via ligation-initiated LAMP reaction. Scientific Reports, 9, 5955.
Gilson, P., Franczak, C., Dubouis, L., Husson, M., Rouyer, M., Demange, J., Perceau, M., Leroux, A., Merlin, J.-L., & Harle, A. (2019). Evaluation of KRAS, NRAS and BRAF hotspot mutations detection for patients with metastatic colorectal cancer using direct DNA pipetting in a fully-automated platform and next-generation sequencing for laboratory workflow optimisation. PLoS One, 14, e0219204.
Guedes, J. G., Veiga, I., Rocha, P., Pinto, P., Pinto, C., Pinheiro, M., Peixoto, A., Fragoso, M., Raimundo, A., & Ferreira, P. (2013). High resolution melting analysis of KRAS, BRAF and PIK3CA in KRAS exon 2 wild-type metastatic colorectal cancer. BMC Cancer, 13, 169.
Hamzehzadeh, L., Khadangi, F., Karimiani, E. G., Pasdar, A., & Kerachian, M. A. (2018). Common KRAS and NRAS gene mutations in sporadic colorectal cancer in Northeastern Iranian patients. Current Problems in Cancer, 42, 572–581.
Ibrahimi, M., Moossavi, M., Mojarad, E. N., Musavi, M., Mohammadoo-khorasani, M., & Shahsavari, Z. (2019). Positive correlation between interleukin-1 receptor antagonist gene 86 bp VNTR polymorphism and colorectal cancer susceptibility: a case-control study. Immunologic Research, 67, 151–156.
Jauhri, M., Bhatnagar, A., Gupta, S., Minhas, S., Shokeen, Y., & Aggarwal, S. (2017). Prevalence and coexistence of KRAS, BRAF, PIK3CA, NRAS, TP53, and APC mutations in Indian colorectal cancer patients: next-generation sequencing–based cohort study. Tumour Biology, 39, 1010428317692265.
Jouini, R., Ferchichi, M., BenBrahim, E., Ayari, I., Khanchel, F., Koubaa, W., Saidi, O., Allani, R., & Chadli-Debbiche, A. (2019). KRAS and NRAS pyrosequencing screening in Tunisian colorectal cancer patients in 2015. Heliyon, 5, e01330.
Khan, I. A., Jahan, P., Hasan, Q., & Rao, P. (2019). Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. Diabetes & Metabolic Syndrome,13(1), 688–694.
Kim, H. S., Heo, J. S., Lee, J., Lee, J. Y., Lee, M.-Y., Lim, S. H., Lee, W. Y., Kim, S. H., Park, Y. A., & Cho, Y. B. (2016). The impact of KRAS mutations on prognosis in surgically resected colorectal cancer patients with liver and lung metastases: a retrospective analysis. BMC Cancer, 16, 120.
Li, W., Qiu, T., Guo, L., Ying, J., & Zhou, A. (2019). NGS-based oncogenic mutations analysis in advanced colorectal cancer patients improves targeted therapy prediction. Pathology, Research and Practice, 215, 483–489.
Lim, S.-R., Gooi, B.-H., Singh, M., & Gam, L.-H. (2011). Analysis of differentially expressed proteins in colorectal cancer using hydroxyapatite column and SDS-PAGE. Applied Biochemistry and Biotechnology, 165, 1211–1224.
Lin, J., Chen, Y., Tang, W., Liu, C., Zhang, S., Guo, Z.-Q., Chen, G., & Zheng, X.-W. (2019). PPARG rs3856806 C> T polymorphism increased the risk of colorectal cancer: a case-control study in eastern Chinese Han Population. Frontiers in Oncology, 9, 63.
Liu, S., Dong, H., Wu, J., & Wang, C. (2019). Association of an miR-502-binding site polymorphism in the 3′-untranslated region of SET8 with colorectal cancer. Oncology Letters, 17, 3960–3964.
Lv, Y., Wang, X., Liang, L., Wang, L., & Lu, J. (2019). SUVmax and metabolic tumor volume: surrogate image biomarkers of KRAS mutation status in colorectal cancer. OncoTargets and Therapy, 12, 2115.
Modest, D. P., Ricard, I., Heinemann, V., Hegewisch-Becker, S., Schmiegel, W., Porschen, R., Stintzing, S., Graeven, U., Arnold, D., & von Weikersthal, L. F. (2016). Outcome according to KRAS-, NRAS-and BRAF-mutation as well as KRAS mutation variants: pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Annals of Oncology, 27, 1746–1753.
Mohamed Suhaimi, N.-A., Foong, Y. M., Lee, D. Y. S., Phyo, W. M., Cima, I., Lee, E. X. W., Goh, W. L., Lim, W.-Y., Chia, K. S., & Kong, S. L. (2015). Non-invasive sensitive detection of KRAS and BRAF mutation in circulating tumor cells of colorectal cancer patients. Molecular Oncology, 9, 850–860.
Negru, S., Papadopoulou, E., Apessos, A., Stanculeanu, D. L., Ciuleanu, E., Volovat, C., Croitoru, A., Kakolyris, S., Aravantinos, G., & Ziras, N. (2014). KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: a cohort study. BMJ Open, 4, e004652.
Nordström, T., Nourizad, K., Ronaghi, M., & Nyrén, P. (2000). Method enabling pyrosequencing on double-stranded DNA. Analytical Biochemistry, 282, 186–193.
Payandeh, M., Sadeghi, M., Sadeghi, E., & Gholami, F. (2015). Analysis of KRAS, BRAF and NRAS in patients with colorectal cancer: the first report of Western Iran. American Journal of Cancer Prevention, 3, 19–22.
Porru, M., Pompili, L., Caruso, C., Biroccio, A., & Leonetti, C. (2018). Targeting KRAS in metastatic colorectal cancer: current strategies and emerging opportunities. Journal of Experimental & Clinical Cancer Research, 37, 57.
Post, J. B., Hami, N., Mertens, A. E., Elfrink, S., Bos, J. L., & Snippert, H. J. G. (2019). CRISPR-induced RASGAP deficiencies in colorectal cancer organoids reveal that only loss of NF1 promotes resistance to EGFR inhibition. Oncotarget, 10, 1440.
Salama, R. H., Sayed, Z. E.-A. A., Ashmawy, A. M., Elsewify, W. A., Ezzat, G. M., Mahmoud, M. A., Alsanory, A. A., & Alsanory, T. A. (2019). Interrelations of apoptotic and cellular senescence genes methylation in inflammatory bowel disease subtypes and colorectal carcinoma in Egyptians patients. Applied Biochemistry and Biotechnology, 1–14. https://doi.org/10.1007/s12010-019-03017-x.
Tan, C., & Du, X. (2012). KRAS mutation testing in metastatic colorectal cancer. World Journal of Gastroenterology, 18, 5171.
Vacante, M., Borzì, A. M., Basile, F., & Biondi, A. (2018). Biomarkers in colorectal cancer: current clinical utility and future perspectives. World Journal of Clinical Cases, 6, 869.
Xu, K., Park, D., Magis, A. T., Zhang, J., Zhou, W., Sica, G. L., Ramalingam, S. S., Curran, W. J., & Deng, X. (2019). Small molecule KRAS agonist for mutant KRAS cancer therapy. Molecular Cancer, 18, 85.
Yang, T., Li, X., Montazeri, Z., Little, J., Farrington, S. M., Ioannidis, J. P., Dunlop, M. G., Campbell, H., Timofeeva, M., & Theodoratou, E. (2018). Gene-environment interactions and colorectal cancer risk: an umbrella review of systematic reviews and meta-analyses of observational studies. International Journal of Cancer. https://doi.org/10.1002/ijc.32057.
Zhang, J., Zheng, J., Yang, Y., Lu, J., Gao, J., Lu, T., Sun, J., Jiang, H., Zhu, Y., & Zheng, Y. (2015). Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: analysis of 1,110 cases. Scientific Reports, 5, 18678.
Zinsky, R., Bölükbas, S., Bartsch, H., Schirren, J., & Fisseler-Eckhoff, A. (2010). Analysis of KRAS mutations of exon 2 codons 12 and 13 by SNaPshot analysis in comparison to common DNA sequencing. Gastroenterology Research and Practice, 2010, 789363.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Goud, K.I., Matam, K., Madasu, A.M. et al. Positive Correlation Between Somatic Mutations in RAS Gene and Colorectal Cancer in Telangana Population: Hospital-Based Study in a Cosmopolitan City. Appl Biochem Biotechnol 190, 703–711 (2020). https://doi.org/10.1007/s12010-019-03119-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03119-6