Abstract
Hepatitis B virus (HBV) infection is a major public health burden. The mechanisms of immune evasion during chronic HBV (CHB) infection are poorly understood. Human leukocyte antigen (HLA)-G, an immune checkpoint molecule, plays a crucial role in the tolerance mechanisms of various infectious diseases. The 3′ untranslated region (3′UTR), including the HLA-G + 3142 C > G polymorphism (rs1063320) and the 14-pb Ins/Del (rs66554220) has been strongly suggested to influence HLA-G expression. This study conducted a case-control analysis to evaluate the potential correlation between the HLA-G + 3142 C > G polymorphism and HBV infection outcome in a Tunisian cohort. The HLA-G + 3142 C > G polymorphism was analysed by PCR-RFLP in 242 patients with chronic HBV infection (116 males and 126 females), 241 healthy controls (116 males and 125 females), and 100 spontaneously resolved subjects (52 males and 48 females). Patients with chronic HBV infection showed a higher frequency of the + 3142G allele compared to healthy controls and spontaneously resolved subjects (p = 0.001 and p = 0.002, respectively). An association between the + 3142G allele and high HBV DNA levels was observed when HBV patients were stratified based on their HBV DNA levels (p = 0.016). Furthermore, the dominant model (GG + GC vs CC) was associated with liver function parameters, including AST, ALT, and high HBV DNA levels (p = 0.04, p < 0.001 and p = 0.002, respectively). However, there was no significant association found between this polymorphism and the fibrosis stage (p = 0.32). The haplotype analysis, using a subset of previously published data on the HLA-G 14-pb Ins/Del polymorphism, revealed an association between the Ins/G haplotype and chronic HBV infection (H1: InsG, p < 0.001). Our findings suggest that the + 3142G allele is a risk factor for the persistence and progression of HBV infection, while the + 3142C allele serves as a protective allele associated with the spontaneous resolution of the infection. Additionally, the HLA-G 3′UTR haplotype Ins/G is associated with chronic HBV infection in the Tunisian population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatitis B virus (HBV) is a major global public health concern. According to the latest data from the World Health Organization (WHO), approximately 296 million people were living with chronic hepatitis B (CHB) infection in 2019, and around of 820,000 deaths occurred annually due to HBV-related diseases, including cirrhosis, hepatocellular carcinoma (HCC), and liver failure [1]. Tunisia is a country with low HBV endemicity [2]. The prevalence of HBsAg ranges from 2.2 to 2.7% [3], with a predominance of genotype D [4]. To address this public health burden, the WHO has established a goal of eradicating the threat of viral hepatitis by 2030. The majority of immunocompetent adults infected with HBV can spontaneously resolve the virus during the acute phase. However, less than 5% develop chronic HBV infection, which can progress to severe stages of liver disease [5]. The host immune response has been shown to play a pivotal role in the natural history of chronic HBV infection. Experimental evidence shows that robust, polyclonal, and multispecific CD4 + /CD8 + T-cell responses, combined with neutralizing antibody responses against all HBV antigens, are typically associated with viral clearance during acute HBV infection. However, persistent HBV infection is mostly characterized by dysfunctional or exhausted adaptive immune responses, and failure to activate the innate immune system such as dendritic cells (DCs), monocytes and NK cells [6, 7]. These investigations have clearly demonstrated the immunological exhaustion associated with the chronic phase of HBV infection. To achieve a functional cure of chronic HBV infection, a better understanding of the mechanisms leading to immune tolerance and affecting the effector function of immune cells is needed.
HLA-G is a non-classical (class Ib) major histocompatibility complex (MHC) molecule, characterized by a low level of polymorphism and restricted tissue expression. The immune checkpoint HLA-G molecule plays an important role in modulating the immune response [8]. The expression of HLA-G molecules induces immune tolerance function by binding to their specific inhibitory receptors expressed by immune cells, such as Ig-like transcript 2 (ILT-2) is expressed on DCs, B cells, NK cells and T cells, ILT-4 predominantly expressed in myeloid cells, whereas Killer cell immunoglobin-like receptor 2DL4 (KIR2DL4) is restricted to NK cells [8,9,10]. Given the importance of the HLA-G molecule, several studies have explored its involvement in susceptibility to different pathological conditions, including cancer [11], inflammatory [12], and infectious diseases [13].
The HLA-G (3′ UTR) plays a crucial role in the control of transcriptional activity and mRNA stability. Variability in this region can effectively modulate the expression of the HLA-G gene [14]. Interestingly, several single nucleotide polymorphisms (SNPs) sites have been identified in the 3′ UTR that could potentially affect alternative splicing and miRNA/mRNA binding. Among these, two variants have been reported so far in terms of functional studies. First, the 14-bp Insertion/Deletion (Ins/Del) variant (rs66554220) has been associated with alternative splicing, with the removal of 92 nucleotides between positions + 2961 and + 2974, and mRNA stability [15]. The Ins allele has been suggested to be associated with low HLA-G protein production, whereas the Del allele is associated with higher levels of HLA-G expression by providing more stability to the mRNA [16]. Furthermore, the + 3142 C > G variant (rs1063320) has been identified as a target site for specific miRNAs, including miR-148a, miR-148b, and miR-152, which may potentially modulate the transcription of the HLA-G gene [15]. It has been reported that the + 3142G allele enhances the binding affinity of these miRNAs, leading to the degradation of HLA-G mRNA and a subsequent reduction in its expression. Conversely, the + 3142C allele is associated with decreased production of soluble HLA-G [17]. It is noteworthy that the HLA-G + 3142 C > G polymorphism has been associated with several diseases, including cancer [18], autoimmune diseases [19,20,21], and viral infections [22, 23].
In this retrospective study, we aimed to evaluate the potential association between the HLA-G + 3142 C > G variant and the outcomes of HBV infection in a Tunisian population.
Subjects and methods
Study population
A total of 583 participants, including 242 patients who are chronically infected with HBV (CHB) [seropositive for hepatitis B surface antigen (HBsAg) for at least 6 months], attending the Department of Infectious Diseases at Farhat Hached University Hospital, 100 spontaneously resolved (SR) subjects of HBV who were seronegative for HBsAg and seropositive for HBV core antigen (anti-HBC), and 241 healthy blood donors included in the study as healthy controls (HC) who were seronegative for all known serological markers of HBV, hepatitis C virus (HCV), and human immunodeficiency virus (HIV) infection. CHB patients with co-morbidities, including liver disease (such as autoimmune and alcoholic liver disease) or other viral diseases (including HCV, hepatitis delta virus (HDV), and HIV) were excluded from the study.
CHB patients were divided into two subgroups according to their HBV replication status: Subgroup (1) consisted of 137 patients with low HBV replication (HBV DNA levels < 2000 IU/mL) and subgroup (2) consisted of 105 patients with high HBV replication (HBV DNA levels ≥ 2000 IU/mL).
CHB patients, HC and SR were matched for sex, age and for geographical area. Informed written consent was obtained from all participants prior to blood collection in this study. Approval for the present study was obtained from the Ethical Review Board (ERB) of Farhat Hached University Hospital (ERB approval number: 35220228).
Laboratory assays
CHB patients were followed regularly with measurements of liver function tests including aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin (TBil), γ-glutamyltransferase (GGT) and alkaline phosphatase (ALP). Serological markers including HBsAg, HBeAg, anti-HBe and anti-HBc were detected using Microparticle Enzyme Immunoassay technology (AxSYM; Abbott Laboratories, Abbott Park, IL, USA). Serum HBV-DNA levels were quantified by a commercial real-time polymerase chain reaction (RT-PCR) (COBAS AmpliPrep/COBAS TaqMan, Roche Diagnostics), with detection limits ranging from 20 IU/mL at the lower level to 110 × 106 IU/mL at the upper level. For statistical analyses, an arbitrary value of 20 and 110 × 106 IU/mL was assigned to samples with undetectable and uninterruptable HBV-DNA, respectively.
DNA extraction and genotyping assays
Genomic DNA was extracted from whole blood EDTA using the QIAamp DNA blood kit according to the manufacturer’s instructions (Qiagen, Chatsworth, CA, USA). Genotyping assay for the HLA-G + 3142 C > G polymorphism was performed by restriction fragment length polymorphism (RFLP)-PCR with the following primers:
The forward primer (5′-CATGCTGAACTGCATTCCTTCC-3′), and the reverse primer (5′-CTGGTGGGACAAGGTTCTACTG-3′) [22]. PCR was performed in a total volume of 25 µL, including 100 ng genomic DNA, 1.5 mM MgCl2, 0.2 mM concentrations of each dNTP, 1U of Taq DNA polymerase (Promega) and 0.4 µM of each specific primer. The PCR conditions were 94 °C for 5 min, 32 cycles of 94 °C for 30 s, 30 s at 65.5 °C and 60 s at 72 °C, followed by a final extension step at 72 °C for 5 min. RFLP was performed by the incubation of PCR product with 3U of BaeGI restriction enzyme (Biolabs, New England, USA) at 37 °C for 3 h [24]. The restriction fragments (406 bp for allele C; 316 and 90 bp for allele G) were separated by electrophoresis on a 2% agarose gel with ethidium bromide, and observed under ultraviolet illumination using the Gel Doc XR Plus (Bio-Rad, USA) (Fig. 1). Additionally, the 14-pb Ins/Del was amplified by SSP-PCR, exclusively for the SR subjects, such as previously described [25] (Fig. 2).
Statistical analysis
Statistical analysis was conducted using SPSS17.0 software (SPSS Inc., Chicago, IL, USA) and Graphpad prism 9. Allele and genotype frequencies were compared between groups using either the X2 or Fisher’s exact test. Odds ratios (OR) [95% confidence interval (CI)] were calculated to estimate the relative risk. The comparison of quantitative variables between groups was conducted using the Mann–Whitney test. The significance threshold for p-values was set at p < 0.0125 after applying the Bonferroni correction to address multiple comparisons. The Hardy–Weinberg equilibrium (HWE) and haplotype inference were analysed using Haploview software (version 4.2).
Results
Characteristics of the study population
Table 1 summarises the characteristics of the study population. The study enrolled 242 patients (116 males and 126 females), 241 HC (116 males and 125 females) and 100 SR (52 males and 48 females). The mean age [± standard error of the mean (SEM)] was 35.6 ± 0.7 for CHB patients, 34.4 ± 0.8 for HC and 37.5 ± 0.2 years for SR subjects. No significant differences in gender or age were reported between study groups. The clinical profiles of CHB patients with high and low HBV DNA levels are showed a significant difference in the HBV DNA levels between the two groups (p < 0.001). All patients included in our study exhibit HBV genotype D. In addition, high ALT and AST levels were reported in patients with high HBV DNA levels ≥ 2000 UI/mL (p < 0.0001).
Association between the HLA-G rs1063320 and the different outcomes of HBV infection
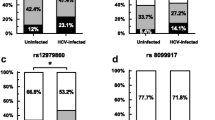
To investigate the role of this polymorphism in the susceptibility to chronic HBV infection, we initially genotyped the rs1063320 in both CHB patients and HC. The corresponding genotype and allele frequencies are presented in Table 2. The genotype distribution of the HLA-G rs1063320 was in Hardy–Weinberg equilibrium in the HC (p > 0.05). The frequency of the G allele was significantly higher in CHB patients as compared to HC (62% vs 52%, p = 0.001), suggesting that CHB patients with the G allele have an increased risk of developing chronic HBV infection (OR = 1.49, 95% CI [1.16–1.93]). In addition, the GG and GC genotypes were overrepresented in CHB patients, conferring a susceptibility to persistent HBV infection (OR = 2.26, 95% CI [1.35–3.78], p = 0.001 and OR = 1.86, 95% CI [1.14–3.03], p = 0.011, respectively). Furthermore, to determine whether the rs1063320 was associated with spontaneously resolved HBV infection, the allele and genotype frequencies were compared between CHB patients and SR subjects. Interestingly, the frequency of the wild-type C allele of the HLA-G rs1063320 was significantly overrepresented in SR subjects (51% vs. 38%, p = 0.002), and was associated with a protective effect that increased the spontaneous clearance of HBV infection (OR = 1.66, 95% CI [1.19–2.32]). Moreover, the frequency of the CC genotype was significantly higher in SR subjects than in CHB patients (30% vs. 15%, respectively; OR = 2.73, 95% CI [1.43–5.21], p = 0.001). However, there was no statistically significant difference in the comparison of SR subjects and HC. These results clearly show that the HLA-G + 3142 C > G polymorphism has an impact on the outcome of HBV infection.
Association between the HLA-G rs1063320 and the HBV DNA level
To investigate the impact of the rs1063320 variant on the regulation of HBV replication, we classified patients into two subgroups according to their HBV DNA levels and compared the allele and genotype frequencies between those with low and high HBV DNA levels. As shown in Table 3, our finding revealed a higher prevalence of the G allele and the GG genotype in patients with high HBV DNA levels compared with those with low HBV DNA levels [(68% vs. 57%, [OR = 1.57, 95% CI [1.08–2.29], p = 0.016) and (45% vs. 32%, [OR = 2.71, IC 95% [1.17–6.26], p = 0.017), respectively]. However, the significance did not survive the Bonferroni correction.
Moreover, the frequencies of the G allele and the GG genotype were significantly higher in CHB patients with high HBV DNA levels than in HC (68% vs. 52%, [OR = 1.95, 95% CI [1.39–2.74], p < 0.001), and similarly when compared with SR subjects (68% vs. 49%, [OR = 2.17, 95% CI [1.45–3.24], p < 0.001). This observation suggested that the presence of the HLA-G + 3142G allele in CHB patients increases the risk of active HBV replication.
Association between the HLA-G rs1063320 and the progression of CHB infection
To assess the influence of the rs1063320 on the progression of CHB infection, we stratified patients according to the stage of liver fibrosis using the METAVIR score. This score distinguishes between mild/moderate (F0–F1) and severe (F2–F4) fibrosis stages. However, we did not find any statistically significant association between the HLA-G rs1063320 polymorphism and the progression of liver disease in CHB patients in our cohort (Table 4).
Correlation of liver functions and virological features according to HLA-G + 3142 C > G genotypes
To evaluate the impact of the rs1063320 on variations of biochemical and virological features in CHB patients, we compared patients with dominant model (GG + GC) to those with CC genotype (Fig. 3). Notably, CHB patients with GG/GC genotypes had significantly elevated levels of transaminases (ALT and AST) and GGT compared to those with CC genotypes (p = 0.04, p < 0.001 and p = 0.002, respectively) (Fig. 3A, B, C). Furthermore, the dominant model showed a correlation with higher HBV DNA levels (p < 0.001, Fig. 3F). In contrast, there was no significant correlation between HLA-G + 3142 C > G genotypes and ALP and T-Bil levels (Fig. 3D, E).
Haplotype analysis of HLA-G 3′UTR polymorphisms of HBV infection
To investigate the impact of HLA-G 3′UTR polymorphisms on the outcome of HBV infection, we constructed haplotypes for two polymorphisms. One polymorphism was derived from actual data of HLA-G + 3142 C < G and the other was based on our previously published study that investigated the HLA-G 14-bp Ins/Del in the same cohort [25]. Furthermore, we genotyped the HLA-G 14-bp Ins/Del specifically for the SR subjects in this study. The comparison of allelic and genotypic frequencies of the 14-bp Ins/Del polymorphism between CHB and HC did not reveal any statistical significance (data not shown).
Four haplotypes were identified, denoted (‘InsG’ (H1), ‘DelC’ (H2), ‘DelG’ (H3) and ‘InsC’ (H4) (Table 5). The most prevalent haplotypes were H1 and H2, with frequencies of 40% and 35%, respectively. The frequency of H1 was significantly higher in the CHB patients compared to HC and SR subjects (CHB vs. HC: p < 0.001 and CHB vs SR: p = 0.05) indicating the association of ‘InsG’ haplotype with chronic HBV infection. However, the frequency of the H4 haplotype was higher in the HC and SR subjects than in the CHB patients (CHB vs. HC: p = 0.002 and CHB vs SR: p = 0.041), implying H4 was suggested to be a protective haplotype against chronic HBV infection. These findings underscore the significant role of HLA-G in influencing disease outcomes.
Discussion
The clinical outcomes of HBV infection are heterogeneous. The host immune response emerges as a pivotal factor influencing the natural course of chronic HBV infection. In fact, efficient and robust immune responses are typically associated with viral clearance during acute HBV infection. However, chronic HBV infection is characterised by dysfunctional or weak immune responses [26]. The precise mechanisms that promote immune tolerance and the persistence of HBV infection are not fully understood. In this context, the HLA-G molecule continues to attract considerable interest as a molecule involved in immune tolerance in several diseases, such as cancer and infectious diseases [13, 27]. Previously, we have shown, as have several other studies, that the soluble HLA-G (sHLA-G) molecule is involved in the progression of HBV infection [28,29,30,31]. Given the substantial influence of the HLA-G + 3142 C > G polymorphism on the regulation of sHLA-G expression [32, 33], this study aims to investigate the potential association of this variant with the outcome and the progression of HBV infection in the Tunisian population.
Our data showed that a higher frequency of the G allele of the HLA-G + 3142 C > G polymorphism and the Ins/G haplotype were associated with an increased risk of developing chronic HBV infection. However, the C allele and the Ins/C haplotype appear to play a protective role during the acute phase and are associated with spontaneous resolution of HBV infection in our population. This finding seems to be in line with a recent study conducted by Okumura et al., showing a significant association of the G allele with susceptibility to chronic HBV infection in the Japanese population [34]. Interestingly, our results are supported by a previous study by Cordero et al. [22]. The study suggests that the presence of the G allele and the Ins/G haplotype are associated with an increased susceptibility to HCV infection in sickle cell disease patients of African descent. In addition, the G allele and the GG genotype of the HLA-G + 3142 C > G polymorphism have been reported to be associated with several other viral infectious diseases. A recent study conducted by Medeiros et al. revealed that both the G allele, and the GG genotype were associated with a predisposition to HIV infection and the Ins/G haplotype was overrepresented in African-derived patients HIV+ from Southern Brazil [35]. Moreover, Xu et al. showed that the G allele and the Ins/G haplotype were associated with a high risk of human papillomavirus (HPV) infection in Chinese Han women [36].Likewise, Da Silva et al. also showed an increased prevalence of the Ins/G haplotype in HIV patients of African descent [37]. This consistent pattern emphasises the association between the + 3142G allele and the insertion of 14-pb of HLA-G 3′UTR and the susceptibility to various viral infectious diseases, particularly in individuals with African ancestry. This not only strengthens our findings regarding the effect of these variants in the HBV infection, but also underscores the pivotal role of host genetic contributions in determining HBV outcomes.
Furthermore, to determine the correlation of the HLA-G + 3142 C > G polymorphism with the progression of chronic HBV infection, we stratified CHB patients according to their viral load (patients with low HBV DNA levels < 2000 IU/mL and patients with high HBV DNA levels ≥ 2000 IU/mL). This threshold is clinically important because it differentiates between active and inactive viral replication. It also serves as a critical indicator to assess the progression of inflammation during HBV infection, as recognised by healthcare professionals [38]. Interestingly, our results showed a significant association between the G allele and the GG genotype with elevated levels of HBV DNA in patients with chronic hepatitis B (CHB). We also observed an increased frequency of the GG + GC model, which correlated with elevated levels of inflammatory liver markers, including AST, ALT and GGT. To the best of our knowledge, the present study is the first to assess the association between HLA-G + 3142 C > G polymorphism and the progression of chronic HBV infection. Previous studies have investigated the level of sHLA-G with different phases of chronic HBV infection. Han et al. showed that serum of sHLA-G levels correlated with ALT levels in CHB patients [29]. Subsequently, increased expression of sHLA-G was correlated with HBV DNA levels [28, 39]. However, our cohort did not show a consistent association between the HLA-G + 3142 C > G polymorphism and liver fibrosis. Notably, our findings have revealed for the first time a potential involvement of the G allele in activating HBV replication and triggering an inflammatory response that may contribute to the progression of HBV infection to liver failure. It’s worth noting that we could not conclusively demonstrate this association in the progression of liver fibrosis within our cohort, possibly due to the limited sample size. Similarly, Okumura et al. also reported a lack of correlation between HLA-G + 3142 C > G polymorphism and progression to HCC [34].
Overall, several lines of evidence suggest that the G allele and/or GG genotype of the HLA-G + 3142 C > G polymorphism was strongly associated with the susceptibility to several infectious diseases [35,36,37]. In the context of HBV infection, both the recent study by Okumura et al. [34] and our current research have confirmed the following findings. In addition, elevated levels of sHLA-G expression have been consistently reported in CHB patients across different cohorts, underscoring the important role of the HLA-G molecule in influencing the outcome of this disease [28,29,30,31]. On the other hand, the HLA-G + 3142 C > G polymorphism is suggested to influence HLA-G expression through post-transcriptional mechanisms by affecting the binding of specific miRNAs, in particular the miR-148/152 family (miR-148a, miR-148b, and miR-152) [17, 33, 40]. Tan et al. showed that the presence of guanine at position + 3142 reduces the expression of soluble HLA-G through mRNA degradation following a high affinity for miR148/152 [33]. Furthermore, Rousseau et al. suggest that the deletion of 14-bp in the 3′UTR of HLA-G correlates with high sHLA-G expression by stabilising the mRNA [41].
Several hypotheses may explain this discrepancy observed between functional studies and genetic and molecular studies carried out on HBV infection. To address this paradox, we hypothesise that the persistence of HBV infection and the elevated levels of sHLA-G, despite the presence of the + 3142G allele, may be due to strategies used by the virus to evade the immune response. Notably, Bian et al. showed that the expression of sHLA-G protein is upregulated by HBV, but not the mRNA, through the downregulation of miR148/152, which serves to attenuate NK cell cytotoxicity [42]. Likewise, Sartorius et al. showed that the HBx protein of HBV downregulates the miR-148/152 to supress the immune response against HBV infection [43]. Recently, the miR-152 has been reported to be associated with dendritic cell dysfunction during acute and chronic hepatitis B virus infection [44].
A further hypothesis can be proposed in relation to the effect of the HLA-G + 3142 C > G variant on the expression of the HLA-G molecule. Veit et al. suggest that the HLA-G + 3142 C > G polymorphism alone may not be the sole determinant that controls the expression of HLA-G and the 14-pb polymorphism is not the main element on the regulation of gene expression at the RNA level [45]. Furthermore, a previous study investigated the impact of the most frequent variation sites described in the HLA-G 3′UTR on plasma sHLA-G levels. The study showed that the UTR-2 haplotype ((InsTCCCGAG), which contains the 14-pb insertion and + 3142G allele, is correlated with medium production of sHLA-G [46]. It is noteworthy that UTR-2 is among the most frequent haplotypes in the African population [47], which may explain the increased susceptibility of individuals of African origin to various types of infectious diseases [20, 35, 48]. Furthermore, Hviid et al. showed a significant correlation between the 14-pb ins/ins genotype and increased IL-10 levels [49]. IL-10 has recently been identified as a crucial mediator of T-cell dysfunction during chronic HBV infection and HIV-1/HCV coinfection diseases [50, 51]. Altogether, these observations highlight the important role of the 14-pb Ins/Del and + 3142 C > G polymorphisms of HLA-G 3′UTR in the susceptibility to various infectious diseases including the HBV infection. Hence, it is crucial to take into account other polymorphisms in the 3′UTR and 5′URR to fully understand the complexity of genetic factors that regulate the HLA-G molecule and its involvement in susceptibility to infectious diseases, including HBV infection.
Several studies have demonstrated elevated expression of the HLA-G molecule in chronic HBV patients [28,29,30,31]. On the other hand, it has been reported that chronic HBV patients are characterized by lower levels and dysfunction of NK cells [52] and a depletion of HBV specific CD4 + and CD8 + T cells [53]. Furthermore, a number of studies have indicated that immune system cells in patients with CHB, including NK cells, T lymphocytes, and dendritic cells, express inhibitory receptors of the HLA-G molecule (ILT2, ILT4 and KIR2DL4) during chronic HBV infection [54, 55]. These findings suggest that the interaction between the HLA-G molecule and its inhibitory receptors is implicated as a mechanism of immune evasion by HBV. This occurs through the protection of infected cells, achieved by modulating the expression of specific cytokines associated with HBV infection, such as IL-1β, IL-10, IL-12, INF-γ, and TNF-α [56]. Moreover, research conducted by our team and other investigators on genetic polymorphisms influencing HLA-G expression, such as the 14-bp Ins/Del and + 3142 C/G in the context of HBV infection could provide valuable insights into the genetic mechanisms regulating HLA-G expression during chronic HBV infection [25, 39]. This underscores the significant role of host genetics as a key factor controlling the expression of immune system genes.
In conclusion, this study is the first to demonstrate an association between the HLA-G + 3142 C > G polymorphism and the outcome and progression of HBV infection in the Tunisian population. The + 3142G allele and GG genotype were found to be linked to susceptibility to chronic HBV infection and elevated levels of HBV DNA. Conversely, the C allele and CC genotype exhibited a protective effect, correlating with the spontaneous resolution of HBV infection. Furthermore, the Ins/G haplotype located within the HLA-G 3′UTR was found to be linked with persistent HBV infection. Our study has certain limitations, including a small number of patients with fibrosis and HCC, as well as a limited number of individuals who spontaneously resolved the infection. Considering the crucial role of the HLA-G molecule in the immune response to HBV infection outcomes, it would be interesting to investigate the association of the entire HLA-G gene, including the 5′URR, coding region, and 3′UTR, with susceptibility to HBV infection in future studies.
References
WHO. World health statistics 2023: monitoring health for the SDGs, Sustainable Development Goals. Geneva, World Health Organization. 2023.
Lahchaichi A, et al. Prevalence and risk factors of hepatitis B in Tunisia. Eur J Public Health. 2019;29(Supplement_4),ckz187.076.
GBD 2019 Hepatitis B Collaborators. Global, regional, and national burden of hepatitis B, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol Hepatol. 2022;7(9):796–829.
Chaouch H, et al. Naturally occurring surface antigen variants of hepatitis B virus in Tunisian patients. Intervirology. 2016;59(1):36–47.
Jeng WJ, Papatheodoridis GV, Lok ASF. Hepatitis B. Lancet. 2023;401(10381):1039–52.
Bertoletti A, Ferrari C. Adaptive immunity in HBV infection. J Hepatol. 2016;64(1 Suppl):S71–83.
Guidotti LG, Isogawa M, Chisari FV. Host-virus interactions in hepatitis B virus infection. Curr Opin Immunol. 2015;36:61–6.
Attia JVD, et al. The molecular and functional characteristics of HLA-G and the interaction with its receptors: where to intervene for cancer immunotherapy? Int J Mol Sci. 2020;21(22):8678.
Gao A, Sun Y, Peng G. ILT4 functions as a potential checkpoint molecule for tumor immunotherapy. Biochim Biophys Acta Rev Cancer. 2018;2:278–85.
Rohn H, et al. Effect of HLA-G5 Immune Checkpoint Molecule on the Expression of ILT-2, CD27, and CD38 in Splenic B cells. J Immunol Res. 2022;12(4829227).
Gan J, et al. HLA-G 3'UTR polymorphism diplotypes and soluble HLA-G plasma levels impact cervical cancer susceptibility and prognosis. Front Immunol. 2022;13(1076040).
Morandi F, et al. Recent advances in our understanding of HLA-G biology: lessons from a wide spectrum of human diseases. J Immunol Res. 2016;4326495(10):29.
Beltrami S, et al. Non-classical HLA class I molecules and their potential role in viral infections. Hum Immunol. 2023;84(8):384–92.
Amodio G, Gregori S. HLA-G genotype/expression/disease association studies: success, hurdles, and perspectives. Front Immunol. 2020;11(1178):1178.
Castelli EC, et al. HLA-G genetic diversity and evolutive aspects in worldwide populations. Sci Rep. 2021;11(1):23070.
de Almeida BS, et al. Genetic association between HLA-G 14-bp polymorphism and diseases: a systematic review and meta-analysis. Hum Immunol. 2018;79(10):724–35.
Porto IO, et al. MicroRNAs targeting the immunomodulatory HLA-G gene: a new survey searching for microRNAs with potential to regulate HLA-G. Mol Immunol. 2015;65(2):230–41.
Bertol BC, et al. HLA-G gene variability is associated with papillary thyroid carcinoma morbidity and the HLA-G protein profile. Int J Mol Sci. 2023;24(16):12858.
Hashemi M, et al. Evaluation of HLA-G 14 bp Ins/Del and +3142G>C polymorphism with susceptibility and early disease activity in rheumatoid arthritis. Adv Med. 2016;2016(10):4985745.
Poomarimuthu M, et al. HLA-G 3’UTR gene polymorphisms and rheumatic heart disease: a familial study among South Indian population. Pediatr Rheumatol Online J. 2017;15(1):10.
de Oliveira-Caramez ML, et al. Evidence for epistatic interaction between HLA-G and LILRB1 in the pathogenesis of nonsegmental vitiligo. Cells. 2023;12(4):630.
Cordero EA, et al. HLA-G polymorphism influences the susceptibility to HCV infection in sickle cell disease patients. Tissue Antigens. 2009;74(4):308–13.
Guberina H, et al. Recipient HLA-G +3142 CC Genotype and Concentrations of Soluble HLA-G Impact on Occurrence of CMV Infection after Living-Donor Kidney Transplantation. Int J Mol Sci. 2017;18(11):2338.
Zambra FM, et al. Response to Bortolotti et al. 2012–a re-evaluation of our polymerase chain reaction-restriction fragment length polymorphism genotyping method. Tissue Antigens. 2013;82(4):286–7.
Laaribi AB, et al. Association of an HLA-G 14-bp insertion/deletion polymorphism with high HBV replication in chronic hepatitis. J Viral Hepat. 2015;22(10):835–41.
Chisari FV, Isogawa M, Wieland SF. Pathogenesis of hepatitis B virus infection. Pathol Biol. 2010;58(4):258–66 (Paris).
Jiang N, et al. HLA and tumour immunology: immune escape, immunotherapy and immune-related adverse events. J Cancer Res Clin Oncol. 2023;149(2):737–47.
Laaribi AB, et al. Increased levels of soluble HLA-G molecules in Tunisian patients with chronic hepatitis B infection. J Viral Hepat. 2017;24(11):1016–22.
Han Q, et al. Association of serum soluble human leukocyte antigen-G levels with chronic hepatitis B virus infection. Clin Exp Med. 2014;14(1):35–43.
Park Y, et al. Soluble human leukocyte antigen-G expression in hepatitis B virus infection and hepatocellular carcinoma. Tissue Antigens. 2012;79(2):97–103.
Shi WW, et al. Plasma soluble human leukocyte antigen-G expression is a potential clinical biomarker in patients with hepatitis B virus infection. Hum Immunol. 2011;72(11):1068–73.
Castelli EC, et al. In silico analysis of microRNAS targeting the HLA-G 3’ untranslated region alleles and haplotypes. Hum Immunol. 2009;70(12):1020–5.
Tan Z, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81(4):829–34.
Okumura T, et al. HLA-G susceptibility to hepatitis B infection and related hepatocellular carcinoma in the Japanese population. Hum Immunol. 2023;84(8):401–7.
Medeiros FS, et al. Variation sites at the HLA-G 3’ untranslated region confer differential susceptibility to HIV/HPV co-infection and aneuploidy in cervical cell. PLoS ONE. 2018;13(10):e0204679.
Xu HH, et al. Association of HLA-G 3’ UTR polymorphism and expression with the progression of cervical lesions in human papillomavirus 18 infections. Infect Agent Cancer. 2018;13(42):42.
da Silva GK, et al. Influence of HLA-G polymorphisms in human immunodeficiency virus infection and hepatitis C virus co-infection in Brazilian and Italian individuals. Infect Genet Evol. 2014;21:418–23.
European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu; European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370–98.
Souto FJD, et al. Liver HLA-G expression is associated with multiple clinical and histopathological forms of chronic hepatitis B virus infection. J Viral Hepatitis. 2011;18(2):102–5.
Castelli EC, et al. In silico analysis of microRNAS targeting the HLA-G 3′ untranslated region alleles and haplotypes. Hum Immunol. 2009;70(12):1020–5.
Rousseau P, et al. The 14 bp deletion-insertion polymorphism in the 3’ UT region of the HLA-G gene influences HLA-G mRNA stability. Hum Immunol. 2003;64(11):1005–10.
Bian X, et al. Down-expression of miR-152 lead to impaired anti-tumor effect of NK via upregulation of HLA-G. Tumour Biol. 2016;37(3):3749–56.
Sartorius K, et al. The Epigenetic Modulation of Cancer and Immune Pathways in Hepatitis B Virus-Associated Hepatocellular Carcinoma: The Influence of HBx and miRNA Dysregulation. Front Immunol. 2021;12(661204).
Singh AK, et al. Identification of miRNAs associated with dendritic cell dysfunction during acute and chronic hepatitis B virus infection. J Med Virol. 2021;93(6):3697–706.
Veit TD, Chies JA. Tolerance versus immune response – microRNAs as important elements in the regulation of the HLA-G gene expression. Transpl Immunol. 2009;20(4):229–31.
Martelli-Palomino G, et al. Polymorphic sites at the 3' untranslated region of the HLA-G gene are associated with differential hla-g soluble levels in the Brazilian and French population. PLoS One. 2013;8(10):e71742.
Castelli EC, et al. Insights into HLA-G Genetics Provided by Worldwide Haplotype Diversity. Front Immunol. 2014;5(476).
Courtin D, et al. HLA-G 3′ UTR-2 haplotype is associated with Human African trypanosomiasis susceptibility. Infect Genet Evol. 2013;17:1–7.
Hviid TV, et al. Polymorphism in the 5’ upstream regulatory and 3’ untranslated regions of the HLA-G gene in relation to soluble HLA-G and IL-10 expression. Hum Immunol. 2006;67(1–2):53–62.
Hackstein CP, et al. Interferon-induced IL-10 drives systemic T-cell dysfunction during chronic liver injury. J Hepatol. 2023;79(1):150–66.
Caraballo Cortés K, et al. T-cell Exhaustion in HIV-1/Hepatitis C Virus Coinfection Is Reduced After Successful Treatment of Chronic Hepatitis C. Open Forum Infect Dis. 2023;10(11):ofad514.
Sun C, et al. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol Immunol. 2015;12(3):292–302.
Gehring AJ, Protzer U. Targeting innate and adaptive immune responses to cure chronic HBV infection. Gastroenterology. 2019;156(2):325–37.
Zhang Y, et al. Increased ILT2 expression contributes to dysfunction of CD56(dim)CD16(+)NK cells in chronic hepatitis B virus infection. Antiviral Res. 2022;205(105385):30.
Fisicaro P, et al. Pathogenetic Mechanisms of T Cell Dysfunction in Chronic HBV Infection and Related Therapeutic Approaches. Front Immunol. 2020;11(849).
Rashidi S, et al. The potential role of HLA-G in the pathogenesis of HBV infection: immunosuppressive or immunoprotective? Infect Genet Evol. 2020;85:104580.
Funding
This research was supported by the Ministry of Higher Education and Scientific Research of Tunisia. Programme d’Encouragement à l’Excellence Scientifique (P2ES), Grant Number: P2ES2020-D5P2.
Author information
Authors and Affiliations
Contributions
A.B Laaribi: Conceptualization, Formal analysis, Investigation and Writing—original draft. A. Letaif: Resources. N. Hannachi and J. Boukadida: Resources, Validation, Supervision and Writing—review & editing. A. Mehri, H. Ben Yahia, H. Chaouch, W. Babay: Formal analysis, Investigation and Data curation. H.I Ouzari: Supervision, Writing—review & editing and Funding acquisition. I. Zidi: Conceptualization, Supervision, Writing—review & editing, Project administration.
Corresponding author
Ethics declarations
Ethics approval
This research was approved by the ethical review board (ERB) of Farhat Hached University Hospital. ERB approval number: 35220228.
Consent to participate
Informed written consent was obtained from all participants.
Consent for publication
All authors reviewed and approved this manuscript for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laaribi, A.B., Mehri, A., Yahia, H.B. et al. Association of HLA-G 3′UTR polymorphisms with hepatitis B virus infection in Tunisian population. Immunol Res (2024). https://doi.org/10.1007/s12026-024-09516-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12026-024-09516-2