Abstract
Background
The objective of this study was to define clinically meaningful phenotypes of intracerebral hemorrhage (ICH) using machine learning.
Methods
We used patient data from two US medical centers and the Antihypertensive Treatment of Acute Cerebral Hemorrhage-II clinical trial. We used k-prototypes to partition patient admission data. We then used silhouette method calculations and elbow method heuristics to optimize the clusters. Associations between phenotypes, complications (e.g., seizures), and functional outcomes were assessed using the Kruskal–Wallis H-test or χ2 test.
Results
There were 916 patients; the mean age was 63.8 ± 14.1 years, and 426 patients were female (46.5%). Three distinct clinical phenotypes emerged: patients with small hematomas, elevated blood pressure, and Glasgow Coma Scale scores > 12 (n = 141, 26.6%); patients with hematoma expansion and elevated international normalized ratio (n = 204, 38.4%); and patients with median hematoma volumes of 24 (interquartile range 8.2–59.5) mL, who were more frequently Black or African American, and who were likely to have intraventricular hemorrhage (n = 186, 35.0%). There were associations between clinical phenotype and seizure (P = 0.024), length of stay (P = 0.001), discharge disposition (P < 0.001), and death or disability (modified Rankin Scale scores 4–6) at 3-months’ follow-up (P < 0.001). We reproduced these three clinical phenotypes of ICH in an independent cohort (n = 385) for external validation.
Conclusions
Machine learning identified three phenotypes of ICH that are clinically significant, associated with patient complications, and associated with functional outcomes. Cerebellar hematomas are an additional phenotype underrepresented in our data sources.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Spontaneous intracerebral hemorrhage (ICH) affects nearly 100,000 Americans each year, and acute management improves the prognosis [1]. Although severity of ICH is easily summarized [2] and standing order sets are common, presentation of ICH is not monolithic. Different etiologies of spontaneous ICH, such as hypertension or cerebral amyloid angiopathy [3], result in different clinical phenotypes [4]. Each clinical phenotype might be expected to have a different clinical course, complications, and potentially different patient outcomes. Thus, optimal management likely differs between phenotypes [5]. For example, patients with lobar hematomas and altered consciousness are at increased risk for seizures and thus would be more likely to benefit from antiseizure medication and electroencephalography monitoring compared to patients with deep hematomas and intact consciousness [6].
Although prior work has sought to identify differences in risk factors, features, and outcomes based on ICH location [7, 8] and characterize the locations and etiologies of spontaneous ICH in specific populations [9], to our knowledge, no study has sought to agnostically identify clinical phenotypes using unsupervised machine learning. As a form of artificial intelligence, machine learning algorithms recognize previously unknown patterns in data structure [4]. Unsupervised machine learning algorithms are agnostic to expert-defined labels and prespecified assumptions, which allows for the discovery of novel patterns [10]. These algorithms have been broadly applied to phenotyping cardiovascular diseases and aortic stenosis [11,12,13,14,15]; distinguishing patterns of end-of-life care delivery in the intensive care unit (ICU) [16]; and grouping stroke symptoms, biomarkers, and complex patient outcomes [17,18,19,20]. Unsupervised methods reduce human bias introduced from classifications based on clinical expertise and thus allow us to validate conventional clinical wisdom and discover phenotypes that may not have previously been characterized. Better characterization of clinical phenotypes could allow for the development of more precise management and, potentially, improved outcomes.
We tested the hypothesis that unsupervised machine learning could identify clinical phenotypes in patients with acute ICH. We also explored if these clinical phenotypes were clinically meaningful based on associations with complications (e.g., seizures) and functional outcomes.
Methods
Patients
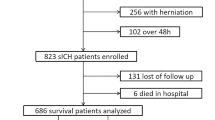
We conducted a retrospective analysis of deidentified prospectively collected patient data obtained from three sources: (1) the Northwestern University Brain Attack Registry (NUBAR), a prospectively collected registry of electronic health records with detailed information on patient outcomes after stroke; (2) a cohort of patients from the Johns Hopkins Hospital and Johns Hopkins Bayview Medical Center, part of Johns Hopkins Medicine [21]; and (3) Antihypertensive Treatment of Acute Cerebral Hemorrhage-II (ATACH-II) [22], a clinical trial data set of patients with ICH that we used for validation of the phenotypes [23]. For consistency, we included only patients from ATACH-II enrolled in the United States because of variability in treatment response internationally (e.g., regional differences in treatments and outcomes for patients from Asia) [24, 25]. Across the three data sources, we analyzed complete patient records [17, 20].
Variable selection was limited because of the availability of data that could be harmonized across all three data sources. Harmonized patient data collected at admission included age, sex, race, ICH volume, ICH location, Glasgow Coma Scale (GCS) score, international normalized ratio (INR), systolic blood pressure (SBP), intraventricular hemorrhage (IVH), infratentorial location, history of diabetes, history of hypertension, and hematoma expansion. ICH location was dichotomized as lobar hematoma location versus thalamus, basal ganglia, brainstem, caudate, cerebellar, lentiform nucleus, or other location of hemorrhage. GCS score was categorized as < 5, 5–12, and 13–15. INR was dichotomized as 1.4 and lower (“normal”) and 1.5 and greater (“abnormal”). All patients had a diagnostic computerized tomography (CT) scan and a standard-of-care follow-up CT scan conducted around the 24-h mark [26, 27]. Hematoma expansion was calculated as the hematoma volume on the second or subsequent CT scan minus the hematoma volume on the initial CT scan. We defined hematoma expansion as growth of 6 mL or greater across all data sets [28]. Hematoma expansion was recoded to a binary true/false variable for harmonization across the three data sources.
Hematoma volumes at Northwestern were measured with validated, semiautomated, voxel-based techniques from CT scans. We previously established the high interrater reliability of this hematoma volume measurement technique in patients with ICH and reported excellent correlation between two separate evaluators (Spearman ρ = 0.99, P < 0.001) [29]. These validated methods account for voxel-by-voxel measurements (three-dimensional representations of volume that have the density of acute hemorrhage). Hematoma volumes for ATACH-II were adjudicated by a central reader. Hematoma volumes for the Hopkins data set were calculated using the ABC/2 method. When IVH was next to ICH, an expert adjudicated where the IVH began and the intracranial hematoma ended.
Outcomes
The patient outcomes assessed included seizure, hospital length of stay (LOS) in days, ICU LOS in days, discharge disposition, and the modified Rankin Scale (mRS; a global functional outcomes scale) score at 3-months’ follow-up. Seizures were defined based on characteristic clinical presentation observed during hospitalization by a clinician and reviewed by a study neurologist or electroencephalography monitoring per protocol [30]. Disposition at discharge was harmonized across the three data sources by recategorizing as died, inpatient (e.g., rehabilitation, acute care, nursing facility), outpatient (e.g., home), and other. The mRS score was dichotomized to mRS scores 0–3 (“good outcome,” independence or better) and mRS scores 4–6 (“poor outcome,” dependence or death) [31,32,33]. Follow-up mRS scores at 3 months were available for patients in the ATACH-II and NUBAR data sets.
Machine Learning
We used unsupervised k-prototype cluster analysis to group patients into clinical phenotypes because this algorithm performs well with mixed categorical and continuous data [34]. The algorithm generated mutually exclusive groups from the 13 independent admission variables using a combination of means for continuous variables and modes for categorical variables. This unsupervised cluster analysis was performed independent of the patient complications or outcomes data. Both elbow method heuristics and average silhouette method calculations were used to determine the optimal number of clusters [17]. Each cluster represents a clinical phenotype composed by maximizing similarities within and differences between clusters according to select admission data [16]. We generated a two-dimensional visualization of the clinical phenotypes using the uniform manifold approximation and projection (UMAP) [35]. The UMAP algorithm employs a nonlinear approach for dimension reduction.
Validation
We validated the k-prototype clustering algorithm in the independent cohort of ATACH-II data [11, 23]. We trained the k-prototype model using the aggregate data from the two US medical centers. We used the same 13 variables identified in the derivation cohort to assign phenotypes in the external validation cohort [11]. Our model was tested in this external cohort to validate the generalizability of the k-prototype clustering algorithm [15].
Statistical Analysis
Continuous data between phenotypes were compared using analysis of variance for normally distributed data or the Kruskal–Wallis H-test for nonnormally distributed data. Categorical data were tested for an association with phenotype using χ2 statistics. A P value of 0.05 was used as the threshold for statistical significance. Analysis was performed in R v4.2.2, package “clustMixType” (RStudio: Integrated Development for R. RStudio, PBC, Boston, MA, 2020. www.rstudio.com) [36, 37].
Results
The 13 patient admission data variables clustered into three clinical phenotypes of ICH (Fig. 1). Elbow method heuristics and average silhouette method calculations suggested the optimal number of clusters (k) was three (Supplementary Figs. 1 and 2). Demographics of the three clinical phenotypes are shown in Table 1. Illustrative head CT scans for the three clinical phenotypes are shown in Supplementary Fig. 3. Demographics of incomplete patient records not included in the analysis are documented in Supplementary Table 1.
Although there was no human labeling of the clusters of patients, the three distinct phenotypes were clinically meaningful. Of the 531 patients from the two US medical centers, 141 (26.6%) were assigned to phenotype 1, 204 (38.4%) were assigned to phenotype 2, and 186 (35.0%) were assigned to phenotype 3 (Table 1). Clinical phenotype 1 included patients with small hematomas, high blood pressure, and GCS scores greater than 12. Clinical phenotype 2 included individuals with hematoma expansion and elevated INR. Clinical phenotype 3 included individuals with larger median hematoma volumes (24.0 [interquartile range 8.2–59.5] mL), who were more commonly Black or African American, and who had IVH (Fig. 2). This phenotype had the greatest proportion (18.8%) of patients with GCS scores less than 5 (Table 1).
Patient complications and outcomes varied with the three clinical phenotypes (Table 1, Fig. 3). Seizures were more common in patients with phenotype 2 (P = 0.024). Patients in phenotype 3 had the longest durations for ICU and hospital LOS, more frequently died during hospitalization (38.2%), and had a greater proportion of poor mRS outcomes at 3-months’ follow-up (83.7%) (Table 1). The three clinical phenotypes were significantly associated with LOS (P = 0.001), discharge disposition (P < 0.001), and poor outcome on the mRS at 3-months’ follow-up (P < 0.001) (Fig. 3). Cause of death (e.g., death by neurological criteria, cardiac arrest, withdrawal of life support) was only available for the Northwestern cohort and was not associated with clinical phenotype (P = 0.6).
We separately validated the three clinical phenotypes in the independent ATACH-II data (Supplementary Fig. 4). Of the 385 patients, 184 (47.8%) were assigned to phenotype 1, 130 (33.8%) were assigned to phenotype 2, and 71 (18.4%) were assigned to phenotype 3. As in the derivation cohort, validation cohort phenotypes differentiated between patients with small hematomas, elevated blood pressure, and GCS scores > 12 (phenotype 1); coagulopathic patients with hematoma expansion (phenotype 2); and patients with large hematomas and IVH (phenotype 3) (Supplementary Table 2). There were associations between the clinically validated phenotypes and LOS (P < 0.001), discharge disposition (P = 0.001), and death or disability (mRS scores 4–6) at 3-months’ follow-up (P < 0.001). In the validation cohort, seizure was not significantly associated with clinical phenotype (P = 0.5), potentially a function of the low seizure incidence in the independent cohort of ATACH-II data (1.0% versus 8.1%, P < 0.001) (Supplementary Table 3).
Discussion
In this multicenter study analyzing data from 916 patients with ICH, we found that unsupervised machine learning clustered patient presentations into three clinically distinct phenotypes. In turn, these data-derived clinical phenotypes had different risk factors for ICH (e.g., chronic hypertension, anticoagulation), different rates of seizures, and different likelihoods of poor functional outcome at follow-up. These results suggest that unsupervised machine learning could be a useful technique to identify clinical phenotypes, anticipate complications (e.g., need for ventricular drainage), and potentially inform protocols for treatment (e.g., prophylactic seizure medication).
Of the three clinical phenotypes, the one characterized by Black or African American race, large hematomas, and IVH was associated with the worst outcomes (phenotype 3). Although the clinical implications of larger hematomas are in line with previous work documenting the strong association between increased ICH volume and poor outcomes [38,39,40], we documented a new pattern of patient race alongside the clinical presentation and functional outcomes for patients in phenotype 3. This finding contributes to the expansive body of work on social determinants of health and ICH incidence, treatment, and outcomes [41,42,43,44]. Phenotype 1 and phenotype 2 were distinctly separated by SBP, an underlying factor that drives care management for patients with ICH [45]. The use of a phenotype may be complementary to standard measures of severity (e.g., ICH score) to anticipate potential complications and treatments.
The clinical phenotypes we present in this study may have potential implications in the targeted care of patients with ICH [15]. Accounting for clinical phenotype may allow for more precisely targeted assessment and treatment for subgroups of patients with ICH, a key step to achieving the goal of precision medicine [4]. Accounting for these phenotypes could promote targeted risk assessment and personalized treatment [10]. The data-derived clinical phenotypes may contribute to targeted therapy (e.g., prophylactic seizure medication, antithrombotic reversal, antihypertensive treatment) for each of the identified phenotypes [46].
These phenotypes could promote attention to targeted prevention of seizures, antihypertensive medication, and hemostatic medication. Seizures worsen outcomes in patients with ICH [30, 47,48,49,50]. The use of prophylactic seizure medication is common (~ 40% of patients) [51], yet indiscriminate administration to patients is associated with worse mRS scores and reduced health-related quality of life, particularly cognitive function [52, 53]. Targeting the use of antiseizure medications to patients at high risk due to seizures is more likely to achieve the intended benefit (preventing seizures) while minimizing the risk of side effects in patients at low risk for seizures. Prothrombotic agents (e.g., activated factor VII) [54] are likely to be beneficial in a subset of patients [54,55,56,57,58] but have potential adverse effects (e.g., venous thromboembolism) [59]. Improved patient selection is needed to precisely identify patients most likely to benefit from prothrombotic medications. Some patients require more antihypertensive medications even though there are increased adverse effects, particularly acute kidney injury [60]. The clinical phenotypes we present here could support judicious administration of prophylactic seizure medication, prothrombotic agents, and antihypertensive treatment.
Strengths of our approach include the large sample size from multiple centers, which could improve generalization. In addition, we reproduced three clinical phenotypes of ICH in an independent cohort. This external validation supports our results and lays the groundwork for the generalizability of our findings [10, 11].
Our agnostic subtyping approach used clinical variables to describe clinical phenotypes of ICH without prespecified assumptions or human-derived heuristics. Clinical phenotypes of ICH may be most useful in consort with human understanding and as a complement to standard measures of severity (e.g., ICH score). However, there are several limitations to this analysis. Although we were able to analyze data from multiple sources, we limited the scope of our analysis to patient data from the United States. International data (particularly Asia) were available from the ATACH-II trial, but we used domestic patients for consistency with the two other US cohorts. Patients from Asia also have regional differences in treatments and outcomes after ICH [25]. These differences could furnish different clinical phenotypes depending on individual patient populations or country of origin. Additionally, the ATACH-II trial experienced a relatively low risk of seizures, potentially diminishing our power to detect a difference in the external validation cohort. Future research may attempt to replicate our findings in cohorts representing different demographic groups and patients from other institutions. Another limitation was the lack of baseline mRS data for comparison with the reported mRS scores at 3-months’ follow-up. However, these seem unlikely to meaningfully change our analysis.
Although our leveraging of routine clinical data enhances the practical utility of understanding the natural segmentation of patients with ICH, our analysis was limited by the inclusion of only basic radiographic descriptors and the exclusion of more diverse imaging, biomarker, and clinical features data. Future research may attempt to replicate these phenotypes and incorporate other clinical features that were not available in the three data sources we harmonized (e.g., presence of abnormal vascular lesions, renal function, pretreatment with antiplatelet medications, levels of LDL, toxicology screen results, microbleeds, presence of dementia, etc.). These diverse data sets may further enable the detection of novel patterns and additional clinical phenotypes of ICH with unsupervised machine learning.
We used a machine learning method to identify clinical phenotypes from the data because existing etiologic classifications of intracranial hemorrhage (e.g., Structural lesion, Medication, Amyloid angiopathy, Systemic/other disease, Hypertension, Undetermined (SMASH-U)) regarding structural lesions criteria (e.g., vascular malformations) remain unclear in > 20% of patients [61]. Although the clinical phenotypes we describe can be mapped in part onto SMASH-U, they have the advantage of accounting for multiple variables present on admission. The variables we used to define phenotypes (e.g., hematoma location, blood pressure) are routinely collected. Even so, not all clinical phenotypes are represented. For example, cerebellar hematoma with brainstem compression is well described, but patients with cerebellar hematomas were excluded from ATACH-II, so the algorithm could not identify it. The clinical phenotypes we describe may be useful for guiding targeted assessments and treatments, even if they do not include every conceivable phenotype. It is possible that other clinical phenotypes of ICH might be discerned from additional data sets, a topic for future research.
Conclusions
In summary, we identified phenotypes of ICH admission data using unsupervised machine learning. These clinical phenotypes were clinically meaningful, associated with complications, and associated with functional outcomes at follow-up. Identifying clinical phenotypes at admission could lead to more targeted patient care by anticipating risk factors, complications, and outcomes.
Data Availability
Data from the two medical center registries in the United States are not publicly available. Qualified investigators wishing to reproduce the analysis may request data access from the corresponding author. Data from the ATACH-II trial are available from the National Institute of Neurological Disorders and Stroke clinical trials archive online.
References
Virani SS, Alonso A, Aparicio HJ, Benjamin EJ, Bittencourt MS, Callaway CW, Heart disease and stroke statistics-2021 update: a report from the American Heart Association, et al. Heart disease and stroke statistics-2021 update: a report from the American Heart Association. Circulation. 2021;143(8):e254-743.
Hemphill JC 3rd, Bonovich DC, Besmertis L, Manley GT, Johnston SC. The ICH score: a simple, reliable grading scale for intracerebral hemorrhage. Stroke. 2001;32(4):891–7.
Graff-Radford J, Rabinstein AA. Cerebral amyloid angiopathy criteria: the next generation. Lancet Neurol. 2022;21(8):674–6.
DeMerle KM, Angus DC, Baillie JK, Brant E, Calfee CS, Carcillo J, et al. Sepsis subclasses: a framework for development and interpretation. Crit Care Med. 2021;49(5):748–59.
Leasure AC, Qureshi AI, Murthy SB, Kamel H, Goldstein JN, Woo D, et al. Association of intensive blood pressure reduction with risk of hematoma expansion in patients with deep intracerebral hemorrhage. JAMA Neurol. 2019;76(8):949–55.
Naidech AM, Beaumont J, Muldoon K, Liotta EM, Maas MB, Potts MB, et al. Prophylactic seizure medication and health-related quality of life after intracerebral hemorrhage. Crit Care Med. 2018;46(9):1480–5.
Martini SR, Flaherty ML, Brown WM, Haverbusch M, Comeau ME, Sauerbeck LR, et al. Risk factors for intracerebral hemorrhage differ according to hemorrhage location. Neurology. 2012;79(23):2275–82.
Samarasekera N, Fonville A, Lerpiniere C, Farrall AJ, Wardlaw JM, White PM, Lothian Audit of the Treatment of Cerebral Haemorrhage Collaborators, et al. Influence of intracerebral hemorrhage location on incidence, characteristics, and outcome: population-based study. Stroke. 2015;46(2):361–8.
Ruíz-Sandoval JL, Cantú C, Barinagarrementeria F. Intracerebral hemorrhage in young people: analysis of risk factors, location, causes, and prognosis. Stroke. 1999;30(3):537–41.
Potter KM, Kennedy JN, Onyemekwu C, Prendergast NT, Pandharipande PP, Ely EW, et al. Data-derived subtypes of delirium during critical illness. EBioMedicine. 2024;100:104942.
Kwak S, Lee Y, Ko T, Yang S, Hwang IC, Park JB, et al. Unsupervised cluster analysis of patients with aortic stenosis reveals distinct population with different phenotypes and outcomes. Circ Cardiovasc Imaging. 2020;13(5):e009707.
Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, et al. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131(3):269–79.
Shah RV, Yeri AS, Murthy VL, Massaro JM, D’Agostino R Sr, Freedman JE, et al. Association of multiorgan computed tomographic phenomap with adverse cardiovascular health outcomes: the Framingham Heart Study. JAMA Cardiol. 2017;2(11):1236–46.
Katz DH, Deo RC, Aguilar FG, Selvaraj S, Martinez EE, Beussink-Nelson L, et al. Phenomapping for the identification of hypertensive patients with the myocardial substrate for heart failure with preserved ejection fraction. J Cardiovasc Transl Res. 2017;10(3):275–84.
Williams MC, Bednarski BP, Pieszko K, Miller RJ, Kwiecinski J, Shanbhag A, et al. Unsupervised learning to characterize patients with known coronary artery disease undergoing myocardial perfusion imaging. Eur J Nucl Med Mol Imaging. 2023;50(9):2656–68.
Kruser JM, Aaby DA, Stevenson DG, Pun BT, Balas MC, Barnes-Daly MA, et al. Assessment of variability in end-of-life care delivery in intensive care units in the United States. JAMA Netw Open. 2019;2(12):e1917344.
Murphy J, Shin HJ, Wang H, Luo Y, Jahromi B, Bleck TP, et al. Clusters Across multiple domains of health-related quality of life reveal complex patient outcomes after subarachnoid hemorrhage. Crit Care Explor. 2021;3(9):e0533.
Sucharew H, Khoury J, Moomaw CJ, Alwell K, Kissela BM, Belagaje S, et al. Profiles of the National Institutes of Health Stroke Scale items as a predictor of patient outcome. Stroke. 2013;44(8):2182–7.
Ding L, Mane R, Wu Z, Jiang Y, Meng X, Jing J, et al. Data-driven clustering approach to identify novel phenotypes using multiple biomarkers in acute ischaemic stroke: a retrospective, multicentre cohort study. EClinicalMedicine. 2022;53:101639.
Fridman S, Bres Bullrich M, Jimenez-Ruiz A, Costantini P, Shah P, Just C, et al. Stroke risk, phenotypes, and death in COVID-19: systematic review and newly reported cases. Neurology. 2020;95(24):e3373–85.
Faigle R, Marsh EB, Llinas RH, Urrutia VC, Gottesman RF. Novel score predicting gastrostomy tube placement in intracerebral hemorrhage. Stroke. 2015;46(1):31–6.
Qureshi AI, Palesch YY, Barsan WG, Hanley DF, Hsu CY, Martin RL, et al. Intensive blood-pressure lowering in patients with acute cerebral hemorrhage. N Engl J Med. 2016;375(11):1033–43.
Ullmann T, Hennig C, Boulesteix AL. Validation of cluster analysis results on validation data: a systematic framework. Wiley Interdiscip Rev Data Min Knowl Discov. 2022;12(3):e1444.
van Asch CJ, Luitse MJ, Rinkel GJ, van der Tweel I, Algra A, Klijn CJ. Incidence, case fatality, and functional outcome of intracerebral haemorrhage over time, according to age, sex, and ethnic origin: a systematic review and meta-analysis. Lancet Neurol. 2010;9(2):167–76.
Toyoda K, Palesch YY, Koga M, Foster L, Yamamoto H, Yoshimura S, et al. Regional differences in the response to acute blood pressure lowering after cerebral hemorrhage. Neurology. 2021;96(5):e740–51.
Lindholm PF, Kwaan HC, Naidech AM. Hemostasis, hematoma expansion, and outcomes after intracerebral hemorrhage. Blood. 2019;134(Suppl 1):4886.
Qureshi AI, Palesch YY. Antihypertensive treatment of acute cerebral hemorrhage (ATACH) II: design, methods, and rationale. Neurocrit Care. 2011;15(3):559–76.
Dowlatshahi D, Demchuk AM, Flaherty ML, Ali M, Lyden PL, Smith EE, VISTA Collaboration. Defining hematoma expansion in intracerebral hemorrhage: relationship with patient outcomes. Neurology. 2011;76(14):1238–44.
Naidech AM, Jovanovic B, Liebling S, Garg RK, Bassin SL, Bendok BR, et al. Reduced platelet activity is associated with early clot growth and worse 3-month outcome after intracerebral hemorrhage. Stroke. 2009;40(7):2398–401.
Bunney G, Murphy J, Colton K, Wang H, Shin HJ, Faigle R, et al. Predicting early seizures after intracerebral hemorrhage with machine learning. Neurocrit Care. 2022;37(Suppl 2):322–7.
Hanley DF, Lane K, McBee N, Ziai W, Tuhrim S, Lees KR, et al. Thrombolytic removal of intraventricular haemorrhage in treatment of severe stroke: results of the randomised, multicentre, multiregion, placebo-controlled CLEAR III trial. Lancet. 2017;389(10069):603–11.
Luong CQ, Nguyen AD, Nguyen CV, Mai TD, Nguyen TA, Do SN, et al. Effectiveness of combined external ventricular drainage with intraventricular fibrinolysis for the treatment of intraventricular haemorrhage with acute obstructive hydrocephalus. Cerebrovasc Dis Extra. 2019;9(2):77–89.
Hall AN, Weaver B, Liotta E, Maas MB, Faigle R, Mroczek DK, et al. Identifying modifiable predictors of patient outcomes after intracerebral hemorrhage with machine learning. Neurocrit Care. 2021;34(1):73–84.
Preudhomme G, Duarte K, Dalleau K, Lacomblez C, Bresso E, Smaïl-Tabbone M, et al. Head-to-head comparison of clustering methods for heterogeneous data: a simulation-driven benchmark. Sci Rep. 2021;11(1):4202.
McInnes L, Healy J, Melville J. UMAP: uniform manifold approximation and projection for dimension reduction. arXiv. 2018. https://doi.org/10.48550/arXiv.1802.03426. Accessed 1 Feb 2018.
R: A Language and Environment for Statistical Computing [computer program]. Vienna, Austria: R Foundation for Statistical Computing; 2021.
RStudio: Integrated development for R [computer program]. Boston, MA: PBC; 2020.
Broderick JP, Brott TG, Duldner JE, Tomsick T, Huster G. Volume of intracerebral hemorrhage: a powerful and easy-to-use predictor of 30-day mortality. Stroke. 1993;24(7):987–93.
Kuohn LR, Witsch J, Steiner T, Sheth KN, Kamel H, Navi BB, et al. Early deterioration, hematoma expansion, and outcomes in deep versus lobar intracerebral hemorrhage: the FAST Trial. Stroke. 2022;53(8):2441–8.
Teo KC, Fong SM, Leung WC, Leung IY, Wong YK, Choi OM, et al. Location-specific hematoma volume cutoff and clinical outcomes in intracerebral hemorrhage. Stroke. 2023;54(6):1548–57.
WHO Commission on Social Determinants of Health. Closing the gap in a generation: health equity through action on the social determinants of health: Commission on Social Determinants of Health Final Report. World Health Organization, Commission on Social Determinants of Health: Genva; 2008.
Leasure AC, King ZA, Torres-Lopez V, Murthy SB, Kamel H, Shoamanesh A, et al. Racial/ethnic disparities in the risk of intracerebral hemorrhage recurrence. Neurology. 2020;94(3):e314–22.
Kittner SJ, Sekar P, Comeau ME, Anderson CD, Parikh GY, Tavarez T, et al. Ethnic and racial variation in intracerebral hemorrhage risk factors and risk factor burden. JAMA Netw Open. 2021;4(8):e2121921.
Stansbury JP, Jia H, Williams LS, Vogel WB, Duncan PW. Ethnic disparities in stroke: epidemiology, acute care, and postacute outcomes. Stroke. 2005;36(2):374–86.
Greenberg SM, Ziai WC, Cordonnier C, Dowlatshahi D, Francis B, Goldstein JN, American Heart Association/American Stroke Association, et al. 2022 guideline for the management of patients with spontaneous intracerebral hemorrhage: a guideline from the American Heart Association/American Stroke Association. Stroke. 2022;53(7):e282-361.
Fujiwara G, Okada Y, Shiomi N, Sakakibara T, Yamaki T, Hashimoto N. Derivation of coagulation phenotypes and the association with prognosis in traumatic brain injury: a cluster analysis of nationwide multicenter study. Neurocrit Care. 2024;40(1):292–302.
Naidech AM, Weaver B, Maas M, Bleck TP, VanHaerents S, Schuele SU. Early seizures are predictive of worse health-related quality of life at follow-up after intracerebral hemorrhage. Crit Care Med. 2021;49(6):e578–84.
Sheikh ZB, Stretz C, Maciel CB, Dhakar MB, Orgass H, Petroff OA, et al. Deep versus lobar intraparenchymal hemorrhage: seizures, hyperexcitable patterns, and clinical outcomes. Crit Care Med. 2020;48(6):e505–13.
Qureshi AI, Tuhrim S, Broderick JP, Batjer HH, Hondo H, Hanley DF. Spontaneous intracerebral hemorrhage. N Engl J Med. 2001;344(19):1450–60.
Vespa PM, O’Phelan K, Shah M, Mirabelli J, Starkman S, Kidwell C, et al. Acute seizures after intracerebral hemorrhage: a factor in progressive midline shift and outcome. Neurology. 2003;60(9):1441–6.
Sheth KN, Martini SR, Moomaw CJ, Koch S, Elkind MS, Sung G,., et al. Prophylactic antiepileptic drug use and outcome in the ethnic/racial variations of intracerebral hemorrhage study. Stroke. 2015;46(12):3532–5.
Naidech AM, Garg RK, Liebling S, Levasseur K, Macken MP, Schuele SU, et al. Anticonvulsant use and outcomes after intracerebral hemorrhage. Stroke. 2009;40(12):3810–5.
Messé SR, Sansing LH, Cucchiara BL, Herman ST, Lyden PD, Kasner SE, CHANT investigators. Prophylactic antiepileptic drug use is associated with poor outcome following ICH. Neurocrit Care. 2009;11(1):38–44.
Narayan RK, Maas AI, Marshall LF, Servadei F, Skolnick BE, Tillinger MN, rFVIIa Traumatic ICH Study Group. Recombinant factor VIIA in traumatic intracerebral hemorrhage: results of a dose-escalation clinical trial. Neurosurgery. 2008;62(4):776–86.
Mahmood A, Needham K, Shakur-Still H, Harris T, Jamaluddin SF, Davies D, et al. Effect of tranexamic acid on intracranial haemorrhage and infarction in patients with traumatic brain injury: a pre-planned substudy in a sample of CRASH-3 trial patients. Emerg Med J. 2021;38(4):270–8.
CRASH-3 trial collaborators. Effects of tranexamic acid on death, disability, vascular occlusive events and other morbidities in patients with acute traumatic brain injury (CRASH-3): a randomised, placebo-controlled trial. Lancet. 2019;394(10210):1713–23.
Ovesen C, Jakobsen JC, Gluud C, Steiner T, Law Z, Flaherty K, et al. Tranexamic acid for prevention of hematoma expansion in intracerebral hemorrhage patients with or without spot sign. Stroke. 2021;52(8):2629–36.
Sprigg N, Flaherty K, Appleton JP, Al-Shahi-Salman R, Bereczki D, Beridze M, et al. Tranexamic acid for hyperacute primary IntraCerebral Haemorrhage (TICH-2): an international randomised, placebo-controlled, phase 3 superiority trial. Lancet. 2018;391(10135):2107–54.
Girolami A, Cosi E, Tasinato V, Santarossa C, Ferrari S, Girolami B. Drug-induced thrombophilic or prothrombotic states: an underestimated clinical problem that involves both legal and illegal compounds. Clin Appl Thromb Hemost. 2017;23(7):775–85.
Naidech AM, Wang H, Hutch M, Murphy J, Paparello J, Bath P, et al. Blood pressure medication and acute kidney injury after intracerebral haemorrhage: an analysis of the ATACH-II trial. BMJ Neurol Open. 2023;5(2):e000458.
Meretoja A, Strbian D, Putaala J, Curtze S, Haapaniemi E, Mustanoja S, et al. SMASH-U: a proposal for etiologic classification of intracerebral hemorrhage. Stroke. 2012;43(10):2592–7.
Funding
This project was supported in part by grant R36HS028941 from the Agency for Healthcare Research and Quality to JM. Dr. Naidech received research support from National Institutes of Health/National Institute of Neurological Disorders and Stroke (grants R01 NS110779 and U01 NS110772).
Author information
Authors and Affiliations
Contributions
JM performed the analysis and wrote and edited the manuscript; JSPdN, EJH, HW, MH, and YL edited the manuscript; RF provided data and edited the manuscript; AMN provided data, performed the analysis, and wrote and edited the manuscript. All authors read and approved the final manuscript as submitted.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethical Approval/Informed Consent
A waiver of informed consent was obtained from the institutional review boards (Northwestern University STU00213824 and Johns Hopkins University) for this research because patients were not contacted, nor was their care changed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A commentary on this article is available at https://doi.org/10.1007/s12028-024-02069-0.
Supplementary Information
Below is the link to the electronic supplementary material.
12028_2024_2067_MOESM4_ESM.tiff
Supplementary file4. UMAP Model outputs visualized in two dimensions, Three clinical phenotypes of ICH were reproduced in an independent validation cohort (n = 385) (TIFF 2077 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Murphy, J., Silva Pinheiro do Nascimento, J., Houskamp, E.J. et al. Phenotypes of Patients with Intracerebral Hemorrhage, Complications, and Outcomes. Neurocrit Care (2024). https://doi.org/10.1007/s12028-024-02067-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12028-024-02067-2